Abstract
As cells integrate molecular signals from their environment, cell surface receptors require modified proteoglycans for the robust activation of signaling pathways. Heparan sulfate proteoglycans (HSPGs) have long unbranched chains of repetitive disaccharide units that can be sulfated at specific positions by heparan sulfate O-sulfotransferase (OST) families. Here, we show that two members of the 3-OST family are required in distinct signaling pathways to control left-right (LR) patterning through control of Kupffer’s vesicle (KV) cilia length and motility. 3-OST-5 functions in the fibroblast growth factor pathway to control cilia length via the ciliogenic transcription factors FoxJ1a and Rfx2. By contrast, a second 3-OST family member, 3-OST-6, does not regulate cilia length, but regulates cilia motility via kinesin motor molecule (Kif3b) expression and cilia arm dynein assembly. Thus, two 3-OST family members cell-autonomously control LR patterning through distinct pathways that regulate KV fluid flow. We propose that individual 3-OST isozymes create distinct modified domains or ‘glycocodes’ on cell surface proteoglycans, which in turn regulate the response to diverse cell signaling pathways.
Keywords: Heparan sulfate proteoglycans, O-Sulfotransferase, OST, 3-OST, HSPG, Fibroblast growth factors, FGF, Cilia, Fluid flow, Left-right development, Heart asymmetry, Gut asymmetry, Zebrafish, Glycocode
INTRODUCTION
Molecular interactions in the extracellular matrix and at the cell surface are important for dynamic regulation of cell-cell signaling. At the cell surface, heparan sulfate proteoglycans (HSPGs) are known to modulate interactions between receptors and ligands (Rapraeger et al., 1991; Yayon et al., 1991; Bellaiche et al., 1998; Tsuda et al., 1999; Belenkaya et al., 2004; Häcker et al., 2005). Heparan sulfate (HS) chains attached to a core protein consist of alternating glucuronic acid and N-acetylglucosamine residues, which undergo a series of modifications, including sulfation, to produce mature long, unbranched HS chains. HSPG core proteins are either inserted into the cell membrane, attached to the extracellular surface of the cell membrane, or in the extracellular matrix. In vivo, loss of core proteins or HS chains indicate that HSPGs regulate multiple developmental processes, including gastrulation and left-right (LR) patterning (Alexander et al., 2000; Topczewski et al., 2001; Kramer et al., 2002; Kramer and Yost, 2002; Marjoram and Wright, 2011). Much less is known about specific rare modifications to HS chains and their specialized function in development (HajMohammadi et al., 2003; Kamimura et al., 2004; Tecle et al., 2013).
The addition of sulfates to HS chains is mediated by three distinct families of heparan sulfate O-sulfotransferases (OSTs): 2-OSTs, 3-OSTs and 6-OSTs. Members of these families have dynamic spatial and temporal regulation during vertebrate development (Shworak et al., 1999; Habuchi et al., 2000; Xia et al., 2002; Nogami et al., 2004; Xu et al., 2005; Cadwallader and Yost, 2006b; Cadwallader and Yost, 2006a). Within these multigene families in the HS chain biosynthetic pathway, the 3-OST family is the most isozyme-rich, but is responsible for only 0.5% of all HS chain sulfation (Colliec-Jouault et al., 1994; Shworak et al., 1999). Considering that each 3-OST family member performs the same biochemical function, sulfation of the hydroxyl group at the 3-carbon on select glucosamine residues in the HS chain (Colliec-Jouault et al., 1994; Shworak et al., 1999), it is intriguing that there are so many 3-OST family members in vertebrates (Cadwallader and Yost, 2013). Here, we explore the hypothesis that each 3-OST family member has a distinct context-dependent, non-redundant function in development.
