Abstract
Hair follicles cyclically degenerate and regenerate throughout adult life and require regular stem cell activation to drive the cycle. In the resting phase of the hair cycle, hair follicle stem cells are maintained in a quiescent state until they receive signals to proliferate. We found that the forkhead transcription factor Foxp1 is crucial for maintaining the quiescence of hair follicle stem cells. Loss of Foxp1 in skin epithelial cells leads to precocious stem cell activation, resulting in drastic shortening of the quiescent phase of the hair cycle. Conversely, overexpression of Foxp1 in keratinocytes prevents cell proliferation by promoting cell cycle arrest. Finally, through both gain- and loss-of-function studies, we identify fibroblast growth factor 18 (Fgf18) as the key downstream target of Foxp1. We show that exogenously supplied FGF18 can prevent the hair follicle stem cells of Foxp1 null mice from being prematurely activated. As Fgf18 controls the length of the quiescent phase and is a key downstream target of Foxp1, our data strongly suggest that Foxp1 regulates the quiescent stem cell state in the hair follicle stem cell niche by controlling Fgf18 expression.
Keywords: Foxp1, Skin, Stem cells, hair follicle, Fgf18, p57KIP2, Mouse
INTRODUCTION
Adult stem cells (SCs) are essential for normal homeostasis and tissue repair. The skin epithelium is composed of the stratified epidermis, which provides the barrier function of the skin, and its appendages, such as the hair follicle, sebaceous gland and sweat glands (Blanpain and Fuchs, 2006). Hair follicle stem cells (HFSCs) are located in a niche called the bulge, which lies directly below the sebaceous gland. Most cells in the bulge are slow-cycling and maintain the cycling portion of the hair follicle (HF) during normal turnover of the hair (Cotsarelis et al., 1990; Morris et al., 2004; Tumbar et al., 2004). In addition, in response to wounding they can be mobilized to the wound sites and regenerate the stratified epidermis (Tumbar et al., 2004; Ito et al., 2005). The HF is composed of concentric layers of epithelial cells surrounding the hair shaft. Progeny of the bulge SCs give rise to transit amplifying cells in the HF matrix that differentiate into all the inner layers of the HF including the hair shaft.
Throughout adult life, HFs cyclically degenerate and regenerate and require cyclical SC activation to maintain their homeostasis (Schneider et al., 2009). After the growth phase (anagen), HFs enter a destructive phase (catagen) during which cells in the lower part of the HFs undergo apoptosis. HFs then go through a quiescent phase (telogen) before re-entering the next growth phase. At distinct stages of the hair cycle, HFSCs receive either inhibitory or activating signals from both the micro- and macro-environments to either remain quiescent or become proliferative (Lee and Tumbar, 2012; Sennett and Rendl, 2012).
Two signaling pathways that play key roles in driving the hair cycle during telogen and anagen are the bone morphogenetic protein (BMP) and β-catenin/Wnt pathways (Plikus, 2012). During telogen, high BMP signaling maintains the HFSCs in a quiescent state (Plikus et al., 2008). Deletion of the BMP receptor Bmpr1a in adult HFs results in precocious activation of HFSCs (Kobielak et al., 2007). Similarly, overexpression of the BMP antagonist noggin activates HF growth and shortens the telogen phase (Botchkarev et al., 2001; Plikus et al., 2008). Conversely, overexpression of BMP retards HF growth and, hence, prolongs the telogen phase.
At the telogen-anagen transition, the downregulation of BMP coincides with activation of β-catenin/Wnt signaling, which is believed to promote stem cell activation and HF growth (Plikus, 2012). Overexpression of β-catenin induces HFSC proliferation and promotes precocious entry into anagen (Lowry et al., 2005). Conversely, ablation of β-catenin in adult HFs prevents the activation of HFSCs and inhibits entry into anagen (Lowry et al., 2005).
Recently, the Tgfβ2 and FGF signaling pathways have also been shown to regulate the telogen-anagen transition. Activation of Tgfβ2 or Fgf7 signaling can induce anagen initiation (Greco et al., 2009; Oshimori and Fuchs, 2012). By contrast, Fgf18 signaling maintains the HFSCs in the quiescent phase (Blanpain et al., 2004; Hsu et al., 2011). Ablation of Fgf18 drastically shortens the telogen phase in a manner similar to BMP receptor deletion (Kimura-Ueki et al., 2012). Despite the identification of these crucial regulators of the telogen and anagen phase, details of how these regulators are controlled are yet to be elucidated.
Foxp1 is a member of the Foxp subfamily of the Fox (forkhead box) superfamily of transcription factors, which are characterized by the highly conserved forkhead DNA-binding domain (Benayoun et al., 2011). Known to act as both a transcriptional repressor and activator, Foxp1 regulates the development of many tissues, including esophagus, lung, heart, thymus and spinal cord (Wang et al., 2004; Hu et al., 2006; Shu et al., 2007; Dasen et al., 2008; Feng et al., 2010; Zhang et al., 2010; Feng et al., 2011; Li et al., 2012). Ablation of Foxp1 disrupts B cell development (Hu et al., 2006) as well as development of the lung and esophagus (Shu et al., 2007). Loss of Foxp1 increases proliferation of cardiomyocytes (Wang et al., 2004; Zhang et al., 2010) and disrupts the maintenance of the quiescent state of naive T cells (Feng et al., 2010; Feng et al., 2011). Foxp1 is also a suspected tumor suppressor, as its expression is reduced in a variety of tumors (Banham et al., 2001). In addition, its loss is associated with worse outcome in several types of cancer (Feng et al., 2012).
In this study, we identify Foxp1 as a key transcription factor that maintains HFSCs in a quiescent state. Deletion of Foxp1 from skin epithelium results in precocious SC activation, manifested as a failure of knockout hair follicles to remain in the telogen (resting) phase. Through gain- and loss-of-function studies, we found that Foxp1 controls the expression of fibroblast growth factor 18 (Fgf18) and the cyclin-dependent kinase (CDK) inhibitor p57KIP2 (Cdkn1c - Mouse Genome Informatics). As loss of Fgf18 also results in the shortening of the quiescent phase, and exogenous FGF18 rescues the loss of SC quiescence in Foxp1 null animals, our data strongly suggest that Foxp1 is the key regulator of Fgf18 and the quiescent SC state in the HFSC niche.
MATERIALS AND METHODS
Mice and intradermal injection of FGF18
Generation of Foxp1fl/fl and K14-cre has been described (Dassule et al., 2000; Feng et al., 2010). Foxp1fl/fl were crossed with K14-cre mice and the F1 progeny were intercrossed to generate Foxp1fl/fl;K14-cre.
Mice were injected intradermally with 1 μg of human FGF18 (Abcam) or bovine serum albumin (BSA) with dark red FluoSphere beads (Life Technologies) for 5 consecutive days as previously described (Hsu et al., 2011). One day after the last injection, skins were harvested for analysis.
All mice were maintained in the Association for Assessment and Accreditation of Laboratory Animal Care (AAALAC)-accredited animal facility at Baylor College of Medicine (BCM) and were used according to protocols that were approved by the BCM institutional care and use committee.
BrdU labeling and detection
Mice were injected with BrdU (5-bromo-3-deoxy-uridine) (Sigma-Aldrich) in PBS at 50 μg/g body weight 3 hours prior to sacrifice. Skins were embedded and frozen in OCT (Tissue-Tek), sectioned, and fixed in 4% paraformaldehyde (PFA) for 10 minutes. After washing in PBS for 15 minutes, skin sections were treated with 1 N HCl at 37°C for 40 minutes and washed for 15 minutes with PBS, before proceeding with immunofluorescence analysis using an anti-BrdU antibody (1:200, Abcam).
