Abstract
Many fish and salamander species regenerate amputated fins or limbs, restoring the size and shape of the original appendage. Regeneration requires that spared cells retain or recall information encoding pattern, a phenomenon termed positional memory. Few factors have been implicated in positional memory during vertebrate appendage regeneration. Here, we investigated potential regulators of anteroposterior (AP) pattern during fin regeneration in adult zebrafish. Sequence-based profiling from tissues along the AP axis of uninjured pectoral fins identified many genes with region-specific expression, several of which encoded transcription factors with known AP-specific expression or function in developing embryonic pectoral appendages. Transgenic reporter strains revealed that regulatory sequences of the transcription factor gene alx4a activated expression in fibroblasts and osteoblasts within anterior fin rays, whereas hand2 regulatory sequences activated expression in these same cell types within posterior rays. Transgenic overexpression of hand2 in all pectoral fin rays did not affect formation of the proliferative regeneration blastema, yet modified the lengths and widths of regenerating bones. Hand2 influenced the character of regenerated rays in part by elevation of the vitamin D-inactivating enzyme encoded by cyp24a1, contributing to region-specific regulation of bone metabolism. Systemic administration of vitamin D during regeneration partially rescued bone defects resulting from hand2 overexpression. Thus, bone-forming cells in a regenerating appendage maintain expression throughout life of transcription factor genes that can influence AP pattern, and differ across the AP axis in their expression signatures of these and other genes. These findings have implications for mechanisms of positional memory in vertebrate tissues.
Keywords: Regeneration, Anteroposterior patterning, Zebrafish, Fin, Blastema, Hand2, Vitamin D, Positional memory
INTRODUCTION
Many recent studies have employed cell transplantation or genetic fate-mapping approaches to identify the cellular sources of tissues that arise during injury-induced regeneration (Buckingham and Meilhac, 2011; Tanaka and Reddien, 2011). Upon defining sources of regeneration through these experiments, a key priority is then to understand how these stem/progenitor cells and differentiated cell types successfully restore complex tissues of the correct size, pattern and function after organ damage.
Limbs and fins are complex three-dimensional structures composed of numerous tissue types. Remarkably, many fish and salamanders retain the ability to regenerate amputated appendages throughout their adult lives. To do this, cells within the appendage stump must retain and recall detailed patterning information, which is commonly referred to as positional memory. A regulator of regenerative positional memory is expected to possess two main characters: (1) presence in a gradient or restricted pattern within the intact and regenerating adult appendage; and (2) its overexpression or blockade impacts the regenerative pattern. In planarians, which are invertebrates that undergo vigorous cellular turnover and regenerate through a stem cell population known as neoblasts, recent evidence indicates that the adult pattern is actively maintained by the regionalized expression of developmental pathway regulators. Upon injury, these same factors help restore tissue and reinstate pattern (Reddien et al., 2007; Gurley et al., 2008; Petersen and Reddien, 2011; Roberts-Galbraith and Newmark, 2013).
During amphibian limb regeneration, retinoic acid (RA) has been implicated in positional memory, as RA treatment causes wrist-level amputations to sprout shoulder-level regenerates (Maden, 1982). Although this finding is remarkable, the role of endogenous RA in positional memory is unclear as there is no discernible proximodistal (PD) gradient of RA in the intact limb. A second candidate factor is Prod1, a proposed receptor for the newt blastemal mitogen Anterior gradient (da Silva et al., 2002; Kumar et al., 2007b). Prod1 is induced by exogenous RA and expressed at slightly higher levels in proximal intact limb regions as compared with distal regions (Kumar et al., 2007a). Although its in vivo function is unknown, inhibition of Prod1 in cultured blastemas blocks the characteristic in vitro behavior of proximal blastemas, and blastemal cells electroporated with excess Prod1 distribute proximally compared with control electroporations (da Silva et al., 2002; Echeverri and Tanaka, 2005). Potential links between RA and Prod1 could be provided by Meis proteins, which are important for the proximalizing effects of RA on regeneration and may regulate Prod1 (Mercader et al., 2005; Shaikh et al., 2011). In summary, although positional memory is a crucial aspect of regeneration, there remains much to learn about how it is encoded and enacted during vertebrate appendage regeneration.
How pattern is initially established in the developing appendages of vertebrate embryos has been intensely studied (Duboc and Logan, 2009; Towers and Tickle, 2009; Zeller et al., 2009). In particular, several regulators of embryonic limb anteroposterior (AP) patterning have been identified based on their restricted expression patterns and the robust effects of their gain- or loss-of-function on limb patterning (Riddle et al., 1993; Qu et al., 1997; Charité et al., 2000; Zákány et al., 2004). For instance, the transcription factor Hand2 is localized in the posterior region of developing pectoral appendages, and its overexpression causes developmental transformations along the AP axis. Despite abundant experimental data on AP patterning in embryonic limbs, positional memory of regenerating appendages has focused instead on PD regulation.
Adult zebrafish regenerate amputated fins with speed and precision. Recent work has produced cellular and molecular models for blastema formation and regenerative outgrowth during fin regeneration, but positional memory remains largely unexplained (Knopf et al., 2011; Blum and Begemann, 2012). Here, we used RNA sequencing to identify factors with potential roles in positional memory along the AP axis in zebrafish pectoral fins. We found many genes with AP region-specific expression in uninjured fins, several of which are transcription factor genes with known roles in AP patterning of embryonic pectoral appendages. Transgenic overexpression of one of these genes, hand2, modified bone patterning during regeneration, in part through regulation of vitamin D metabolism and signaling. These experiments identify a factor with the characteristics of a positional memory component, and provide evidence that adult zebrafish fins maintain positional information via the sustained regional restriction of key embryonic patterning genes.
MATERIALS AND METHODS
Zebrafish
Males of several zebrafish strains show defects in pectoral fin regeneration (Nachtrab et al., 2011), necessitating the use of females for these experiments. All animals were between 4 and 12 months of age and in an outbred Ekkwill (EK) strain background. Pectoral fins were amputated proximal to the first bifurcation point at approximately one-third of their original length using iridectomy scissors. Fin lengths and widths were measured from images using Leica Application Suite V3.6 software. Widths of the segment proximal to the first bifurcation of the ray were measured in uninjured fins, and the second ray segment distal to the amputation plane was measured in fin regenerates. Heat-shock experiments were performed by giving transgenic and clutchmate controls a daily 38°C heat shock as described (Wills et al., 2008). A Vivo-Morpholino (Gene Tools) was directly microinjected into the posterior region of 3- and 4-dpa pectoral fin regenerates. The translation-blocking hand2 morpholino was described previously (Maves et al., 2009). Fins were collected and dissected 12 hours after the second morpholino injection, at ∼4.5 dpa. 1α,25-dihydroxyvitamin D3 (Sigma, D1530) was dissolved in ethanol to make a 10 μM stock solution and stored at -20°C. For intraperitoneal injections, this stock solution was diluted 1:10 with water and 10 μl was injected per fish. For quantitative PCR assays after heat shocks in transgenic animals during regeneration, fins were collected for RNA isolation 6 hours after the heat shock at 4 dpa unless otherwise indicated. hand2 mutant embryos were collected along with clutchmates from hand2s6 heterozygous crosses and identified visually at 4 dpf for RNA isolation (Yelon et al., 2000).
Construction of transgenic animals
osx:EGFP-CAAX was generated by subcloning an EGFP-CAAX cassette that had been amplified from Tol2kit plasmid #384 (Kwan et al., 2007) downstream of published promoter sequences of medaka osterix (Renn and Winkler, 2009). The full name of this transgenic line is Tg(osterix:EGFP-CAAX)pd51. For alx4a:DsRed2, the first exon of alx4a in the BAC clone CH211-107P11 was replaced with a DsRed2 cassette at the translational initiation site by Red/ET recombineering (GeneBridges). The full name of this transgenic line is Tg(alx4a:DsRed2)pd52. For tbx18:EGFP, the first exon of tbx18 in the BAC clone CH211-197L9 was replaced with an EGFP cassette at the translational initiation site by Red/ET recombineering (GeneBridges). The full name of this transgenic line is Tg(tbx18:EGFP)pd21. For hsp70l:alx4a, hsp70l:id4, hsp70l:lhx9 and hsp70l:hand2, full-length cDNAs were cloned from adult pectoral fins and then subcloned downstream of the inducible hsp70l promoter (Halloran et al., 2000). The full names of these transgenic lines are Tg(hsp70l:alx4a)pd53, Tg(hsp70l:id4)pd54, Tg(hsp70l:lhx9)pd55 and Tg(hsp70l:hand2)pd56. An α-crystallin:EGFP cassette was inserted in reverse orientation to make lens fluorescence an identifier of transgenic animals (Waxman et al., 2008). Purified plasmid or BAC DNA was co-injected with I-SceI into single-cell embryos.