In all vertebrates, the heart, brain and gut develop left-right asymmetries that are crucial for normal organ function (Yost, 2001; Hamada et al., 2002; Bisgrove et al., 2003; Levin, 2005; López-Gracia and Ros, 2007). These organ asymmetries are dependent on a conserved cascade of asymmetric expression in left lateral plate mesoderm (LPM) of the nodal, lefty and pitx2 gene families. Several upstream components contribute to normal asymmetric LPM gene expression, including (1) cilia-dependent fluid flow in a ‘ciliated organ of asymmetry’ at the posterior of the embryonic midline, the node in mouse (Zhou et al., 1993), Kupffer’s vesicle (KV) in zebrafish (Essner et al., 2005) and medaka (Hojo et al., 2007; Soroldoni et al., 2007), and the gastrocoel roof plate in Xenopus laevis (Schweickert et al., 2007), (2) an intact embryonic midline (Danos and Yost, 1995; Danos and Yost, 1996; Chen et al., 1997; Meno et al., 1998; Yamamoto et al., 2003), and (3) in Xenopus at least, molecular asymmetries in cleavage and gastrula stages (Danos and Yost, 1995; Kramer and Yost, 2002; Yamamoto et al., 2003; Adams et al., 2006) that precede the appearance of cilia (Essner et al., 2002). Several important signaling pathways have been implicated in the establishment of LR asymmetries, including Shh, fibroblast growth factor (FGF), bone morphogenetic protein (BMP) and Wnt pathways (Albertson and Yelick, 2005; Levin, 2005; Hong and Dawid, 2009; Neugebauer et al., 2009). Understanding how the signals are mediated by the developing embryo will be crucial to our understanding of LR asymmetry.
In zebrafish, at least five 3-OST family members are normally expressed in and around the ciliated cells of the KV (Cadwallader and Yost, 2006a). To determine whether individual members of the 3-OST family have distinct cellular and developmental functions, we knocked down each family member by injection of morpholinos (MOs; antisense oligonucleotides) and assessed KV cilia and downstream LR patterning in zebrafish. Strikingly, two of the five family members expressed in KV ciliated cells, 3-OST-5 (gene name: hs3st1l2) and 3-OST-6 (gene name: hs3st3l), had distinct roles in regulating motile KV cilia, indicating non-redundant functions within the 3-OST family.
We propose that specific sulfation patterns on HS chains create ‘glycocodes’ that are developmentally regulated, cell-type specific, and ultimately confer specificity to most major cell signaling pathways at the cell surface (Cadwallader and Yost, 2013). Perhaps small subsets of the vast number of possible combinations of HS chain modifications are utilized as gatekeepers for specific molecular interactions, such as ligand-receptor interactions. If so, uncovering the bioinformation potentially embedded in the sequences of HSPG chain modifications will enjoin a substantial leap in our understanding of cell signaling. The first postulate of the glycocode hypothesis is that each member of an OST family performs a distinct function: the same catalytic reaction but in a context-dependent fashion, generating distinct sequences of modifications on HS chains (Cadwallader and Yost, 2013). Here, we test this postulate and find that individual 3-OST family members have distinct functions, modulating distinct pathways within the same cells during development.
MATERIALS AND METHODS
Maintenance of zebrafish stocks and embryo culture
Danio rerio were maintained at 28.5°C on a 14-hour light/10-hour dark cycle. Embryos were collected from natural spawnings, cultured and staged as previously described (Westerfield, 2000). Heart looping was determined in wild-type (WT) embryos by visualization in living embryos, by in situ hybridization or by fluorescence microscopy of GFP expression in cardiomyocytes in Tg(cmlc2:GFP) (Huang et al., 2003). 3-OST-5/Fgf8 epistasis experiments were conducted using the fgf8 hypomorphic allele fgf8ati282a/+, which is predicted to generate a truncated Fgf8 protein (Reifers et al., 1998).
Embryo injections
Antisense morpholinos (MOs) were obtained from Gene Tools. To knock down gene expression in all cells, MO was injected between the one- and four-cell stages as previously described (Nasevicius and Ekker, 2000). For one-four cell injections, 1 nl MO was injected into embryos; MO concentrations used were as listed in Table 1. To deliver MOs specifically to dorsal forerunner cells (DFCs), the precursors of KV ciliated cells, a fluorescein-labeled control MO was co-injected with a gene-specific MO in a 1:1 ratio into the yolk cell between the 500- and 1000-cell stages; embryos with fluorescent DFCs were selected for analysis, as described (Amack and Yost, 2004). The sub-threshold dose of GTAGCATTCGTACCTGTACCAGTCC for fgf8/ace experiments was injected at a concentration of 0.175 mM.