Histology and immunofluorescence
Staining for Hematoxylin and Eosin (H&E) and immunofluorescence were performed as described previously (Nguyen et al., 2006). Back skins were embedded and frozen in OCT. Frozen back skin sections (6-8 μm) were fixed in 4% PFA. For immunofluorescence, the MOM kit (Vector Laboratories) was used for mouse monoclonal antibodies, and the following blocking buffer was used for all other antibodies: 10% normal donkey serum, 2% BSA, 2% fish skin gelatin, 0.2% Triton X-100 in PBS. Primary antibodies were used at the following concentrations: guinea pig anti-Tcf3 (1:300, produced in the Nguyen laboratory), rat anti-cd104 (1:200, BD Biosciences), mouse anti-NFATc1 (1:50, Santa Cruz), goat anti-Lhx2 (1:100, Santa Cruz), rabbit anti-Sox9 (1:100, Santa Cruz), rat anti-BrdU (1:200, Abcam), rabbit anti-Gata3 (1:100, Santa Cruz), anti-trichohyalin clone AE15 (1:100, Santa Cruz), anti-hair shaft cytokeratin clone AE13 (1:100, Abcam), rabbit anti-Lef1 (1:200, Cell Signaling), mouse anti-Foxp1 mAb 1G1 (1:300) (Wang et al., 2003).
Antigens were visualized with FITC- or RRx-conjugated secondary antibodies (Jackson ImmunoResearch). Hoechst dye (Life Technologies) staining was used to mark nuclei. All images were acquired with a Zeiss Axioskop microscope.
Plasmid constructs and lentivirus production
The Gateway entry vector for the Foxp1 gene was generated as follows: The 5′HA-tagged Foxp1 allele was PCR-amplified from the pCDNA3.1-HA-Foxp1A plasmid (Wang et al., 2003) with the addition of BamHI and NotI sites using primers HAtag_Bam_fw and Foxp1_Not_rv. The PCR was conducted using Herculase II Fusion DNA Polymerase (Agilent Technologies). The resulting PCR product was digested and subcloned directly into BamHI/NotI-digested pENTR 1A (Life Technologies). The Emerald GFP (EmGFP) control entry vector was generated by digesting pBABE-EmGFP with HindIII/NheI and subcloning into pENTR 1A digested with XmnI/EcoRV. To generate tetracycline (tet)-inducible lentivirus constructs, the Gateway entry vectors were recombined into pINDUCER20 (Meerbrey et al., 2011) using LR Clonase II (Life Technologies) according to the manufacturer’s instructions. All plasmids were verified by restriction digest and sequencing before use. All cloning primers are listed in supplementary material Table S1.
HEK 293T cells were cultured in Iscove’s Modified Dulbecco’s Medium (IMDM) containing 10% fetal bovine serum (FBS), 2 mM supplemental l-glutamine and penicillin/streptomycin. To produce lentivirus particles, cells were plated at 4×106 cells per 10-cm plate. The following day, they were transfected with 7 μg pINDUCER vector and 750 ng each of pHDM-Hgpm2, pRc/CMV-RaII, pHDM-tat1b and pHDM-VSV-G with 30 μl of TransIT-293 transfection reagent (Mirus Bio). Virus-containing supernatants were collected at 48 and 72 hours post-transfection, pooled, and clarified by 0.45-μm filtration.
Primary cell culture and manipulation
Primary keratinocytes were isolated and cultured as previously described (Nguyen et al., 2009). For the first three passages, primary keratinocytes were cultured on J2 fibroblasts treated with mitomycin C (Roche Applied Sciences). Subsequently, keratinocytes were cultured without feeder cells.
For lentiviral infections, keratinocytes were plated at 1×105 cells per well in 6-well tissue culture plates. The following day, lentiviral supernatants were thawed, diluted with IMDM or used undiluted, and supplemented with 8 μg/ml Polybrene (Sigma-Aldrich). Growth media was aspirated from the keratinocyte-containing plates and replaced with 2 ml of lentivirus. The plates were incubated at 37°C for 15 minutes and then centrifuged for 30 minutes at 1100 g, 32°C. After centrifuging, the cells were washed with 3 ml/well PBS, re-fed with keratinocyte growth media, and returned to 37°C for growth. Starting 3-5 days after transduction, cells were selected with puromycin (1.5 μg/ml) or G418 (100 μg/ml).
For the cell proliferation assay, 2×104 cells were plated onto each well of a 24-well plate. Cells were trypsinized and counted using a Beckman Z2 Coulter counter every 24 hours.
Flow cytometry
Cell isolation by fluorescence-activated cell sorting (FACS) was performed as described using BD FACS Aria II cell sorter (Blanpain et al., 2004; Rendl et al., 2005; Nguyen et al., 2006). Postnatal day (P)28 and P49 skins of control and Foxp1 conditional knockout (cKO) mice were used to isolate bulge HFSCs based on the expression of α6-integrin (CD49f; also known as Itgα6) and CD34. As previously described, fat and underlying subcutis from the back skins were removed with a scalpel. The remaining back skins were floated on trypsin overnight at 4°C. The single cell suspensions were strained (70-μm then 40-μm pores; BD Biosciences) before being incubated with primary antibodies directly coupled with a fluorochrome. Antibodies used for FACS analysis were: anti-CD49f-PE (BD Biosciences), CD34 coupled to biotin (eBioscience), and streptavidin coupled to APC (BD Biosciences). After staining, the cells are washed with PBS and re-suspended in 4% FBS in PBS with 0.5 μg/ml DAPI (Sigma-Aldrich). Cells were gated for single events and viability before being sorted on the basis of α6-integrin and CD34 expression.
Back skins of K14-H2BGFP mice were used to isolate embryonic day (E)13.5 epidermal cells and P4 epidermal, outer root sheath (ORS) and matrix (Mx) cells based on the expression of surface marker α6-integrin and the level of GFP. Single cell suspensions of E13.5 total back skin were isolated by incubating the skins in trypsin at 37°C for 10 minutes. The cells were strained and then incubated with CD49f-PE for 30 minutes, washed, and re-suspended in 4% FBS in PBS with 0.5 μg/ml DAPI. P4 basal, ORS and Mx cells were isolated as follows. P4 epidermis and dermis fractions were first separated by dispase (Roche) incubation at 37°C for 45 minutes. Single cell suspensions of basal cells were enriched by incubating the epidermal fraction in trypsin for 5 minutes at 37°C before straining and staining with CD49f-PE. The basal cells were sorted by FACS based on the expression of α6-integrin and GFP. HF cells were enriched by incubating the dermis with collagenase at 37°C for 40 minutes and centrifuging at 300 g and then 20 g. The enriched HF cells were then trypsinized for 5 minutes at 37°C and strained to yield single cell suspensions. ORS and Mx cells were sorted by FACS as GFPhigh or GFPlow, respectively.
For cell cycle analysis, cultured keratinocytes were trypsinized, after which ∼106-107 cells were pelleted and re-suspended in 0.5 ml of PBS. The cell suspension was added to 4.5 ml of 70% ethanol and incubated on ice for 2 hours before storage at 4°C for up to one month. To perform cell cycle analysis, cells were pelleted, re-suspended in 1 ml of PBS containing 1 μg/ml DAPI, and incubated for 30 minutes before analysis. Raw FACS data were collected using BD LSR II or LSR Fortessa benchtop cytometers equipped with a 355-nm UV laser. Aggregates and doublets were eliminated from the analysis by forward- and side-scatter pulse-width gating. Curve deconvolutions were performed using FlowJo software according to the Jean-Dett-Fox model (Watson et al., 1987).
Real-time PCR
Total RNA was purified with Trizol (Sigma) or using the RNeasy Micro Kit (Qiagen) according to the manufacturer’s instructions. Each RNA sample (100 ng-1 μg) was reverse transcribed with the Superscript III First-Strand Synthesis System (Life Technologies) using Oligo(dT) primers. PCR amplifications of genes of interest were performed using primers located in different exons or spanning intron-exon junctions to obtain amplicons less than 200 bp in length. Real-time PCR was performed with the LightCycler DNA SYBR Green I master mix on a LightCycler 480 real-time PCR system (Roche). Differences between samples and controls were calculated based on the 2-ΔΔCP method, using the Mrpl19 or Gapdh genes as an internal control. Primer sequences are listed in supplementary material Table S1.