RNA isolation and quantitative PCR (qPCR)
For gene expression analysis, fin regions from three fish were dissected and pooled for each sample. RNA was isolated using Tri Reagent (Sigma). cDNA was synthesized from 1 μg total RNA using the Roche First-Strand Synthesis Kit. qPCR was performed using the Roche LightCycler 480 and SYBR Green I Master Mix. All samples were analyzed in biological triplicate and technical duplicate, and all reactions were performed with an annealing temperature of 60°C. The analysis was performed using the ΔΔCT method as previously described (Yin et al., 2008). Primers are listed in supplementary material Table S1.
RNA sequencing (RNA-Seq)
The two most anterior and posterior rays (AP1 and AP5) were collected and pooled from pectoral fins of 20 6- to 8-month-old zebrafish in duplicate. RNA was isolated using Tri Reagent. Samples were then submitted to the Duke Genome Sequencing and Analysis Core for library preparation and run on an Illumina HiSeq2000. The data were analyzed using TopHat, Bowtie and Cufflinks according to described protocols (Trapnell et al., 2012). Ensembl Zv9.70 was used for genome annotation. Gene ontology analysis was performed using the Princeton University Lewis-Sigler Institute for Integrative Genomics website (http://go.princeton.edu/).
Immunofluorescence and BrdU incorporation
Fins were removed and fixed in 4% paraformaldehyde at room temperature for 1 hour. Staining of fin cryosections was performed as described (Johnson and Weston, 1995; Wills et al., 2008) using a p63 (Tp63) antibody (mouse 4A4, Santa Cruz Biotechnology) at 1:200 or Zns-5 (ZIRC) at 1:200. Imaging and colocalization analysis were performed using a Zeiss LSM 700 confocal microscope. Quantification of pectoral fin BrdU incorporation was as described (Nachtrab et al., 2011).
RESULTS
Regionalized gene expression in adult zebrafish pectoral fins
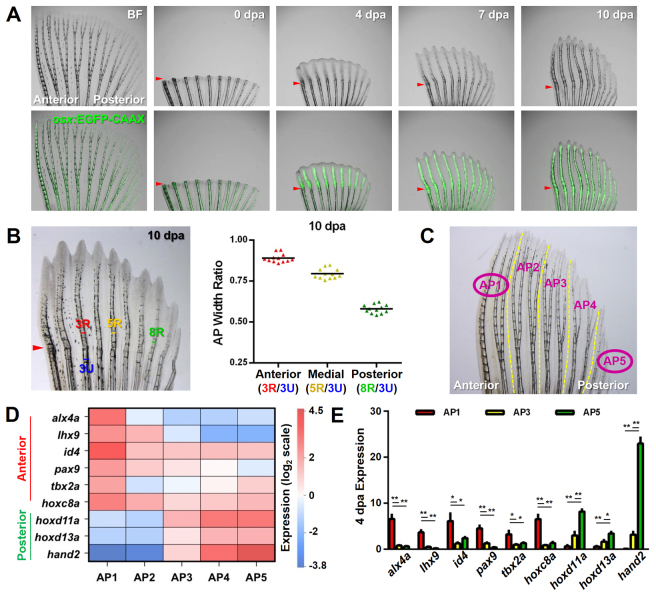
The skeletal components of fins, a set of cylindrical segmented rays, are composed of dermal bone that is formed by the deposition of collagen and other proteins as well as mineral components. Fin rays are connected by intraray mesenchyme and covered by epidermis, and encase fibroblasts, nerves, blood vessels and pigment cells. There are clear differences in fin ray lengths and widths along the AP axes of pectoral fins. For instance, the third (anterior) ray is on average 47% longer and 65% wider than the eighth (posterior) ray at the base where segmentation begins (Fig. 1A; supplementary material Fig. S1). Fin regeneration in zebrafish is initiated by the formation of a blastema at the healed distal tip of each ray by 2-4 days post-amputation (dpa). Fin ray regeneration proceeds by maintenance of proliferative blastemal tissue just distal to a patterning zone, in which osteoblasts align and mineralize bone (Akimenko et al., 2003; Poss et al., 2003). We analyzed ray morphology and osteoblast differentiation events during regeneration, aided by a transgenic reporter strain visualizing expression of the osteoblast transcription factor osterix (osx or sp7). Differences in the patterns of aligned osteoblasts among rays on the AP axis began to manifest during a period from 5-7 dpa, and the AP ray pattern was restored by 10 dpa (Fig. 1A,B).
Fig. 1.
Regeneration of pattern and underlying differential gene expression in pectoral fins. (A) Zebrafish pectoral fins possess and regenerate an anteroposterior (AP) skeletal pattern. Early osteoblasts marked by osx:EGFP-CAAX (green) are present in the regenerate prior to the onset of AP differences, which arise between 5 and 7 days post-amputation (dpa). Arrowheads indicate amputation plane. BF, bright field. (B) Zebrafish robustly regenerate the AP bone pattern. Quantification of the relative ratios between the width of the uninjured portion of anterior ray 3 (blue) and the widths of regenerating rays across the AP axis. n=12; bar indicates mean. (C) Diagram of a pectoral fin defining the two regions used for RNA-Seq (circled AP1 and AP5) and the five regions (AP1-AP5) used for subsequent qPCR validations. (D) Each AP region of uninjured zebrafish pectoral fins expresses a unique AP code of patterning transcription factor genes. Results shown reflect qPCR confirmation of RNA-Seq data, normalized to β-actin 1 (actb1) levels. n=3. (E) The AP regionalized expression of transcription factor genes is maintained during regeneration. qPCR expression profiles at 4 dpa, normalized to actb1 levels. n=3; mean ± s.e.m.; *P<0.05, **P<0.005, Student’s t-test.
To identify genes that might be responsible for AP differences in ray pattern, we sequenced RNAs collected from the most anterior (AP1) and most posterior (AP5) regions of pectoral fins (Fig. 1C; sequencing data are deposited in the NCBI SRA database, accession SRP027598). We found 235 predicted genes with significantly different expression in these two regions, representing ∼0.8% of the transcriptome (supplementary material Fig. S2A). These 235 predicted genes comprised 195 annotated genes and 40 putative genes. Of the 195 AP genes, 105 were elevated in anterior rays and 90 in posterior rays. To assess whether certain categories or classes of genes were enriched in our AP dataset, we performed a gene ontology (GO) search. Using the GO category ‘biological process’, we found our AP genes to be enriched for general terms such as ‘developmental process’ and ‘biological regulation’. However, we also saw enrichment for the more specific terms ‘fin development’, ‘nervous system development’ and ‘regulation of transcription, DNA-dependent’ (supplementary material Fig. S2B).
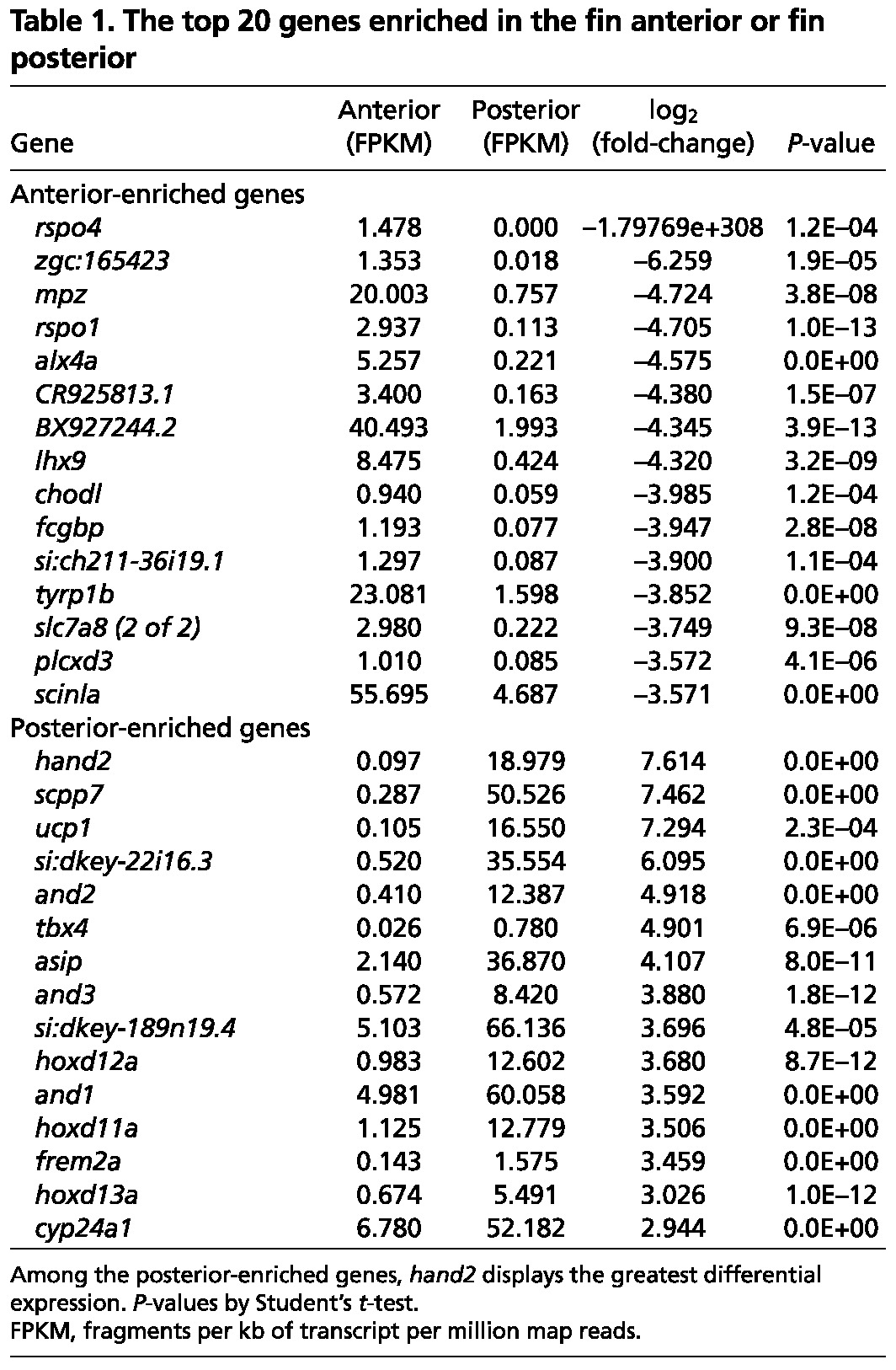
For several reasons, we focused our initial analysis on differentially expressed transcription factor genes. First, transcription factors are central to many developmental programs; thus, their regionalized expression has the potential to impact multiple downstream genes. Second, as mentioned above, a rich field of embryonic limb development has detailed the relationship between transcription factors and AP skeletal patterning (Qu et al., 1997; Charité et al., 2000; Fernandez-Teran et al., 2000; Zákány et al., 2004; Tzchori et al., 2009; Galli et al., 2010; Sheth et al., 2012). Third, our dataset revealed many examples of developmental transcription factors with expression differences along the AP axis. In particular, the transcription factor hand2, which is crucial for pectoral fin development and expressed in the posterior region of developing pectoral fin and forelimb buds (Charité et al., 2000; Fernandez-Teran et al., 2000; Yelon et al., 2000), showed the most polarized expression of all genes enriched in posterior fin rays (Table 1). Additionally, the embryonic anterior patterning factors alx4a and lhx9 were fifth and eighth on the list of genes enriched in anterior rays (Table 1).