Table 1.
Morpholino sequence and injection amounts
mRNA synthesis
The 3-OST-6 ΔTM construct was subcloned from a construct containing the full-length open reading frame (ORF) of 3-OST-6 (Cadwallader and Yost, 2006a) by PCR using the following primers: 5′-CCCATGGACTCCTCCGAAAACTCCCAG-3′ and 5′-CTCTAGAGTCATTGCCATCCGAAGTCCATC-3′. The full-length ORF of 3-OST-5 and 3-OST-6, as well as 3-OST-6 ΔTM were subcloned into pCS2+NMCS. mRNA for injection was synthesized using the mMessage mMachine Kit (Ambion) or the AmpliCap (Epicentre) utilizing the SP6 polymerase. Newly synthesized mRNA was purified by ammonium acetate precipitation.
RT-PCR
RT-PCR was used to determine the effect of MO injections on mRNA splicing (primers listed in supplementary material Figs S1, S2). Total RNA was extracted from embryos using TRIzol Reagent (Invitrogen) and cDNA was synthesized by MMLV-RT primed by random decamers (Ambion Retroscript kit). PfuUltra (Stratagene) was used for amplification of cDNA using 33 cycles of PCR. Semi-quantitative analysis was performed to calculate the percentage decrease of spliced mRNA using ImageQuant TL (GE Healthcare).
Whole-mount in situ hybridization
Zebrafish embryos were fixed in PBS containing 4% paraformaldehyde (PFA) plus sucrose, rinsed in 1×PBS, dehydrated in methanol series and stored at -20°C. To generate antisense RNA probes, linear DNA templates were transcribed using either T3 or T7 polymerase in the presence of digoxigenin labeling mix (Roche) and free nucleotides were removed using LiCl precipitation. In situ hybridizations were carried out as previously described (Essner et al., 2000) using a Biolane HTI in situ machine (Huller and Huttner AG). Embryos were cleared in 70% glycerol in PBST (0.1% Tween-20 in PBS) and photographed using either a Leica MZ12 or a Nikon SMZ1000 dissecting microscope. Digital images were processed using Adobe Photoshop and ACD Systems Canvas. cDNA templates used include spaw (Long et al., 2003), shh (Essner et al., 2005), ntl (Amack and Yost, 2004), sox17 (Amack and Yost, 2004), lefty1 (Essner et al., 2005), dnah9 (Essner et al., 2005), foxJ1a (Tian et al., 2009), rfx2 (Bisgrove et al., 2012), kif3b (B.W.B., unpublished), cmlc2 (myl7) (Yelon et al., 1999), 3-OST-5 (Cadwallader and Yost, 2006a), 3-OST-6 (Cadwallader and Yost, 2006a), sef (il17rd) (Tsang et al., 2002) and fkd2 (foxa3) (Odenthal and Nüsslein-Volhard, 1998).
Immunohistochemistry
Embryos were fixed overnight in 4% PFA at 4°C and dehydrated stepwise into methanol for storage at -20°C. After stepwise re-hydration, embryos were blocked for 1 hour in PBS containing 5% sheep serum (Sigma), 1% bovine serum albumin (BSA), 1% DMSO and 0.1% Triton X-100. Embryos were incubated with mouse anti-acetylated tubulin (1:300, Sigma T-6793) and rabbit anti-PKCζ (1:100, Santa Cruz sc-216) antibodies in blocking solution overnight. After washing embryos in 1% BSA and 1% DMSO in PBST, embryos were incubated with fluorescent secondary antibodies (1:300, Molecular Probes) overnight. Embryos were then washed and mounted in Slow Fade anti-bleaching reagent (Molecular Probes). KV immunostaining was imaged using an Olympus Fluoview FV300 scanning laser confocal microscope with a 60× objective and images were processed using ImageJ and Metamorph software.
Fluorescent bead injections
KV fluid flow was assayed as previously described (Essner et al., 2005). Embryos were removed from their chorions at 6-8 somite stage (SS) and mounted in 1% low melt agarose on glass slides. Fluorescent beads (0.5-2 μm; Polysciences) were injected into KV, and beads were imaged on a Leica DMRA compound microscope using a 40× Plan Apo objective. Movies were generated using a Coolsnap HQ digital camera (Photometrics) and Metamorph (Universal Imaging Corp.) and Quicktime (Apple) software. Beads were tracked using Metamorph.