Western blot
Cultured cells were washed three times in PBS and lysed in plates by addition of radio immunoprecipitation assay (RIPA) buffer. After being incubated on ice for 30 minutes, the lysates were then centrifuged at 20,000 g at 4°C for 15 minutes. Protein concentrations were determined using the BCA assay kit (ThermoFisher Pierce) according to the manufacturer’s instructions. For western blots, protein lysates were diluted to a final concentration of 1 μg/μl in 1× Laemmli sample buffer, heated at 95°C for 5 minutes, and loaded onto Tris-glycine SDS gels for electrophoresis. For Foxp1 detection, lysates were transferred to nitrocellulose membranes (Pall Corporation), and blocked and probed as previously described (Wang et al., 2003). For detection of other proteins, the membrane was blocked with 5% nonfat dry milk (NFDM) in 0.05% Tween-20 in PBS (PBS-T) and antibody incubations were performed in 1% NFDM/PBS-T. For all western blots, the final detection step was performed using a horseradish peroxidase (HRP)-conjugated secondary antibody (Jackson ImmunoResearch) and the SuperSignal West Pico ECL Kit (ThermoFisher Pierce). The following antibodies were used: mouse anti-Foxp1 mAb 1G1 (1:2000) (Wang et al., 2003), mouse anti-β-actin clone AC-15 (Sigma, 1:50,000) and rat anti-HA (1:1000, Roche).
Statistical analyses
Statistical analyses were performed using Microsoft Excel. P values were calculated using the two-tailed Student’s t-test or the Fisher exact test, as appropriate. Differences were regarded as statistically significant or highly significant if P values were <0.05 and <0.01, respectively.
RESULTS
Foxp1 is expressed in embryonic progenitor cells, adult hair follicle stem cells and transit amplifying cells
To address the role of Foxp1 in epidermal SCs, we first analyzed the expression pattern of Foxp1 during skin development. By immunofluorescence analysis using antibodies specific to Foxp1, we found that Foxp1 is expressed uniformly in the epithelium at E13.5 when the epithelium exists as a single layer of multipotent ectodermal cells (Fig. 1A). As the epithelium stratifies to become multilayered and the placode invaginates to form a new hair germ, Foxp1 expression becomes enriched in the hair germ, which develops into the HF. In postnatal HFs, Foxp1 expression is seen in the bulge HFSCs and their immediate progeny, the outer root sheath (ORS) cell, as well as the transit amplifying cells in the matrix (Mx) region. Foxp1 is present in the bulge HFSCs in all phases of the hair cycle (Fig. 1A).
Fig. 1.
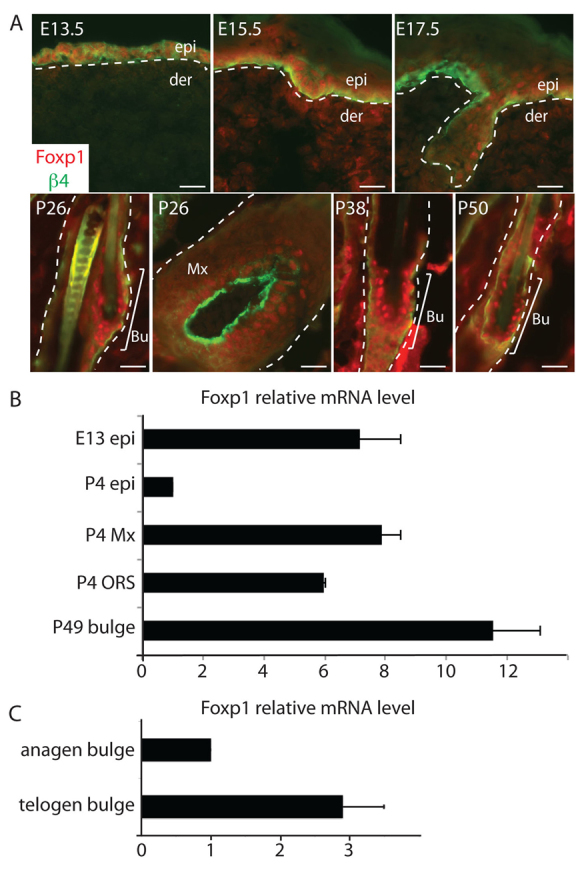
Foxp1 marks embryonic epidermal cells, adult hair follicle stem cells and transit amplifying cells in skin. (A) Immunofluorescence images of skins at different stages of development. Skins of the indicated stage of development were embedded in OCT, sectioned, and immunostained with antibodies against Foxp1 (red) and integrin β4 (green), which marks the basement membrane. White dashed lines denote epidermal-dermal border. Scale bars: 50 μm. (B) Real-time PCR analysis of Foxp1 expression in different compartments of the epidermis at different stages of development. Real-time PCR analysis was performed on cDNA synthesized from mRNA that was isolated from specified epidermal compartments at the indicated time of development. (C) Real-time PCR analysis of Foxp1 expression in anagen and telogen bulge HFSCs. Real-time PCR analysis was performed on cDNA synthesized from total RNA that was isolated from FACS-sorted anagen (P28) and telogen (P49) bulge HFSCs. Error bars represent s.d. Bu, bulge; der, dermis; epi, epidermis; Mx, matrix; ORS, outer root sheath.
We confirmed the expression pattern of Foxp1 by performing quantitative real-time RT-PCR analysis on RNA isolated from different compartments of the skin at different stages of development (Fig. 1B). We isolated E13 basal epidermal cells (epi) and P4 epi, ORS and Mx cells by FACS from skins of K14-H2BGFP mice, as described previously (Nguyen et al., 2006). We also FACS-sorted P49 bulge HFSCs using the surface markers α6-integrin and CD34 as previously described (Blanpain et al., 2004). Real-time PCR analyses confirmed that Foxp1 is expressed in embryonic epidermal cells, adult HFSCs, and their immediate progeny in the ORS and matrix. Although by immunostaining we cannot detect a difference in the expression level of Foxp1 at different stages of the hair cycle, by real-time PCR analysis Foxp1 transcript levels were nearly threefold higher in telogen than in anagen (Fig. 1C).
Ablation of Foxp1 alters the length of the hair shafts and drastically shortens the quiescent phase of the hair cycle
To study the role of Foxp1 in skin epithelial SCs, we assessed the effects of Foxp1 deletion in the skin epithelium. We crossed the Foxp1fl/fl line into a line expressing cre recombinase under the keratin 14 promoter (K14), which is first active in embryonic epithelium and becomes highly active in the basal and ORS cells, including the bulge SCs (Byrne et al., 1994). We then self-crossed the F1 progeny to generate Foxp1fl/fl;K14-cre (Foxp1 cKO) mice, which were born at Mendelian ratios and appeared normal at birth. By immunofluorescence analysis, we confirmed that Foxp1 was completely ablated in skin epithelium of Foxp1 cKO mice (Fig. 2A).
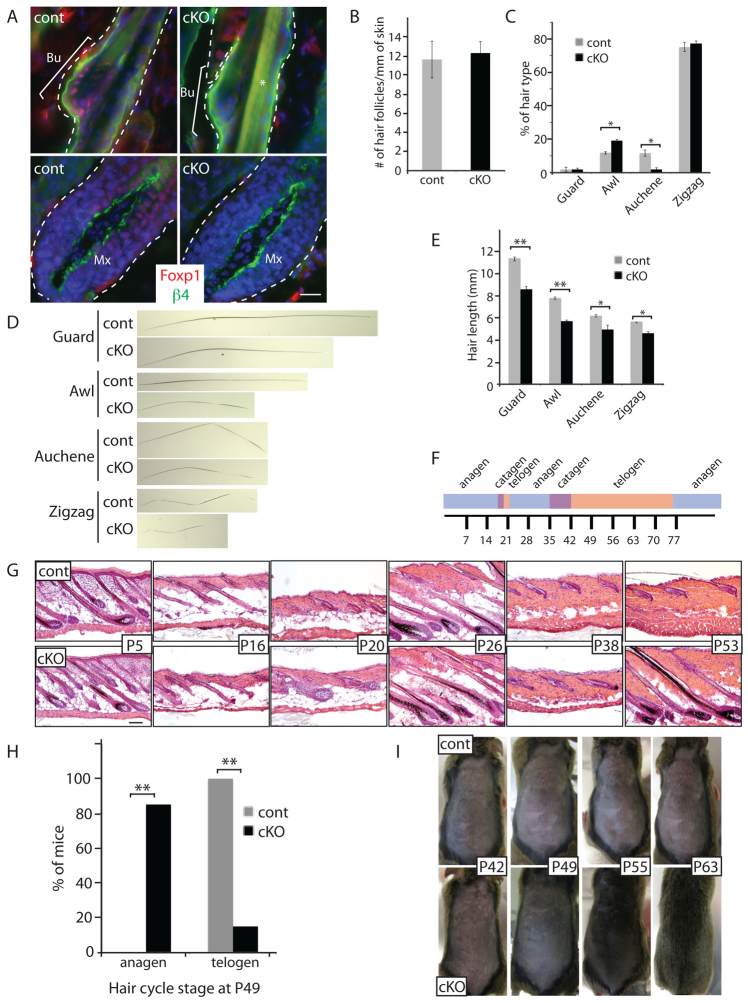
Fig. 2.