Table 1.
The top 20 genes enriched in the fin anterior or fin posterior
To define transcription factor expression signatures in pectoral fins, we performed quantitative RT-PCR (qPCR) using tissues across five different regions of the AP axis. This approach confirmed restriction of hand2 to posterior fin rays, and also verified region-specific expression for eight other transcription factor genes: alx4a, lhx9, id4, pax9, tbx2a, hoxc8a, hoxd11a and hoxd13a. Six of these eight transcription factors have roles in AP patterning during limb or fin development (Qu et al., 1997; Harrelson et al., 2004; Zákány et al., 2004; McGlinn et al., 2005; Tzchori et al., 2009), whereas id4 has been implicated in bone homeostasis (Tokuzawa et al., 2010). The anterior expression of the Hoxc genes has been described during pectoral fin development (Molven et al., 1990), and HOXC8 has specifically been shown to be expressed in the anterior region of developing chick wings (Nelson et al., 1996). Gauged by the expression of these nine factors, each set of two rays across the AP axis displayed a unique gene signature (Fig. 1D). Notably, hand2, hoxd13a, alx4a and lhx9 displayed regional expression in adult fins similar to their reported expression in embryonic forelimb or pectoral fin buds (Qu et al., 1997; Charité et al., 2000; Ahn and Ho, 2008; Wang et al., 2011). If important for patterning regenerating structures, these transcription factors would be expected to retain their regionalized character during regeneration. Indeed, at 4 dpa, all nine factors had a profile similar to that in uninjured fins (Fig. 1E).
In summary, adult zebrafish pectoral fins retain region-specific signatures of transcription factor genes important for AP patterning of embryonic pectoral appendages, consistent with potential roles in patterning and positional memory.
Fin fibroblasts and osteoblasts differ across the AP axis in the expression of patterning transcription factor genes
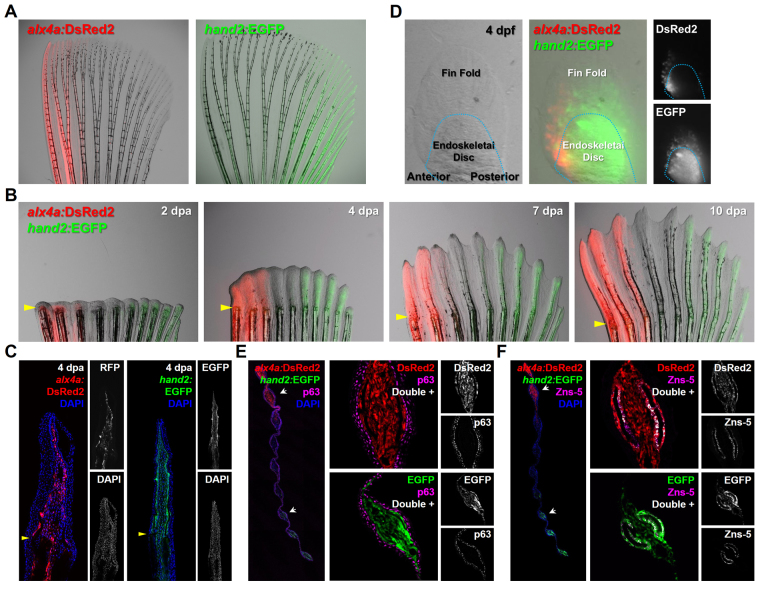
To visualize the differential expression of anterior and posterior factors, we generated a BAC transgenic reporter line Tg(alx4a:DsRed2)pd52 and also assessed reporter expression in the Tg(hand2:EGFP)pd24 BAC transgenic line (Kikuchi et al., 2011). Using these lines, we found that the alx4a region extended from the most anterior to the third rays of the pectoral fin. Conversely, the hand2 domain extended from the sixth ray to the posterior edge of the fin (Fig. 2A). In uninjured fins, both regulatory sequences activated expression along the entire PD axis of the fin, and similarly during regeneration to the near distal tips (Fig. 2A-C). These adult expression domains were reminiscent of their profiles in developing embryonic fin buds (Fig. 2D). We visualized in the same way the expression of a third transcription factor, tbx18, which is known to be expressed along the entire AP axis of developing zebrafish pectoral fins (Liu and Stainier, 2010). The BAC transgenic reporter line Tg(tbx18:EGFP)pd21 revealed fin-wide expression of tbx18 in adult zebrafish pectoral fins, a result verified by qPCR (supplementary material Fig. S3A,B).
Fig. 2.
Visualization of AP region-specific transcription factor expression in fin fibroblasts and osteoblasts. (A) Expression of fluorescent transgenic reporters in adult pectoral fins. The alx4a:DsRed2 domain ranges from the most anterior to the third ray. The hand2:EGFP domain extends from the posterior edge to the sixth ray of the fin. (B) The AP expression characteristics of alx4a:DsRed2 and hand2:EGFP are maintained throughout regeneration. Arrowheads indicate amputation plane. (C) Longitudinal sections at 4 dpa confirm reporter expression to nearly the distal tip of the fin. Arrowheads indicate amputation plane. (D) Expression of fluorescent reporters in embryonic pectoral fins. At 4 days post-fertilization (dpf), both reporters display region-specific expression. The expression is similar to that of the adult, but the double-negative medial region is not yet defined. (E) Transverse sections indicate that the expression of each reporter is restricted to the fin mesenchyme. The antibody against p63 marks fin epidermis adjacent to the mesenchymal compartment. Arrowheads indicate amputation plane. (F) Transverse sections indicating that both alx4a:DsRed2 and hand2:EGFP are expressed in a population of fin osteoblasts identifiable by Zns-5 immunoreactivity. Arrowheads indicate amputation plane.
To assess expression differences among fins, we also examined alx4a-, hand2- and tbx18-driven transgenic reporter expression in other adult zebrafish fins. This revealed a similar pattern of alx4a, hand2 and tbx18 expression in pelvic fins as in pectoral fins. In the anal and dorsal fins, alx4a was expressed in the most anterior marginal ray, whereas hand2 was expressed weakly in the most caudal ray. In the caudal fin, alx4a was expressed only in the most ventral ray, and hand2 expression was not detectable. tbx18 was expressed weakly in these unpaired fin types (supplementary material Fig. S3C,D). These observations indicated that all adult zebrafish fins maintain the expression of transcription factors associated with their initial development, with different expression domains in differentially patterned appendages.
To determine which cells contained AP regionalized expression of hand2 and alx4a, we histologically assessed pectoral fins from the reporter strains. Longitudinal and transverse sections of regenerates revealed that the expression of hand2 and alx4a was limited to the mesenchymal compartment of fins (Fig. 2C,E). Although the majority of cells in the mesenchyme are fibroblasts, we also found that many osteoblasts lining the bone rays were distinctly positive for either of these transcription factors (Fig. 2F). Thus, AP patterning transcription factors are maintained in unique expression domains in the bone-forming cells of uninjured and regenerating fins. Although differentiated osteoblasts from different fin ray regions appear identical and have been considered as such in models of regeneration, it is now clear that they have different gene expression signatures.
hand2 overexpression alters bone lengths and widths in regenerating fins
Next, we examined whether the regional restriction of transcription factors is required for normal patterning during fin regeneration. We generated transgenic zebrafish permitting heat-inducible expression of the anterior genes alx4a, lhx9 or id4 or of the posterior gene hand2. This approach enables inducible expression of a candidate factor across the entire AP axis of the adult pectoral fin. We reasoned that the overexpression of potential positional memory components would alter regenerative patterning and be manifested in changes in the lengths and widths of fin rays. This approach is analogous to the overexpression techniques that defined key regions and factors for embryonic limb patterning in mouse and chick, prior to the development of many conditional knockout models (Maccabe et al., 1973; Riddle et al., 1993; Charité et al., 2000).
We identified inducible transgenic lines for the anterior genes alx4a, id4 and lhx9 [Tg(hsp70l:alx4a)pd53, Tg(hsp70l:id4)pd54 and Tg(hsp70l:lhx9)pd55] that enabled ∼13-, 10- and 13-fold increases, respectively, in the expression of these genes in posterior rays after a single heat shock at 4 dpa (supplementary material Fig. S4A). We then examined whether this overexpression could alter regenerative pattern by amputating pectoral fins in these lines and administering a daily heat shock. At 7 dpa, there was no gross change in the appearance of the fins (supplementary material Fig. S4B). We quantified the lengths and widths of regenerated rays to ascertain whether there were any subtle alterations. Heightened id4 levels during regeneration produced an 11% increase in posterior fin ray lengths (supplementary material Fig. S4C). Elevated alx4a increased relative medial and posterior ray widths by 4% and 6%, respectively, and there was a 6% increase in the anterior and medial regions caused by id4 overexpression (supplementary material Fig. S4D). These modest changes in patterning suggested that the anterior factors we examined are not individually sufficient to influence patterning in regenerating pectoral fin rays.
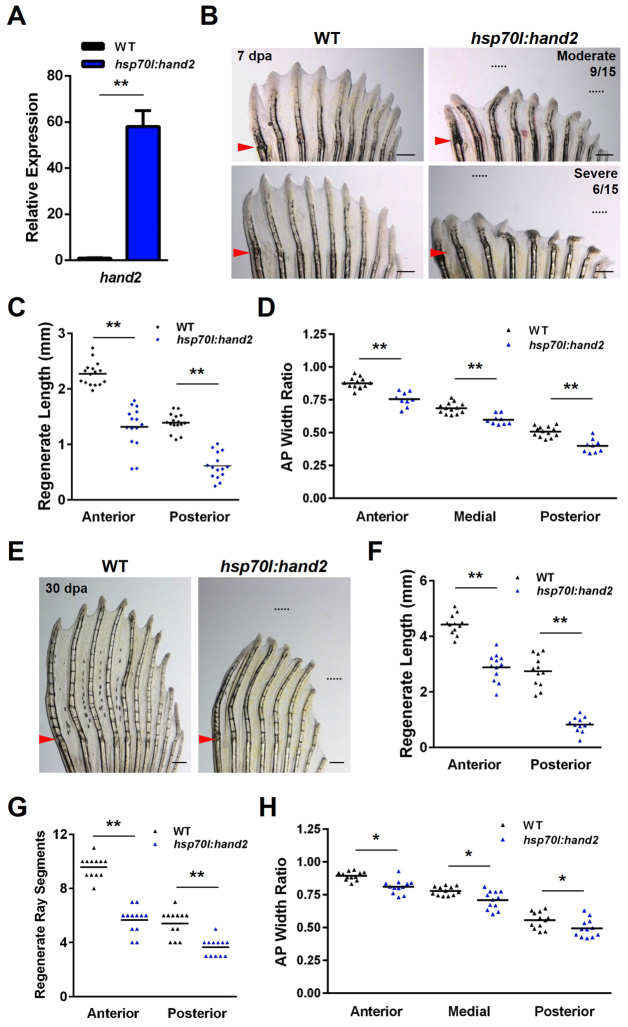
We next assessed the effects of hand2 overexpression [Tg(hsp70l:hand2)pd56], which was increased 58-fold in anterior fin regions after a single heat shock at 4 dpa (Fig. 3A), a change that reflects more the negligible endogenous levels of hand2 mRNA in anterior rays, rather than the sheer magnitude of hand2 induction (which is similar to that of alx4a, lhx9 or id4 after heat-shock induction of the respective transgenes) (Fig. 1D,E; supplementary material Fig. S5A). Seven days of hand2 induction initiated after amputation was sufficient to produce moderate to severe defects in the patterns of regenerating fin rays (Fig. 3B). Overall, there was a 42% decrease in the lengths of regenerated anterior rays at 7 dpa and a 56% decrease in lengths of posterior rays (Fig. 3C). We also found reductions in width ratios across pectoral fins ranging from 14 to 22% (Fig. 3D). To assess whether the hand2 overexpression phenotype was the result of a transient delay in regeneration, we extended the experiment to 30 dpa, approximately twice the time within which regeneration is normally completed. We observed shortened regenerates with smaller rays and fewer segments in the hsp70l:hand2 transgenic fish at 30 dpa, similar to phenotypes at 7 dpa (Fig. 3E-G), indicating that the patterning phenotype was not the result of a transient regenerative delay.
Fig. 3.
Overexpression of hand2 during fin regeneration alters ray patterning. (A) hand2 expression is induced 58-fold in anterior regions of hsp70l:hand2 pectoral fins 4 hours after a single heat shock at 4 dpa. Values are normalized to actb1 levels and relative to wild-type controls. n=3; mean ± s.e.m. (B) Appearance of hsp70l:hand2 and wild-type clutchmate fins at 7 dpa after a series of daily heat shocks. hand2 overexpression generates shorter rays with a reduced number of bone segments. Phenotypes range from moderate (upper right) to severe (lower right). Representative regenerative growths of wild-type rays 3 and 8 are denoted by dotted lines. (C) Overexpression of hand2 reduces the lengths of regenerating rays across the AP axis of pectoral fins. n=16 (wild type) and n=15 (hsp70l:hand2). (D) Overexpression of hand2 during regeneration reduces the widths of regenerating fin rays across the AP axis. Transgenic fish that displayed a moderate phenotype were quantified. n=13 (wild type) and n=6 (hsp70l:hand2). (E) Appearance of hsp70l:hand2 and wild-type clutchmate fins at 30 dpa after daily heat shocks. hsp70l:hand2 regenerates remain stunted with shorter and thinner fin rays. Representative regenerative growths of wild-type rays 3 and 8 are denoted by dotted lines. (F) Quantification of hsp70l:hand2 ray lengths at 30 dpa. n=12. (G) Quantification of hsp70l:hand2 segment numbers at 30 dpa. n=12. (H) Quantification of hsp70l:hand2 ray widths at 30 dpa. n=12. *P<0.05, **P<0.005, Student’s t-test; bar indicates mean. Arrowheads (B,E) indicate amputation plane. Scale bars: 0.5 mm.
Overexpression of hand2 in developing mammalian limbs results in striking phenotypes, including polydactyly and anterior-to-posterior digit conversions (Charité et al., 2000). However, overexpression of hand2 during zebrafish pectoral fin regeneration caused a less striking phenotype - an increase in the posterior character of all fin rays. There are several reasons that might explain this result. First, unlike an embryonic fin bud, mammalian limb bud or newt limb blastema, in which a single primordium is patterned by signaling molecules, an adult fin regenerate is a composite of individual ray primordia. The overall fin shape is a cumulative readout of the patterning of the individual ray blastemas to give each ray a length and width, and it is unlikely that the overexpression of a single factor such as hand2 would produce a phenotype similar to that seen in other contexts. Second, hand2 is only one of several transcription factors with differential AP expression in adult pectoral fins, and overexpression of hand2 did not cause similar significant alternations in the expression of other AP transcription factors (supplementary material Fig. S5B). Third, heat shock raises expression without altering the underlying endogenous asymmetry, leading to a shallower but still significant AP gradient of hand2 across the AP axis (supplementary material Fig. S5C). Finally, as many adult fin rays have no detectable hand2 expression, there is unlikely to be a striking change in length and width caused by changing only Hand2 levels.