Ablation of Foxp1 alters the length of hair shafts and drastically shortens the quiescent phase of the hair cycle. (A) Foxp1 is ablated in epithelial cells of Foxp1 cKO skin. Immunostaining of Foxp1 shows nuclear Foxp1 (red) expression in the HFSCs of control (Foxp1fl/fl) but not Foxp1 cKO (K14-cre;Foxp1fl/fl) skins. Blue Hoechst staining marks DNA in the nuclei. Integrin β4 (green) marks the epithelial cells bordering the dermis. Dashed lines outline the HF. Bu, bulge; Mx, matrix. Scale bar: 20 μm. (B) Quantification of number of HFs shows that the number of HFs is comparable between Foxp1 cKO and control skins. The number of HFs was counted in five different fields of H&E sections of control and Foxp1 cKO skins. Data are mean ± s.e.m. from two mice. P=0.35. (C) Quantification of hair types shows that the number of awl and auchene hair types are altered in Foxp1 cKO skins. n=115 hairs per mouse. Data are mean ± s.e.m. from three mice. **P<0.01. (D) Representative images of control and Foxp1 cKO hair shafts of four hair types at P18 show reduced length of all four hair types in cKO. (E) Quantification of hair shaft lengths of P18 plucked hairs shows that the length of hair shafts of all four hair types were shorter in the Foxp1 cKO mice. n=20 hairs per hair type. Data are mean ± s.e.m. from three mice. **P<0.01. (F) Scheme illustrating approximate durations of the different phases of the hair cycle in the first and second hair cycles in our mixed background Foxp1 cKO mice. Numbers indicate the postnatal day number. (G) H&E-stained skin sections of control (K14-cre;Foxp1+/fl) and Foxp1 cKO (K14-cre;Foxp1fl/fl) isolated at the indicated day after birth demonstrate that Foxp1 cKO HFs have shortened quiescent phase and hence enter the growth phase precociously. Scale bar: 100 μm. (H) Quantification of the number of mice at the specified hair cycle at P49. The data indicate that whereas the skins of 100% of control mice (n=29) are in telogen, only 15% of Foxp1 cKO are in telogen stage (n=28). Skins of the majority of the Foxp1 cKO mice are in anagen. ***P<0.001. (I) Photographs of mice taken at P42 and 1, 2 and 3 weeks later following shaving of dorsal skins. Foxp1 cKO skin grew hair whereas the control skins did not, indicating that whereas the control HFs still remain in the quiescent phase, the Foxp1 cKO HFs are already in the growth phase.
We next quantified the number of HFs and the relative number of HF types in the Foxp1 cKO and the control back skins. During embryonic life, consecutive waves of HF development yield guard, awl/auchene and zigzag HF types, which differ in length and structure (Duverger and Morasso, 2009). The first wave initiated at E14.5 gives rise to guard hairs, which are straight and long. The second wave generates awl hairs, which are shorter than the guard hairs; and auchene hairs, which are marked by one bend. The third wave gives rise to zigzag hairs, which are the most abundant and contain several sharp bends. We found that the number of HFs in the Foxp1 cKO mice was comparable to that of the control mice (Fig. 2B); however, the frequency of awl hairs was increased and the frequency of auchene hairs was decreased in the Foxp1 cKO (Fig. 2C). Measurement of the hair shaft length in different hair types also revealed that the average length of all hair types was shorter in the Foxp1 cKO (Fig. 2E).
At the end of morphogenesis, the first telogen phase is short, beginning at ∼P19 and lasting for several days. In wild-type littermates of our mixed-background Foxp1 cKO mice, the second telogen started at ∼P42 and lasts several weeks. The duration of different phases of the hair cycle is illustrated in Fig. 2F. H&E staining of dorsal skin sections revealed that Foxp1 ablation did not affect the onset and duration of catagen. It only drastically shortened the telogen phase of the hair cycle (Fig. 2G). At P20, the control (Krt14-cre;Foxp1+/fl) hair follicles were in the telogen phase, whereas the Foxp1 cKO hair follicles were already in anagen. Similarly, at P53 the control hair follicles were still in telogen whereas the Fox1cKO hair follicles were fully in anagen. Using H&E staining of tissue sections to categorize the hair follicle stages, we found that only 15% of Foxp1 cKO mice, compared with 100% of controls, were in telogen at P49 (Fig. 2H).
To confirm that Foxp1 cKO HFs enter anagen prematurely, we shaved the dorsal skins of Foxp1 cKO and littermate control mice at P42 and monitored hair regrowth. At the time of shaving, both Foxp1 cKO and control mice had light pink-gray skin, characteristic of telogen. Within one week of shaving, Foxp1 cKO mice had entered anagen, manifested by a change in skin color to dark brown or black; and within three weeks they had regrown a full coat of hair. Control mice, as expected, remained in telogen and did not regrow hair for the duration of the experiment (Fig. 2I). Together, our data indicate that loss of Foxp1 drastically shortens the quiescent phase of the hair cycle.
Ablation of Foxp1 does not change the number or identity of HFSCs
Because HFs of Foxp1 cKO mice behaved differently to the HFs of the controls, we next sought to determine whether loss of Foxp1 alters the number or the identity of HFSCs. Using P28 skins, when both control and Foxp1 cKO mice are in anagen, we isolated HFSCs by FACS using the HFSC markers α6-integrin and CD34 (Fig. 3A). We found that HFSCs from Foxp1 cKO skins still express these established SC surface markers. The number of α6+, CD34+ HFSCs was marginally but not significantly increased in Foxp1 cKO compared with control skin (Fig. 3B).
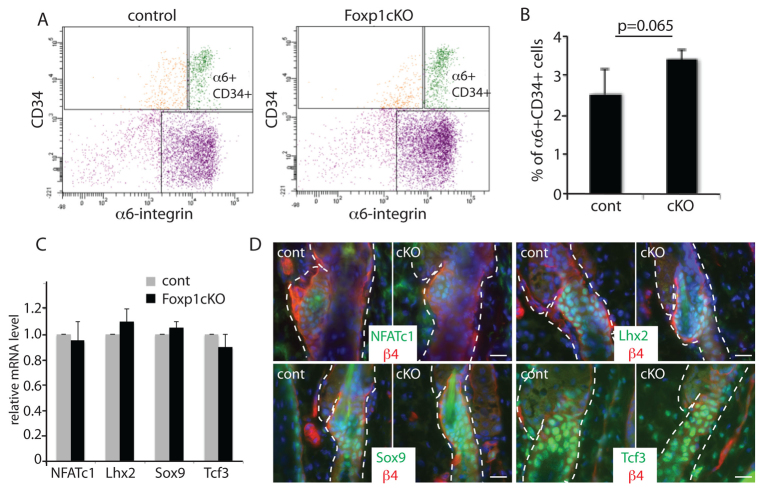
Fig. 3.