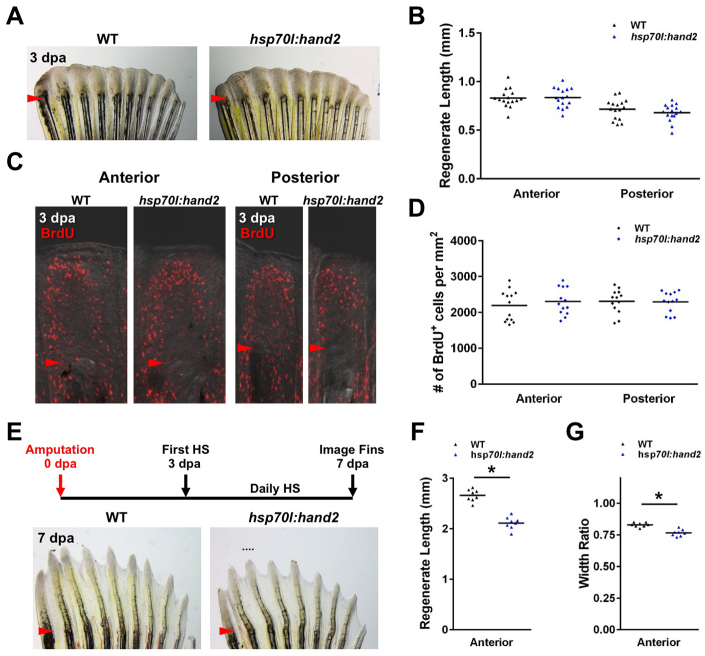
To identify a critical window for the effects of hand2 overexpression, we controlled the timing and number of heat shocks with respect to amputation. When hand2 was induced only during the first 3 days after amputation and prior to significant bone deposition, there was no significant change in the lengths of regenerating fins (Fig. 4A,B). In agreement with this, daily hand2 induction had no significant effect on indices of blastemal cell proliferation at 3 dpa, as compared with heat-shocked wild-type zebrafish (Fig. 4C,D). However, when daily hand2 induction was initiated at 3 dpa, there was a 20% decrease in the lengths of regenerated anterior rays and an 8% decrease in ray widths by 7 dpa (Fig. 4E-G). These results indicated that elevating the levels of the normally posterior-restricted hand2 throughout the fin does not affect early regenerative growth, but alters ray patterning by reducing ray lengths and widths.
Fig. 4.
hand2 overexpression exerts its effects during later stages of regeneration. (A,B) No difference in the appearance of hsp70l:hand2 and wild-type clutchmate fins at 3 dpa after a series of daily heat shocks. n=16. (C,D) Blastemal proliferation as measured by BrdU incorporation is also unaffected by hand2 overexpression. n=13. (E) Stunted regeneration in an hsp70l:hand2 pectoral fin compared with a wild-type clutchmate fin at 7 dpa after a series of daily heat shocks beginning at 3 dpa. Representative regenerative growth of wild-type ray 3 is denoted by a dotted line. (F,G) Significant reductions in anterior ray lengths and widths manifest when hand2 is overexpressed during later stages of regeneration. n=8. *P<0.05, Student’s t-test; bar indicates mean. Arrowheads (A,C,E) indicate amputation plane.
Together, our data indicate that the maintenance of hand2 in the mesenchyme of posterior rays contributes to the restoration of posterior ray characteristics during regeneration.
Hand2 effects on vitamin D metabolism in zebrafish fins
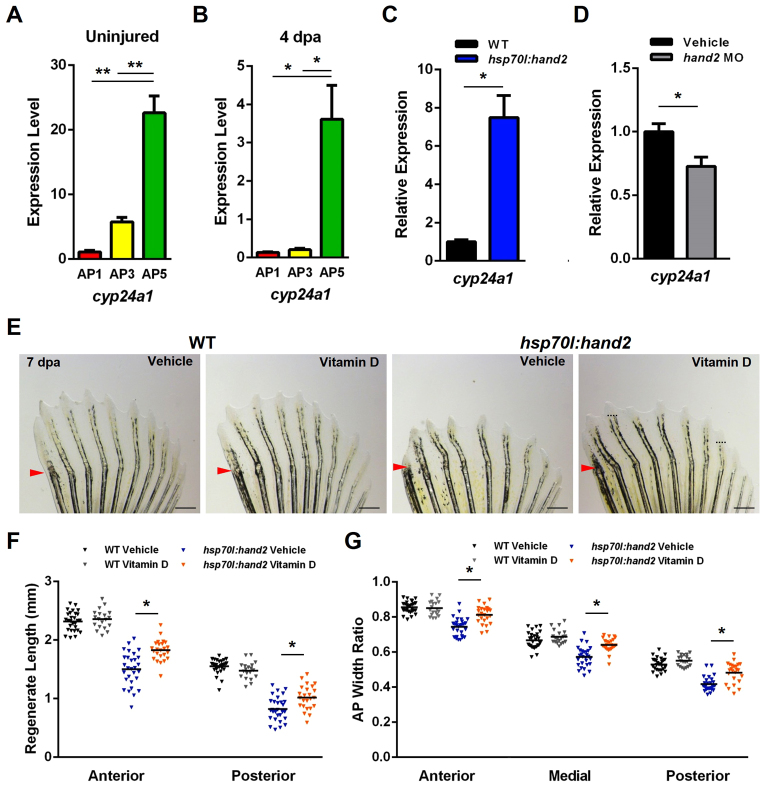
hand2 regulates the localization of sonic hedgehog (shh) in the posterior region of developing embryonic forelimbs, wings or pectoral fins (Charité et al., 2000; Fernandez-Teran et al., 2000; Yelon et al., 2000; Galli et al., 2010). However, shha is expressed in epidermal cells at the distal tip of each uninjured and regenerating adult fin ray, regardless of whether that ray expresses hand2 (Quint et al., 2002; Lee et al., 2009). We examined our RNA-Seq dataset for possible alternative targets of hand2 during regeneration, with a focus on genes known to be involved in bone formation or homeostasis. A significant AP asymmetry was observed for cyp24a1, which encodes a vitamin D-inactivating enzyme (Knutson and DeLuca, 1974), which was expressed at higher levels in posterior rays, like hand2 (Table 1, Fig. 5A). Vitamin D is a known regulator of calcium and bone homeostasis in mammals and has also been shown to influence bone formation in zebrafish larvae (Gardiner et al., 2000; Fleming et al., 2005; Baldock et al., 2006). Although levels of cyp24a1 expression were reduced during regeneration, the AP distribution of expression was maintained (Fig. 5B). To test whether hand2 regulates the posterior expression domain of cyp24a1 during fin regeneration, we measured the effects of transgenic hand2 elevation at 4 dpa. Increased hand2 expression raised anterior cyp24a1 expression 7.5-fold, while also elevating posterior cyp24a1 levels and maintaining an AP gradient (Fig. 5C; supplementary material Fig. S6A). Experimental induction of alx4a, lhx9 or id4 during regeneration had no detectable effect on posterior cyp24a1 expression (supplementary material Fig. S6B).
Fig. 5.
AP regionalization of vitamin D signaling controlled by Hand2. (A) cyp24a1, which encodes a vitamin D-inactivating enzyme, is more highly expressed in posterior regions of uninjured pectoral fins. (B) Posterior enrichment of cyp24a1 is maintained during regeneration at 4 dpa. (C) hand2 induction daily during regeneration elevates anterior cyp24a1 expression ∼7.5-fold at 4 dpa relative to heat-shocked wild-type clutchmate controls. (D) hand2 morpholino injection into pectoral fin regenerates reduces posterior cyp24a1 expression by ∼28% relative to vehicle-injected clutchmate controls. (A-D) Expression is normalized to actb1. *P<0.05, **P<0.005, Student’s t-test. n=3; mean ± s.e.m. (E) Appearance of 7-dpa fins from hsp70l:hand2 and wild-type clutchmates given a daily heat shock and either daily vehicle or vitamin D injections. Vitamin D injection had little effect on wild-type regeneration, but partially suppressed the effects on bone patterning caused by hand2 overexpression. Representative regenerative growth of hsp70l:hand2 vehicle-injected rays 3 and 8 is denoted by dotted lines. Arrowheads indicate amputation plane. (F) Quantification of the effects of daily vitamin D injection on ray lengths, indicating improvement in hsp70l:hand2 animals. (G) Daily vitamin D injection increases ray widths in regenerating hsp70l:hand2 fins. (F,G) n=20-28; bar indicates mean. **P<0.05, Student’s t-test. Scale bars: 0.6 mm.
To further test this relationship, we injected a translation-blocking morpholino known to phenocopy hand2 loss-of-function mutations into 3- and 4-dpa regenerates (Maves et al., 2009). These injections caused a 28% decrease in cyp24a1 levels, consistent with Hand2 regulation of cyp24a1 during fin regeneration (Fig. 5D). We also observed a 32% decrease in cyp24a1 levels in 4-dpf hand2 mutant embryos compared with their clutchmates (supplementary material Fig. S6C).
We next examined whether posterior cyp24a1 expression levels correlated with differential vitamin D signaling across the AP axis of pectoral fins. calb2a, bglap and sparc, which are known targets of vitamin D signaling in mammals (McDonnell et al., 1989; Wasserman and Fullmer, 1989; zur Nieden et al., 2003), were induced in uninjured pectoral fins after intraperitoneal injection of vitamin D, indicating that they are also vitamin D-responsive in zebrafish (supplementary material Fig. S6D). The expression of each of these genes was significantly higher in the cyp24a1-low anterior regions of uninjured pectoral fins (supplementary material Fig. S6E), suggesting differential vitamin D signaling across the fin AP axis. Additionally, hand2 overexpression in regenerating pectoral fins reduced calb2a, bglap and sparc levels by 64%, 57% and 22%, respectively, consistent with a model in which Hand2 regulates vitamin D signaling in pectoral fin rays (supplementary material Fig. S6F).
We also examined whether Hand2 influences a second class of posterior genes involved in bone formation and fin ray pattern. The actinodin genes are structural components of actinotrichia, which are unmineralized fibrils found in developing pectoral fins (Zhang et al., 2010). Unexpectedly, the entire gene family, and1-4, was elevated in the posterior regions of uninjured adult pectoral fins, as detected by RNA-Seq and qPCR (Table 1; supplementary material Fig. S7A). In contrast to cyp24a1, daily induction of hand2 during regeneration did not significantly increase the levels of actinodin family genes in the anterior regions of pectoral fins at 4 dpa (supplementary material Fig. S7B). This result indicates that Hand2 controls a subset of genes that show AP regionalized expression and potentially contribute to posterior ray characteristics.
To define the significance of vitamin D signaling as a target of Hand2, we supplemented zebrafish with vitamin D and assessed pectoral fin regeneration. Daily vitamin D injection had no effects on pectoral fin regeneration in wild-type fish. By contrast, this systemic regimen led to 24% and 20% increases in the lengths of regenerating anterior and posterior rays, respectively, during hand2 overexpression (Fig. 5E,F). Accompanying the length increase was an ∼11% increase in the relative widths of the rays across the AP axis (Fig. 5G).
These gene expression and pharmacological rescue experiments together support a model in which Hand2 controls patterning in regenerating pectoral fins by modulating vitamin D metabolism and signaling (Fig. 6).
Fig. 6.
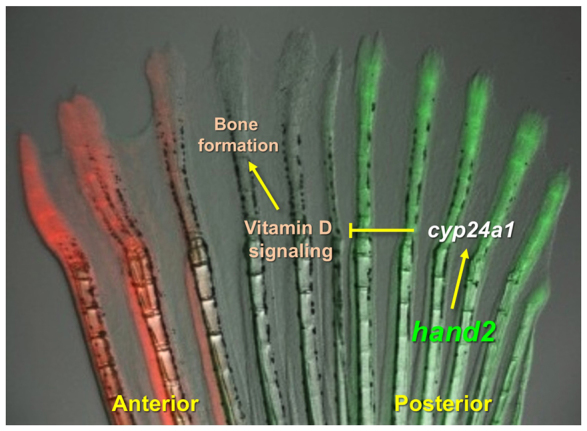
Model for Hand2 influences on positional memory. Adult zebrafish pectoral fins maintain region-specific expression of transcription factor genes. Levels of the posterior transcription factor gene hand2 can help control bone patterning during pectoral fin regeneration. Hand2 levels regulate bone formation during regeneration by direct or indirect regulation of the vitamin D-inactivating enzyme encoded by cyp24a1, restricting bone formation in the posterior rays of pectoral fins.
DISCUSSION
A defining feature of appendage regeneration is the maintenance and recall of positional information, evident in the restoration of correctly patterned structures. Here we discovered a transcription factor gene expression signature and a downstream signaling pathway that might underlie AP positional memory in zebrafish pectoral fins. Many genes displayed region-specific expression across the AP axis of uninjured adult pectoral fins, and we examined the impact of four of them on patterning during regeneration. One of these transcription factor genes, hand2, displayed the expected aspects of a regulator of positional memory. First, hand2 expression is maintained in an AP region-specific manner in bone-forming cells of uninjured and regenerating fins. That is, anterior ray osteoblasts have a different transcription factor signature to posterior ray osteoblasts, as defined minimally for hand2 and alx4 expression. Second, hand2 overexpression changes the AP patterning of bone during fin regeneration. Furthermore, our data indicate that hand2 regulates posterior bone formation in part through local control of the activity of a systemic signal - vitamin D.
Our findings reveal that zebrafish pectoral fins maintain a basal level of transcription factors in preferential domains throughout all life stages, as opposed to turning off expression after patterning and differentiation. We suggest that this concept is likely to be central to positional memory in zebrafish fins, and potentially other regenerative vertebrate tissues. The expression of embryonic signaling factors in uninjured adult structures has been indicated in other examples of appendage regeneration (Nicolas et al., 2003; Schnapp et al., 2005; Wills et al., 2008; Poss, 2010), and mammalian fibroblasts are known to express region-specific Hox gene codes (Chang et al., 2002; Rinn et al., 2006).
Although positional memory factors are likely to recapitulate aspects of the embryonic developmental program, they are also likely to regulate pattern by mechanisms that are distinct from embryogenesis. Relevant to this second idea, our data do not indicate that Hand2 influences pattern via Shh signaling in adult pectoral fins as it does in embryonic pectoral appendages. Rather, they implicate the control of vitamin D signaling as an important regulatory mechanism, an interaction that to our knowledge has not previously been reported. This mechanism might be relevant to other tissues in which Hand2 is of influence, such as the developing craniofacial structures. Previous work has demonstrated the complex role of vitamin D in mammalian bone homeostasis (Lieben and Carmeliet, 2013). Although implanting vitamin D-soaked beads was shown to have somewhat minor effects on axolotl limb regeneration (Washabaugh and Tsonis, 1995), to our knowledge there have been no reports that vitamin D signaling helps define AP patterning in regenerating appendages. This lack of evidence can be explained in part by the fact that only when signaling was compromised by hand2 overexpression was a vitamin D-related phenotype observed (Fig. 5E-G). Aside from appendage regeneration, vitamin D has been shown to influence liver, axon and skeletal muscle regeneration (Ethier et al., 1990; Chabas et al., 2008; Stratos et al., 2013). It will be interesting to examine whether the regulation of vitamin D signaling is important in additional regenerative contexts.
By what mechanism(s) do adult cells maintain the regionalized expression of patterning transcription factors? There are at least two important gene regulatory components of regeneration that might on the surface appear contradictory. First, cells must be capable of rapid gene expression flux to enact major changes such as de-differentiation in response to injury. Second, as shown here, cells also lock in the expression of key developmental regulators in a region-specific manner throughout life. Novel epigenetic regulation, possibly at the chromatin level, is likely to underlie this versatility.
Although we found differential AP expression of numerous transcription factors, functional manipulation of just one of these produced clear patterning effects during regeneration. This might reflect the limitations of the overexpression technologies currently available for use in adult zebrafish fins. Alternatively, it is possible that multiple transcription factors act in concert as a code to specify positional information (Gebelein et al., 2004; Dasen et al., 2005; Tümpel et al., 2009). Consistent with this notion, we also found posterior-enriched genes that were not influenced by the overexpression of hand2 in anterior rays. Moreover, complete reprogramming of positional memory is likely to require simultaneous increases and decreases in gene expression affecting multiple genes. Emerging genetic toolsets for zebrafish and other highly regenerative vertebrates should enable the discovery of core modules of positional memory (Meng et al., 2008; Bedell et al., 2012; Dahlem et al., 2012), bringing us closer to an understanding of how regeneration recalls and replicates complex tissues.
Acknowledgments
We thank J. Burris, A. Eastes, P. Williams and N. Blake for zebrafish care; the Duke Genome Sequencing and Analysis Core for RNA sequencing; and K.D.P. laboratory members for comments on the manuscript.
Footnotes
Funding
V.A.T. was supported by a Graduate Research Fellowship [1106401] from the National Science Foundation. This work was supported by a grant from the National Institutes of Health [GM074057] to K.D.P. K.D.P. is a Howard Hughes Medical Institute Early Career Scientist. Deposited in PMC for release after 6 months.
Competing interests statement
The authors declare no competing financial interests.
Author contributions
G.N. conducted the experiments and analyzed the data, G.N. and K.D.P. designed the experiments and wrote the paper, K.K. generated the hand2:EGFP and tbx18:EGFP reporter lines, and V.A.T. performed RT-PCR experiments.