Ablation of Foxp1 does not change HFSC number or identity. (A) FACS analysis of P28 skins show that cell surface markers α6-integrin and CD34, which mark the HFSCs, are present in Foxp1 cKO skin cells. (B) Quantification of the frequency of the a6-integrin+CD34+ HFSCs in control (Foxp1fl/fl) and in Foxp1 cKO (K14-cre; Foxp1fl/fl) skins. The percentage of HFSCs was comparable between the two skins (P=0.065). (C) Real-time PCR analysis of P28 HFSCs of control and Foxp1 cKO skins show comparable expression levels of established markers of HFSCs. (D) Immunofluorescence images of P26 skins of Foxp1 cKO and control mice show that Foxp1 cKO HFSCs express Nfatc1, Lhx2, Sox9 and Tcf3, which are established markers of HFSCs. Dashed lines outline HF. Scale bars: 20 μm. Error bars represent s.d.
In addition, real-time PCR analysis of established markers of bulge HFSC, such as Nfatc1, Lhx2, Sox9 and Tcf3, showed that these transcription factors are abundantly expressed in Foxp1 cKO HFSCs at wild-type levels (Fig. 3C). Immunofluorescence analyses further confirmed that the expression pattern of these HFSC genes is similar between Foxp1 cKO and control bulge cells (Fig. 3D). We also found that ablation of Foxp1 does not disrupt terminal differentiation of any of the hair follicle lineages (supplementary material Fig. S1).
Based on these observations, we conclude that ablation of Foxp1 does not alter the number or the identity of the HFSCs sufficiently to cause the dramatic hair cycle phenotype we observed.
Ablation of Foxp1 results in precocious stem cell activation
Foxp1 cKO HFs enter the growth phase prematurely, suggesting that the HFSCs in Foxp1 cKO skin are activated precociously to proliferate. To examine the proliferative state of the HFs, we immunostained skin sections for the proliferative marker Ki67. At P21, P42 and P60, most of the control HFs are still in telogen and display little Ki67 staining. By contrast, most HFs of Foxp1 cKO skin are positive for Ki67 (Fig. 4A). We obtained similar results using a 3-hour BrdU pulse-chase to detect proliferating cells (Fig. 4B). Notably, we detected BrdU labeling in the HF bulge of Foxp1 cKO skins, indicating advanced stem cell activation.
Fig. 4.
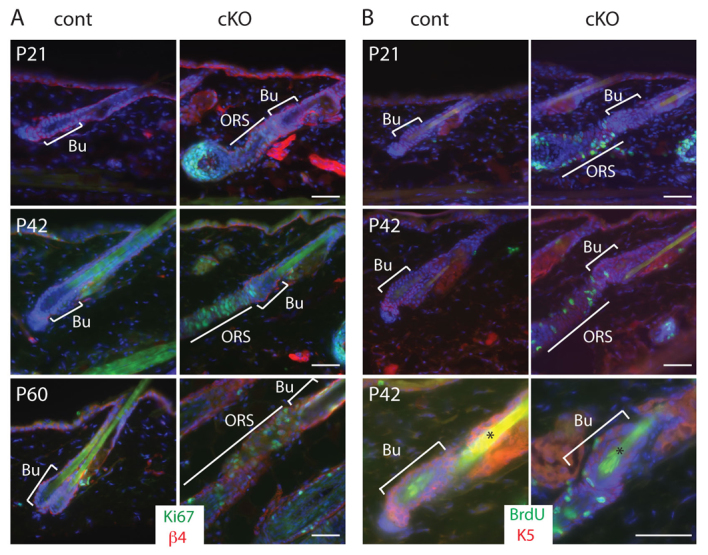
Ablation of Foxp1 promotes precocious HFSC activation. (A) Immunostaining of the proliferative marker Ki67 (green) in P21, P42 and P60 skins shows that HFs of Foxp1 cKO skins display Ki67 expression, indicating that their HFSCs were prematurely activated. (B) Immunostaining of BrdU (green) in skin sections of P21 and P42 mice that were given a 3-hour BrdU pulse prior to sacrifice. HFs of Foxp1 cKO skins incorporate BrdU labeling whereas HFs of control skins do not. Asterisks indicate autofluorescence of the hair shaft. Bu, bulge; ORS, outer root sheath. Scale bars: 50 μm.
Overexpression of Foxp1 leads to cell cycle arrest
Because loss of Foxp1 results in precocious proliferation of HFSCs, we wanted to determine whether overexpression of Foxp1 would negatively affect proliferation. We transduced wild-type keratinocytes with tet-inducible lentiviruses expressing HA-tagged Foxp1A or GFP (as a negative control), and selected transduced cells using G418. We verified that Foxp1 expression is tightly regulated and strongly inducible with the tetracycline derivative doxycycline (dox) in transduced cells (Fig. 5A,B). We then measured the effect of dox-induced Foxp1 overexpression on cell proliferation. We found that within 48 hours of dox addition, Foxp1-overexpressing keratinocytes ceased to proliferate whereas the control cells were still proliferating exponentially (Fig. 5C). Similar results were also obtained with overexpression of Foxp1 using a constitutively active retroviral vector (supplementary material Fig. S2). Cell cycle analysis showed that Foxp1 induction resulted in a higher percentage of cells in the G1 phase, and a reduced number of cells in the G2 and S phases (Fig. 5D). Together, these data suggest that overexpression of Foxp1 induces cell cycle arrest, which causes a cessation of cell growth.
Fig. 5.
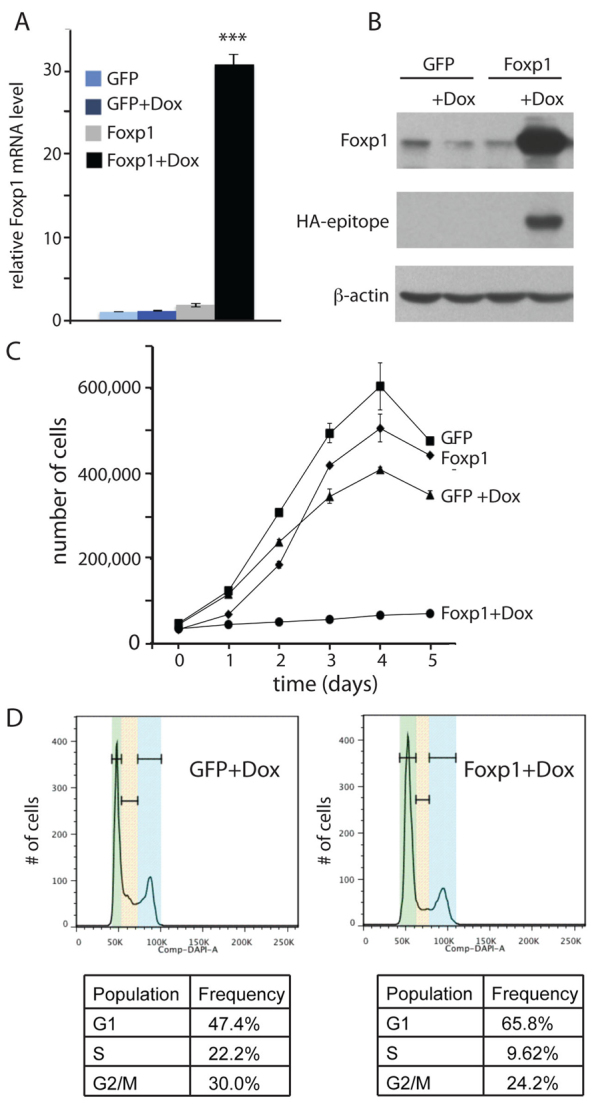
Overexpression of Foxp1 leads to cell cycle arrest. (A) Real-time PCR analysis shows Foxp1 is induced in tet-regulatable manner in tet-inducible Foxp1 keratinocytes. Keratinocytes were transduced to express tet-inducible HA-tagged Foxp1 or GFP and were selected with G418. Real-time PCR was performed on cDNA synthesized from RNA that was isolated from the transduced cells that were grown in media with doxycycline (Dox) or vehicle for 24 hours. Data are mean ± s.e.m. ***P<0.001. (B) Western analysis verifies that tet-inducible HA-Foxp1 was induced in a Dox-dependent manner. Keratinocytes were transduced to express tet-inducible HA-tagged Foxp1 or GFP and were selected with G418. With antibodies against Foxp1 or HA-epitope, western analysis was performed on protein lysates that were isolated from the transduced cells that were grown in media with Dox or vehicle for 24 hours. (C) Growth curve demonstrates that expression of Foxp1 results in a cessation of growth. Twenty thousand tet-inducible Foxp1- or GFP-expressing cells were plated onto each well of a 24-well plate. A day later, cells were counted as t=0 and Dox or vehicle were added to the cells. Cells were counted in duplicate every 24 hours subsequently. Error bars represent s.d. (D) Cell cycle profile of cells induced to express Foxp1 shows that overexpression of Foxp1 results in an increased number of cells in the G1 phase. Cell cycle analysis was performed on keratinocytes expressing tet-inducible Foxp1 or GFP that were grown with Dox or vehicle for 24 hours. Green, yellow and blue bands represent G1, S and G2/M, respectively.