Supplementary material
Supplementary material available online at http://dev.biologists.org/lookup/suppl/doi:10.1242/dev.098798/-/DC1
References
- Ahn D., Ho R. K. (2008). Tri-phasic expression of posterior Hox genes during development of pectoral fins in zebrafish: implications for the evolution of vertebrate paired appendages. Dev. Biol. 322, 220–233 [DOI] [PubMed] [Google Scholar]
- Akimenko M. A., Marí-Beffa M., Becerra J., Géraudie J. (2003). Old questions, new tools, and some answers to the mystery of fin regeneration. Dev. Dyn. 226, 190–201 [DOI] [PubMed] [Google Scholar]
- Baldock P. A., Thomas G. P., Hodge J. M., Baker S. U., Dressel U., O’Loughlin P. D., Nicholson G. C., Briffa K. H., Eisman J. A., Gardiner E. M. (2006) Vitamin D action and regulation of bone remodeling: suppression of osteoclastogenesis by the mature osteoblast. J. Bone Miner. Res. 21, 1618–1626 [DOI] [PubMed] [Google Scholar]
- Bedell V. M., Wang Y., Campbell J. M., Poshusta T. L., Starker C. G., Krug R. G., II, Tan W., Penheiter S. G., Ma A. C., Leung A. Y., et al. (2012). In vivo genome editing using a high-efficiency TALEN system. Nature 491, 114–118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blum N., Begemann G. (2012). Retinoic acid signaling controls the formation, proliferation and survival of the blastema during adult zebrafish fin regeneration. Development 139, 107–116 [DOI] [PubMed] [Google Scholar]
- Buckingham M. E., Meilhac S. M. (2011). Tracing cells for tracking cell lineage and clonal behavior. Dev. Cell 21, 394–409 [DOI] [PubMed] [Google Scholar]
- Chabas J. F., Alluin O., Rao G., Garcia S., Lavaut M. N., Risso J. J., Legre R., Magalon G., Khrestchatisky M., Marqueste T., et al. (2008). Vitamin D2 potentiates axon regeneration. J. Neurotrauma 25, 1247–1256 [DOI] [PubMed] [Google Scholar]
- Chang H. Y., Chi J. T., Dudoit S., Bondre C., van de Rijn M., Botstein D., Brown P. O. (2002). Diversity, topographic differentiation, and positional memory in human fibroblasts. Proc. Natl. Acad. Sci. USA 99, 12877–12882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charité J., McFadden D. G., Olson E. N. (2000). The bHLH transcription factor dHAND controls Sonic hedgehog expression and establishment of the zone of polarizing activity during limb development. Development 127, 2461–2470 [DOI] [PubMed] [Google Scholar]
- da Silva S. M., Gates P. B., Brockes J. P. (2002). The newt ortholog of CD59 is implicated in proximodistal identity during amphibian limb regeneration. Dev. Cell 3, 547–555 [DOI] [PubMed] [Google Scholar]
- Dahlem T. J., Hoshijima K., Jurynec M. J., Gunther D., Starker C. G., Locke A. S., Weis A. M., Voytas D. F., Grunwald D. J. (2012). Simple methods for generating and detecting locus-specific mutations induced with TALENs in the zebrafish genome. PLoS Genet. 8, e1002861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dasen J. S., Tice B. C., Brenner-Morton S., Jessell T. M. (2005). A Hox regulatory network establishes motor neuron pool identity and target-muscle connectivity. Cell 123, 477–491 [DOI] [PubMed] [Google Scholar]
- Duboc V., Logan M. P. (2009). Building limb morphology through integration of signalling modules. Curr. Opin. Genet. Dev. 19, 497–503 [DOI] [PubMed] [Google Scholar]
- Echeverri K., Tanaka E. M. (2005). Proximodistal patterning during limb regeneration. Dev. Biol. 279, 391–401 [DOI] [PubMed] [Google Scholar]
- Ethier C., Kestekian R., Beaulieu C., Dubé C., Havrankova J., Gascon-Barré M. (1990). Vitamin D depletion retards the normal regeneration process after partial hepatectomy in the rat. Endocrinology 126, 2947–2959 [DOI] [PubMed] [Google Scholar]
- Fernandez-Teran M., Piedra M. E., Kathiriya I. S., Srivastava D., Rodriguez-Rey J. C., Ros M. A. (2000). Role of dHAND in the anterior-posterior polarization of the limb bud: implications for the Sonic hedgehog pathway. Development 127, 2133–2142 [DOI] [PubMed] [Google Scholar]
- Fleming A., Sato M., Goldsmith P. (2005). High-throughput in vivo screening for bone anabolic compounds with zebrafish. J. Biomol. Screen. 10, 823–831 [DOI] [PubMed] [Google Scholar]
- Galli A., Robay D., Osterwalder M., Bao X., Bénazet J. D., Tariq M., Paro R., Mackem S., Zeller R. (2010). Distinct roles of Hand2 in initiating polarity and posterior Shh expression during the onset of mouse limb bud development. PLoS Genet. 6, e1000901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardiner E. M., Baldock P. A., Thomas G. P., Sims N. A., Henderson N. K., Hollis B., White C. P., Sunn K. L., Morrison N. A., Walsh W. R., et al. (2000). Increased formation and decreased resorption of bone in mice with elevated vitamin D receptor in mature cells of the osteoblastic lineage. FASEB J. 14, 1908–1916 [DOI] [PubMed] [Google Scholar]
- Gebelein B., McKay D. J., Mann R. S. (2004). Direct integration of Hox and segmentation gene inputs during Drosophila development. Nature 431, 653–659 [DOI] [PubMed] [Google Scholar]
- Gurley K. A., Rink J. C., Sánchez Alvarado A. (2008). Beta-catenin defines head versus tail identity during planarian regeneration and homeostasis. Science 319, 323–327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halloran M. C., Sato-Maeda M., Warren J. T., Su F., Lele Z., Krone P. H., Kuwada J. Y., Shoji W. (2000). Laser-induced gene expression in specific cells of transgenic zebrafish. Development 127, 1953–1960 [DOI] [PubMed] [Google Scholar]
- Harrelson Z., Kelly R. G., Goldin S. N., Gibson-Brown J. J., Bollag R. J., Silver L. M., Papaioannou V. E. (2004). Tbx2 is essential for patterning the atrioventricular canal and for morphogenesis of the outflow tract during heart development. Development 131, 5041–5052 [DOI] [PubMed] [Google Scholar]
- Johnson S. L., Weston J. A. (1995). Temperature-sensitive mutations that cause stage-specific defects in zebrafish fin regeneration. Genetics 141, 1583–1595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kikuchi K., Holdway J. E., Major R. J., Blum N., Dahn R. D., Begemann G., Poss K. D. (2011). Retinoic acid production by endocardium and epicardium is an injury response essential for zebrafish heart regeneration. Dev. Cell 20, 397–404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knopf F., Hammond C., Chekuru A., Kurth T., Hans S., Weber C. W., Mahatma G., Fisher S., Brand M., Schulte-Merker S., et al. (2011). Bone regenerates via dedifferentiation of osteoblasts in the zebrafish fin. Dev. Cell 20, 713–724 [DOI] [PubMed] [Google Scholar]
- Knutson J. C., DeLuca H. F. (1974). 25-Hydroxyvitamin D3-24-hydroxylase. Subcellular location and properties. Biochemistry 13, 1543–1548 [DOI] [PubMed] [Google Scholar]
- Kumar A., Gates P. B., Brockes J. P. (2007a). Positional identity of adult stem cells in salamander limb regeneration. C. R. Biol. 330, 485–490 [DOI] [PubMed] [Google Scholar]
- Kumar A., Godwin J. W., Gates P. B., Garza-Garcia A. A., Brockes J. P. (2007b). Molecular basis for the nerve dependence of limb regeneration in an adult vertebrate. Science 318, 772–777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwan K. M., Fujimoto E., Grabher C., Mangum B. D., Hardy M. E., Campbell D. S., Parant J. M., Yost H. J., Kanki J. P., Chien C. B. (2007). The Tol2kit: a multisite gateway-based construction kit for Tol2 transposon transgenesis constructs. Dev. Dyn. 236, 3088–3099 [DOI] [PubMed] [Google Scholar]
- Lee Y., Hami D., De Val S., Kagermeier-Schenk B., Wills A. A., Black B. L., Weidinger G., Poss K. D. (2009). Maintenance of blastemal proliferation by functionally diverse epidermis in regenerating zebrafish fins. Dev. Biol. 331, 270–280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lieben L., Carmeliet G. (2013). Vitamin D signaling in osteocytes: Effects on bone and mineral homeostasis. Bone 54, 237–243 [DOI] [PubMed] [Google Scholar]
- Liu J., Stainier D. Y. (2010). Tbx5 and Bmp signaling are essential for proepicardium specification in zebrafish. Circ. Res. 106, 1818–1828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maccabe A. B., Gasseling M. T., Saunders J. W., Jr (1973). Spatiotemporal distribution of mechanisms that control outgrowth and anteroposterior polarization of the limb bud in the chick embryo. Mech. Ageing Dev. 2, 1–12 [DOI] [PubMed] [Google Scholar]
- Maden M. (1982). Vitamin A and pattern formation in the regenerating limb. Nature 295, 672–675 [DOI] [PubMed] [Google Scholar]
- Maves L., Tyler A., Moens C. B., Tapscott S. J. (2009). Pbx acts with Hand2 in early myocardial differentiation. Dev. Biol. 333, 409–418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonnell D. P., Scott R. A., Kerner S. A., O’Malley B. W., Pike J. W. (1989). Functional domains of the human vitamin D3 receptor regulate osteocalcin gene expression. Mol. Endocrinol. 3, 635–644 [DOI] [PubMed] [Google Scholar]