Foxp1 controls expression of the cyclin-dependent kinase inhibitor p57KIP2 and the secreted growth factor Fgf18
As Foxp1 overexpression leads to cell cycle arrest, we sought to determine which cyclin-dependent kinase (CDK) inhibitor is induced by Foxp1 to cause cell cycle arrest. By real-time RT-PCR analysis of a panel of CDK inhibitors, we determined that p57KIP2 was most highly induced by Foxp1 overexpression (Fig. 6A). Conversely, p57KIP2 was the only CDK inhibitor that was downregulated in HFSCs lacking Foxp1 (Fig. 6B). In keratinocytes and HFSCs of both wild-type and cKO mice, p16 (Cdkn2a - Mouse Genome Informatics) was below the limits of qPCR detection.
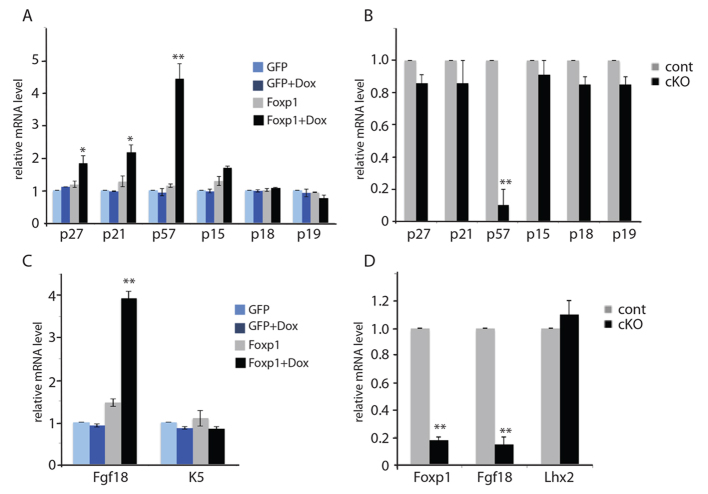
Fig. 6.
Foxp1 controls the expression of p57KIP2 and Fgf18. (A) Real-time PCR analysis shows that overexpression of Foxp1 results in the induction of the CDK inhibitor p57KIP2. Keratinocytes were transduced to express tet-inducible HA-tagged Foxp1 or GFP and were selected by drug selection. Real-time PCR was performed on cDNA synthesized from mRNA that was isolated from the transduced cells that were grown in media with doxycycline (Dox) or vehicle for 24 hours. (B) Real-time PCR analysis shows that loss of Foxp1 results in reduction of p57KIP2 expression. Real-time PCR was performed on cDNA synthesized from RNA that was isolated from HFSCs of Foxp1 cKO or control skins. (C) Real-time PCR analysis shows that overexpression of Foxp1 results in the induction of Fgf18 mRNA expression. Keratinocytes were transduced to express tet-inducible HA-tagged Foxp1 or GFP and were selected with G418. Real-time PCR was performed on cDNA synthesized from RNA that were isolated from the transduced cells that were grown in media with Dox or vehicle for 24 hours. (D) Real-time PCR analysis shows that loss of Foxp1 results in reduction of Fgf18 mRNA expression. Real-time PCR was performed on cDNA synthesized from RNA that was isolated from HFSCs of Foxp1 cKO or control skins. Data are mean ± s.e.m. *P<0.05, **P<0.01. p27, Cdkn1b; p21, Cdkn1a; p57, Cdkn1c; p15, Cdkn2b; p18, Cdkn2c; p19, Cdkn2d.
As loss of Fgf18 in the skin epithelium has been shown to shorten the length of the quiescent phase of the hair cycle, a phenotype similar to that of our Foxp1 cKO mice, we next determined whether Fgf18 is a downstream target of Foxp1. We found that overexpression of Foxp1 results in an induction of Fgf18 mRNA expression in vitro (Fig. 6C). Consistent with our hypothesis that Fgf18 is downstream of Foxp1, HFSCs of Foxp1 cKO skin showed a drastic reduction in Fgf18 mRNA levels (Fig. 6D). Gain and loss of function of Foxp1 did not alter the expression of Bmp6, which also plays a role in maintaining quiescence in the bulge, suggesting that the Foxp1 knockout effect is specifically due to its effects on Fgf18 and not Bmp6 (supplementary material Fig. S3).
Foxp1 controls the quiescent phase of the hair cycle through regulation of Fgf18
To determine whether the reduction of Fgf18 is the underlying cause of the premature entry of the Foxp1 cKO HFs into anagen, we evaluated whether exogenously delivered FGF18 can counteract the acceleration. At the beginning of telogen, we injected Foxp1 cKO mice intradermally with FGF18 or BSA control together with fluorescent beads on 5 consecutive days. At day 6 after the first injection, when control HFs are still in telogen, cKO HFs throughout the back skin entered anagen prematurely including at the BSA injection sites, as shown by proliferation marker Ki67 staining of the bulge and hair germ. By contrast, HFs at the sites of FGF18 injections remained in telogen, as shown by the absence of Ki67 (Fig. 7A,B; supplementary material Fig. S4).
Fig. 7.

Foxp1 controls the quiescence phase through regulation of Fgf18. (A) Immunofluorescence images of Foxp1 cKO skins that were injected with FGF18 or BSA show that FGF18 can prevent the precocious anagen entry that is promoted by the loss of Foxp1. Foxp1 cKO mice were injected with 1 μg FGF18 or BSA together with fluorescent beads daily for 5 days at the beginning of the second telogen. One day after the last injection, skins were embedded and immunostained for the proliferative marker Ki67 (green). Bu, bulge; DP, dermal papilla; HG, hair germ; SG, sebaceous gland. Scale bar: 20 μm. (B) Quantification of the number of Ki67-positive cells per Bu and HG in Foxp1 cKO skins that were injected with FGF18 or BSA together with fluorescent beads. HFs immediately above the fluorescent beads were analyzed for Ki67 staining. Box-and-whisker plots show number of events per Bu and HG: mid-line is the median; box, 25th to 75th percentiles; whiskers, minimum and maximum. ***P<0.001. Twenty-five HFs were analyzed for each experiment in duplicate.
That exogenous FGF18 prevented Foxp1 cKO HFs from entering anagen prematurely further indicated that Fgf18 acts downstream of Foxp1. Together, our loss-and gain-of-function studies suggest that Foxp1 is a crucial regulator of HFSC quiescence by controlling Fgf18 expression.
DISCUSSION
The forkhead transcription factor Foxp1 regulates many different developmental processes, from spinal cord, lung and heart to B and T cell development (Wang et al., 2004; Hu et al., 2006; Shu et al., 2007; Dasen et al., 2008; Feng et al., 2010; Zhang et al., 2010; Feng et al., 2011; Li et al., 2012). In this study, we show that Foxp1 is essential for the maintenance of the quiescent state of HFSCs. Mice that lack Foxp1 expression in skin epithelial cells still develop normal HFs during morphogenesis, and possess an intact stem cell population expressing normal markers. In Foxp1 cKO mice, however, HFSCs are not maintained in a quiescent phase during the resting phase of the hair cycle, and consequently re-enter the anagen phase shortly following catagen.
Consistent with a role for Foxp1 in maintaining the quiescent state of stem cells, we found that overexpression of Foxp1 in keratinocytes induces cell cycle arrest, with increased expression of the CDK inhibitor p57KIP2. Conversely, HFSCs of Foxp1 cKO skins show decreased levels of p57KIP2. Our observation that Foxp1 regulates quiescence is consistent with previous reports in other tissues. For instance, ablation of Foxp1 results in an increase in cardiomyocyte proliferation, which is accompanied by a decrease in the CDK inhibitor p27KIP1 (Wang et al., 2004). Similarly, loss of Foxp1 leads to the loss of quiescence in thymocytes and naive mature T cells (Feng et al., 2010).
A role of p57KIP2 in maintaining cells in the G0/G1 phase of the cell cycle is well established (Sherr and Roberts, 1995). Our finding that the level of p57KIP2 is decreased in HFSCs of Foxp1 cKO skins and is increased in keratinocytes following overexpression of Foxp1 suggests that Foxp1 might control the quiescent state of HFSCs by regulating the level of p57KIP2. Most p57KIP2-null mice die shortly after birth with altered cell proliferation and differentiation, which results in developmental defects of various organs (Yan et al., 1997; Zhang et al., 1997). Although some p57KIP2-deficient mice survive, their skin phenotype has not been documented.
In addition to p57KIP2, we found that Fgf18 was also a downstream target of Foxp1. Fgf18 was profoundly downregulated in HFSCs of Foxp1 cKO skins whereas its level was increased in Foxp1-overexpressing cells. Fgf18 has previously been established to be a crucial regulator of quiescence in HFSCs. Fgf18 is expressed in the bulge, especially enriched in the inner layer (Blanpain et al., 2004). Removal or ablation of the Fgf18-expressing inner bulge cells causes follicles to enter anagen, but this effect is blocked by administration of exogenous FGF18 (Hsu et al., 2011). Similarly, genetic deletion of Fgf18 in skin epithelium drastically shortens the telogen phase (Kimura-Ueki et al., 2012). As Fgf18 is downstream of Foxp1 and ablation of either Foxp1 or Fgf18 results in shortened telogen, we propose that Foxp1 maintains a quiescent state in HFSCs by regulation of Fgf18.
Although changes of Foxp1 expression lead to changes in Fgf18 expression, we anticipate that Fgf18 is only an indirect target of Foxp1. Bioinformatic analysis revealed no conserved Foxp1-binding sites in the Fgf18 promoter (data not shown). Given that Foxp family members are primarily known to be transcriptional repressors, we suggest that Foxp1 induces Fgf18 expression by repressing negative regulator(s) of Fgf18.
Although our data show that Foxp1 maintains the quiescent state of HFSCs, the fact that Foxp1 is also expressed in proliferative transit amplifying cells in the hair matrix suggests that Foxp1 expression alone is not a universal determinant of quiescence. It seems likely that other factors present in one or the other cell type determine the functional outcome of Foxp1 activity. Similar conclusions have been drawn for Foxp1 regulation of motoneurons (Dasen et al., 2008). Differential response to loss of Foxp1 in different lineages of the same tissue has already been reported in the heart; Foxp1 deletion causes decreased proliferation in the endocardium and increased proliferation in cardiomyocytes (Zhang et al., 2010). In the endocardium, Foxp1 directly represses Sox17 to regulate expression of Fgf16 and Fgf20, whereas in cardiomyocytes Foxp1 regulates expression of Nkx2.5 (Zhang et al., 2010).
In addition to its effects on HFSC quiescence, we also observed changes in the numbers of the different hair types in the Foxp1 cKO mice. We currently do not know how the loss of Foxp1 leads an altered frequency of awl and auchene hair types, because little is known about how awl and auchene hair are differentially specified. Auchene hair is distinguished from awl hair only by a distinct kink. We also do not know how deletion of Foxp1 results in a reduction of hair shaft length, although we suspect that it involves the role of Foxp1 in matrix cells. The length of the hair shaft is thought to be specified by the duration of the anagen phase or by the rate of proliferation/differentiation in the hair bulb. As the duration of anagen was similar between Foxp1 cKO and control skins, it is possible that loss of Foxp1 in the matrix altered the balance of proliferation and differentiation of matrix cells. We are currently investigating how the loss of Foxp1 affects matrix cells.
In summary, we have identified Foxp1 as a crucial regulator of quiescence in hair follicle stem cells, acting indirectly via activation of Fgf18 and p57KIP2. We are currently undertaking further studies to clarify the mechanism by which Foxp1 activates expression of these factors.
Supplementary Material
Acknowledgments
We would like to thank Drs H. A. Dierick and M. Rendl for their critical reading of our manuscript. We are grateful for the expert assistance of Joel M. Sederstrom and June Harriss.
Footnotes
Funding
This work was supported in part by research grants to H.N. from the Dan L. Duncan Cancer Scholar Award; The Naman Family Fund for Basic Research and Caroline Wiess Law Fund for Molecular Medicine; the Curtis and Doris K. Hankamer Foundation; and the National Institutes of Health (NIH) [R01-AR059122]; and by grants to H.T. from the NIH [R01-CA31534]; and the Marie Betzner Morrow Centennial Endowment. This project was also supported by the Cytometry and Cell Sorting Core at BCM with funding from the National Institutes of Health (NIH) [NIAID P30AI036211, NCI P30CA125123 and NCRR S10RR024574]. Deposited in PMC for release after 12 months.
Competing interests statement
The authors declare no competing financial interests.
Author contributions
H.N. designed experiments. E.L., J.M.H. and H.N. performed the experiments and analyzed the data. G.E.G., Q.M., A.T.K. assisted with the experiments. J.D.D. and H.T generated the Foxp1fl/fl mice. H.N. and J.M.H. wrote the manuscript.