- McGlinn E., van Bueren K. L., Fiorenza S., Mo R., Poh A. M., Forrest A., Soares M. B., Bonaldo M. F., Grimmond S., Hui C. C., et al. (2005). Pax9 and Jagged1 act downstream of Gli3 in vertebrate limb development. Mech. Dev. 122, 1218–1233 [DOI] [PubMed] [Google Scholar]
- Meng X., Noyes M. B., Zhu L. J., Lawson N. D., Wolfe S. A. (2008). Targeted gene inactivation in zebrafish using engineered zinc-finger nucleases. Nat. Biotechnol. 26, 695–701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mercader N., Tanaka E. M., Torres M. (2005). Proximodistal identity during vertebrate limb regeneration is regulated by Meis homeodomain proteins. Development 132, 4131–4142 [DOI] [PubMed] [Google Scholar]
- Molven A., Wright C. V., Bremiller R., De Robertis E. M., Kimmel C. B. (1990). Expression of a homeobox gene product in normal and mutant zebrafish embryos: evolution of the tetrapod body plan. Development 109, 279–288 [DOI] [PubMed] [Google Scholar]
- Nachtrab G., Czerwinski M., Poss K. D. (2011). Sexually dimorphic fin regeneration in zebrafish controlled by androgen/GSK3 signaling. Curr. Biol. 21, 1912–1917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson C. E., Morgan B. A., Burke A. C., Laufer E., DiMambro E., Murtaugh L. C., Gonzales E., Tessarollo L., Parada L. F., Tabin C. (1996). Analysis of Hox gene expression in the chick limb bud. Development 122, 1449–1466 [DOI] [PubMed] [Google Scholar]
- Nicolas S., Papillon D., Perez Y., Caubit X., Le Parco Y. (2003). The spatial restrictions of 5’HoxC genes expression are maintained in adult newt spinal cord. Biol. Cell 95, 389–394 [DOI] [PubMed] [Google Scholar]
- Petersen C. P., Reddien P. W. (2011). Polarized notum activation at wounds inhibits Wnt function to promote planarian head regeneration. Science 332, 852–855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poss K. D. (2010). Advances in understanding tissue regenerative capacity and mechanisms in animals. Nat. Rev. Genet. 11, 710–722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poss K. D., Keating M. T., Nechiporuk A. (2003). Tales of regeneration in zebrafish. Dev. Dyn. 226, 202–210 [DOI] [PubMed] [Google Scholar]
- Qu S., Niswender K. D., Ji Q., van der Meer R., Keeney D., Magnuson M. A., Wisdom R. (1997). Polydactyly and ectopic ZPA formation in Alx-4 mutant mice. Development 124, 3999–4008 [DOI] [PubMed] [Google Scholar]
- Quint E., Smith A., Avaron F., Laforest L., Miles J., Gaffield W., Akimenko M. A. (2002). Bone patterning is altered in the regenerating zebrafish caudal fin after ectopic expression of sonic hedgehog and bmp2b or exposure to cyclopamine. Proc. Natl. Acad. Sci. USA 99, 8713–8718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddien P. W., Bermange A. L., Kicza A. M., Sánchez Alvarado A. (2007). BMP signaling regulates the dorsal planarian midline and is needed for asymmetric regeneration. Development 134, 4043–4051 [DOI] [PubMed] [Google Scholar]
- Renn J., Winkler C. (2009). Osterix-mCherry transgenic medaka for in vivo imaging of bone formation. Dev. Dyn. 238, 241–248 [DOI] [PubMed] [Google Scholar]
- Riddle R. D., Johnson R. L., Laufer E., Tabin C. (1993). Sonic hedgehog mediates the polarizing activity of the ZPA. Cell 75, 1401–1416 [DOI] [PubMed] [Google Scholar]
- Rinn J. L., Bondre C., Gladstone H. B., Brown P. O., Chang H. Y. (2006). Anatomic demarcation by positional variation in fibroblast gene expression programs. PLoS Genet. 2, e119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts-Galbraith R. H., Newmark P. A. (2013). Follistatin antagonizes activin signaling and acts with notum to direct planarian head regeneration. Proc. Natl. Acad. Sci. USA 110, 1363–1368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnapp E., Kragl M., Rubin L., Tanaka E. M. (2005). Hedgehog signaling controls dorsoventral patterning, blastema cell proliferation and cartilage induction during axolotl tail regeneration. Development 132, 3243–3253 [DOI] [PubMed] [Google Scholar]
- Shaikh N., Gates P. B., Brockes J. P. (2011). The Meis homeoprotein regulates the axolotl Prod 1 promoter during limb regeneration. Gene 484, 69–74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheth R., Marcon L., Bastida M. F., Junco M., Quintana L., Dahn R., Kmita M., Sharpe J., Ros M. A. (2012). Hox genes regulate digit patterning by controlling the wavelength of a Turing-type mechanism. Science 338, 1476–1480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stratos I., Li Z., Herlyn P., Rotter R., Behrendt A. K., Mittlmeier T., Vollmar B. (2013). Vitamin D increases cellular turnover and functionally restores the skeletal muscle after crush injury in rats. Am. J. Pathol. 182, 895–904 [DOI] [PubMed] [Google Scholar]
- Tanaka E. M., Reddien P. W. (2011). The cellular basis for animal regeneration. Dev. Cell 21, 172–185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tokuzawa Y., Yagi K., Yamashita Y., Nakachi Y., Nikaido I., Bono H., Ninomiya Y., Kanesaki-Yatsuka Y., Akita M., Motegi H., et al. (2010). Id4, a new candidate gene for senile osteoporosis, acts as a molecular switch promoting osteoblast differentiation. PLoS Genet. 6, e1001019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Towers M., Tickle C. (2009). Growing models of vertebrate limb development. Development 136, 179–190 [DOI] [PubMed] [Google Scholar]
- Trapnell C., Roberts A., Goff L., Pertea G., Kim D., Kelley D. R., Pimentel H., Salzberg S. L., Rinn J. L., Pachter L. (2012). Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat. Protoc. 7, 562–578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tümpel S., Wiedemann L. M., Krumlauf R. (2009). Hox genes and segmentation of the vertebrate hindbrain. Curr. Top. Dev. Biol. 88, 103–137 [DOI] [PubMed] [Google Scholar]
- Tzchori I., Day T. F., Carolan P. J., Zhao Y., Wassif C. A., Li L., Lewandoski M., Gorivodsky M., Love P. E., Porter F. D., et al. (2009). LIM homeobox transcription factors integrate signaling events that control three-dimensional limb patterning and growth. Development 136, 1375–1385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z., Young R. L., Xue H., Wagner G. P. (2011). Transcriptomic analysis of avian digits reveals conserved and derived digit identities in birds. Nature 477, 583–586 [DOI] [PubMed] [Google Scholar]
- Washabaugh C. H., Tsonis P. A. (1995). Effects of vitamin D metabolites on axolotl limb regeneration. Dev. Growth Differ. 37, 497–503 [DOI] [PubMed] [Google Scholar]
- Wasserman R. H., Fullmer C. S. (1989). On the molecular mechanism of intestinal calcium transport. Adv. Exp. Med. Biol. 249, 45–65 [DOI] [PubMed] [Google Scholar]
- Waxman J. S., Keegan B. R., Roberts R. W., Poss K. D., Yelon D. (2008). Hoxb5b acts downstream of retinoic acid signaling in the forelimb field to restrict heart field potential in zebrafish. Dev. Cell 15, 923–934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wills A. A., Kidd A. R., 3rd, Lepilina A., Poss K. D. (2008). Fgfs control homeostatic regeneration in adult zebrafish fins. Development 135, 3063–3070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yelon D., Ticho B., Halpern M. E., Ruvinsky I., Ho R. K., Silver L. M., Stainier D. Y. (2000). The bHLH transcription factor hand2 plays parallel roles in zebrafish heart and pectoral fin development. Development 127, 2573–2582 [DOI] [PubMed] [Google Scholar]
- Yin V. P., Thomson J. M., Thummel R., Hyde D. R., Hammond S. M., Poss K. D. (2008). Fgf-dependent depletion of microRNA-133 promotes appendage regeneration in zebrafish. Genes Dev. 22, 728–733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zákány J., Kmita M., Duboule D. (2004). A dual role for Hox genes in limb anterior-posterior asymmetry. Science 304, 1669–1672 [DOI] [PubMed] [Google Scholar]
- Zeller R., López-Ríos J., Zuniga A. (2009). Vertebrate limb bud development: moving towards integrative analysis of organogenesis. Nat. Rev. Genet. 10, 845–858 [DOI] [PubMed] [Google Scholar]
- Zhang J., Wagh P., Guay D., Sanchez-Pulido L., Padhi B. K., Korzh V., Andrade-Navarro M. A., Akimenko M. A. (2010). Loss of fish actinotrichia proteins and the fin-to-limb transition. Nature 466, 234–237 [DOI] [PubMed] [Google Scholar]
- zur Nieden N. I., Kempka G., Ahr H. J. (2003). In vitro differentiation of embryonic stem cells into mineralized osteoblasts. Differentiation 71, 18–27 [DOI] [PubMed] [Google Scholar]