Supplementary material
Supplementary material available online at http://dev.biologists.org/lookup/suppl/doi:10.1242/dev.097477/-/DC1
References
- Banham A. H., Beasley N., Campo E., Fernandez P. L., Fidler C., Gatter K., Jones M., Mason D. Y., Prime J. E., Trougouboff P., et al. (2001). The FOXP1 winged helix transcription factor is a novel candidate tumor suppressor gene on chromosome 3p. Cancer Res. 61, 8820–8829 [PubMed] [Google Scholar]
- Benayoun B. A., Caburet S., Veitia R. A. (2011). Forkhead transcription factors: key players in health and disease. Trends Genet. 27, 224–232 [DOI] [PubMed] [Google Scholar]
- Blanpain C., Fuchs E. (2006). Epidermal stem cells of the skin. Annu. Rev. Cell Dev. Biol. 22, 339–373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanpain C., Lowry W. E., Geoghegan A., Polak L., Fuchs E. (2004). Self-renewal, multipotency, and the existence of two cell populations within an epithelial stem cell niche. Cell 118, 635–648 [DOI] [PubMed] [Google Scholar]
- Botchkarev V. A., Botchkareva N. V., Nakamura M., Huber O., Funa K., Lauster R., Paus R., Gilchrest B. A. (2001). Noggin is required for induction of the hair follicle growth phase in postnatal skin. FASEB J. 15, 2205–2214 [DOI] [PubMed] [Google Scholar]
- Byrne C., Tainsky M., Fuchs E. (1994). Programming gene expression in developing epidermis. Development 120, 2369–2383 [DOI] [PubMed] [Google Scholar]
- Cotsarelis G., Sun T. T., Lavker R. M. (1990). Label-retaining cells reside in the bulge area of pilosebaceous unit: implications for follicular stem cells, hair cycle, and skin carcinogenesis. Cell 61, 1329–1337 [DOI] [PubMed] [Google Scholar]
- Dasen J. S., De Camilli A., Wang B., Tucker P. W., Jessell T. M. (2008). Hox repertoires for motor neuron diversity and connectivity gated by a single accessory factor, FoxP1. Cell 134, 304–316 [DOI] [PubMed] [Google Scholar]
- Dassule H. R., Lewis P., Bei M., Maas R., McMahon A. P. (2000). Sonic hedgehog regulates growth and morphogenesis of the tooth. Development 127, 4775–4785 [DOI] [PubMed] [Google Scholar]
- Duverger O., Morasso M. I. (2009). Epidermal patterning and induction of different hair types during mouse embryonic development. Birth Defects 87, 263–272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng X., Ippolito G. C., Tian L., Wiehagen K., Oh S., Sambandam A., Willen J., Bunte R. M., Maika S. D., Harriss J. V., et al. (2010). Foxp1 is an essential transcriptional regulator for the generation of quiescent naive T cells during thymocyte development. Blood 115, 510–518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng X., Wang H., Takata H., Day T. J., Willen J., Hu H. (2011). Transcription factor Foxp1 exerts essential cell-intrinsic regulation of the quiescence of naive T cells. Nat. Immunol. 12, 544–550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng J., Zhang X., Zhu H., Wang X., Ni S., Huang J. (2012). High expression of FoxP1 is associated with improved survival in patients with non-small cell lung cancer. Am. J. Clin. Pathol. 138, 230–235 [DOI] [PubMed] [Google Scholar]
- Greco V., Chen T., Rendl M., Schober M., Pasolli H. A., Stokes N., Dela Cruz-Racelis J., Fuchs E. (2009). A two-step mechanism for stem cell activation during hair regeneration. Cell Stem Cell 4, 155–169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu Y. C., Pasolli H. A., Fuchs E. (2011). Dynamics between stem cells, niche, and progeny in the hair follicle. Cell 144, 92–105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu H., Wang B., Borde M., Nardone J., Maika S., Allred L., Tucker P. W., Rao A. (2006). Foxp1 is an essential transcriptional regulator of B cell development. Nat. Immunol. 7, 819–826 [DOI] [PubMed] [Google Scholar]
- Ito M., Liu Y., Yang Z., Nguyen J., Liang F., Morris R. J., Cotsarelis G. (2005). Stem cells in the hair follicle bulge contribute to wound repair but not to homeostasis of the epidermis. Nat. Med. 11, 1351–1354 [DOI] [PubMed] [Google Scholar]
- Kimura-Ueki M., Oda Y., Oki J., Komi-Kuramochi A., Honda E., Asada M., Suzuki M., Imamura T. (2012). Hair cycle resting phase is regulated by cyclic epithelial FGF18 signaling. J. Invest. Dermatol. 132, 1338–1345 [DOI] [PubMed] [Google Scholar]
- Kobielak K., Stokes N., de la Cruz J., Polak L., Fuchs E. (2007). Loss of a quiescent niche but not follicle stem cells in the absence of BMP signaling. Proc. Natl. Acad. Sci. USA 104, 10063–10068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J., Tumbar T. (2012). Hairy tale of signaling in hair follicle development and cycling. Semin. Cell Dev. Biol. 23, 906–916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S., Wang Y., Zhang Y., Lu M. M., DeMayo F. J., Dekker J. D., Tucker P. W., Morrisey E. E. (2012). Foxp1/4 control epithelial cell fate during lung development and regeneration through regulation of anterior gradient 2. Development 139, 2500–2509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowry W. E., Blanpain C., Nowak J. A., Guasch G., Lewis L., Fuchs E. (2005). Defining the impact of beta-catenin/Tcf transactivation on epithelial stem cells. Genes Dev. 19, 1596–1611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meerbrey K. L., Hu G., Kessler J. D., Roarty K., Li M. Z., Fang J. E., Herschkowitz J. I., Burrows A. E., Ciccia A., Sun T., et al. (2011). The pINDUCER lentiviral toolkit for inducible RNA interference in vitro and in vivo. Proc. Natl. Acad. Sci. USA 108, 3665–3670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris R. J., Liu Y., Marles L., Yang Z., Trempus C., Li S., Lin J. S., Sawicki J. A., Cotsarelis G. (2004). Capturing and profiling adult hair follicle stem cells. Nat. Biotechnol. 22, 411–417 [DOI] [PubMed] [Google Scholar]
- Nguyen H., Rendl M., Fuchs E. (2006). Tcf3 governs stem cell features and represses cell fate determination in skin. Cell 127, 171–183 [DOI] [PubMed] [Google Scholar]
- Nguyen H., Merrill B. J., Polak L., Nikolova M., Rendl M., Shaver T. M., Pasolli H. A., Fuchs E. (2009). Tcf3 and Tcf4 are essential for long-term homeostasis of skin epithelia. Nat. Genet. 41, 1068–1075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oshimori N., Fuchs E. (2012). Paracrine TGF-β signaling counterbalances BMP-mediated repression in hair follicle stem cell activation. Cell Stem Cell 10, 63–75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plikus M. V. (2012). New activators and inhibitors in the hair cycle clock: targeting stem cells’ state of competence. J. Invest. Dermatol. 132, 1321–1324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plikus M. V., Mayer J. A., de la Cruz D., Baker R. E., Maini P. K., Maxson R., Chuong C. M. (2008). Cyclic dermal BMP signalling regulates stem cell activation during hair regeneration. Nature 451, 340–344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rendl M., Lewis L., Fuchs E. (2005). Molecular dissection of mesenchymal-epithelial interactions in the hair follicle. PLoS Biol. 3, e331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider M. R., Schmidt-Ullrich R., Paus R. (2009). The hair follicle as a dynamic miniorgan. Curr. Biol. 19, R132–R142 [DOI] [PubMed] [Google Scholar]
- Sennett R., Rendl M. (2012). Mesenchymal-epithelial interactions during hair follicle morphogenesis and cycling. Semin. Cell Dev. Biol. 23, 917–927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherr C. J., Roberts J. M. (1995). Inhibitors of mammalian G1 cyclin-dependent kinases. Genes Dev. 9, 1149–1163 [DOI] [PubMed] [Google Scholar]
- Shu W., Lu M. M., Zhang Y., Tucker P. W., Zhou D., Morrisey E. E. (2007). Foxp2 and Foxp1 cooperatively regulate lung and esophagus development. Development 134, 1991–2000 [DOI] [PubMed] [Google Scholar]
- Tumbar T., Guasch G., Greco V., Blanpain C., Lowry W. E., Rendl M., Fuchs E. (2004). Defining the epithelial stem cell niche in skin. Science 303, 359–363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang B., Lin D., Li C., Tucker P. (2003). Multiple domains define the expression and regulatory properties of Foxp1 forkhead transcriptional repressors. J. Biol. Chem. 278, 24259–24268 [DOI] [PubMed] [Google Scholar]
- Wang B., Weidenfeld J., Lu M. M., Maika S., Kuziel W. A., Morrisey E. E., Tucker P. W. (2004). Foxp1 regulates cardiac outflow tract, endocardial cushion morphogenesis and myocyte proliferation and maturation. Development 131, 4477–4487 [DOI] [PubMed] [Google Scholar]
- Watson J. V., Chambers S. H., Smith P. J. (1987). A pragmatic approach to the analysis of DNA histograms with a definable G1 peak. Cytometry 8, 1–8 [DOI] [PubMed] [Google Scholar]
- Yan Y., Frisén J., Lee M. H., Massagué J., Barbacid M. (1997). Ablation of the CDK inhibitor p57Kip2 results in increased apoptosis and delayed differentiation during mouse development. Genes Dev. 11, 973–983 [DOI] [PubMed] [Google Scholar]
- Zhang P., Liégeois N. J., Wong C., Finegold M., Hou H., Thompson J. C., Silverman A., Harper J. W., DePinho R. A., Elledge S. J. (1997). Altered cell differentiation and proliferation in mice lacking p57KIP2 indicates a role in Beckwith-Wiedemann syndrome. Nature 387, 151–158 [DOI] [PubMed] [Google Scholar]
- Zhang Y., Li S., Yuan L., Tian Y., Weidenfeld J., Yang J., Liu F., Chokas A. L., Morrisey E. E. (2010). Foxp1 coordinates cardiomyocyte proliferation through both cell-autonomous and nonautonomous mechanisms. Genes Dev. 24, 1746–1757 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.