Electron microscopy
Electron microscopy protocol was modified from a previously published protocol to examine cilia in zebrafish KV (Kreiling et al., 2007). Live 8 SS embryos were dechorinated by hand and then fixed in 4% glutaraldehyde in 0.1 M sodium cacodylate buffer overnight at 4°C. Embryos were washed three times (15 minutes per wash) in 0.1 M sodium cacodylate buffer and incubated for 1 hour on ice in 1% osmium tetroxide in 0.1 M sodium cacodylate buffer. After osmium treatment, embryos were washed three times (15 minutes per wash) in 0.1 M sodium cacodylate buffer. Embryos were embedded in 1% agarose (in H2O) to allow orientation during sectioning. Embryos embedded in agarose were dehydrated in an EtOH series ending in acetone washes. Embryos were then embedded in Epon through washes with 3:1, 1:1 and 1:3 Acetone:Epon for 2 hours each at room temperature. Finally, the embryos were placed in 100% Epon with at least two changes (2 hours per wash) before placing them at 60°C overnight to solidify epoxy. Embedded samples were sectioned and stained with uranyl acetate and lead citrate. Electron micrographs were taken using a FEI Tecnai T12 at 80 kV. Images were de-identified and blindly scored by three individuals for presence or absence of inner and outer dynein arms.
High resolution melt analysis
After confocal analysis, embryos were removed from microscope slides and rinsed in 1× PBST with one change of buffer over 2 hours. Embryos were placed at 55°C overnight in DNA extraction buffer containing 20 mM Tris pH 8.0, 50 mM KCl, 1.5 mM MgCl2 0.3% Tween-20, 0.3% NP-40 and 15 μg/ml of ProteinaseK. After overnight incubation, ProteinaseK was deactivated by 10 minutes at 95°C. A PCR reaction was set up in 96-well plates (Bio-Rad, HSP9665) using 1 μl DNA in a 10 μl reaction and included LCGreen (Idaho Technology). PCR primers were designed to create a 73-base fragment flanking the G→A transition mutation causing the acerebellar phenotype (forward, 5′-TGTATGAACAGGAGGGGGA-3′; reverse, 5′-CAGTTTTAGTAAGTCACAAAAGTG-3′). The following PCR conditions were used: 95°C for 10 seconds; 40 cycles of 95°C for 8 seconds, 52°C for 8 seconds, 72°C for 8 seconds; 95°C for 8 seconds then 45°C for 10 seconds. PCR samples were analyzed for different melting curves on a Lightscanner (Idaho Technology) over a 65-90°C range as previously described (Parant et al., 2009).
Statistical analysis
Each experiment was repeated at least three times in order to assess statistical significance. Means, standard deviations and standard errors were calculated using R Statistical Software platform. Student’s t-test was performed for analysis of cilia length, and heart and gut looping. Fisher’s exact test was used to analyze changes in in situ expression categories. A one-way ANOVA followed by a Tukey analysis was performed on fluorescent bead velocity. A multi-way ANOVA followed by a Tukey analysis was performed for cilia length analysis in fgf8/ace embryos injected with 3-OST-5 MO1. P values below 0.05 were considered significant.
RESULTS
3-OST-5 is required for LR development
Injection of a translation-blocking MO specific to 3-OST-5 (3-OST-5 MO1) resulted in embryos with a curved body axis at 48 hours post-fertilization (hpf) (Fig. 1A,B) and altered LR patterning, with high frequencies of reversed heart (39%, n=120) and gut (41%, n=120) looping (Fig. 1C-E; supplementary material Fig. S1A). Injection of a second 3-OST-5 MO (3-OST-5 MO2), designed to inhibit 3-OST-5 mRNA splicing, resulted in similar LR defects, including frequent heart (29%, n=121) and gut (29%, n=121) reversals (Fig. 1E). 3-OST-5 MO2 morphants had increased accumulation of unspliced RNA and corresponding reduction of mRNA (supplementary material Fig. S1B,C).
Fig. 1.
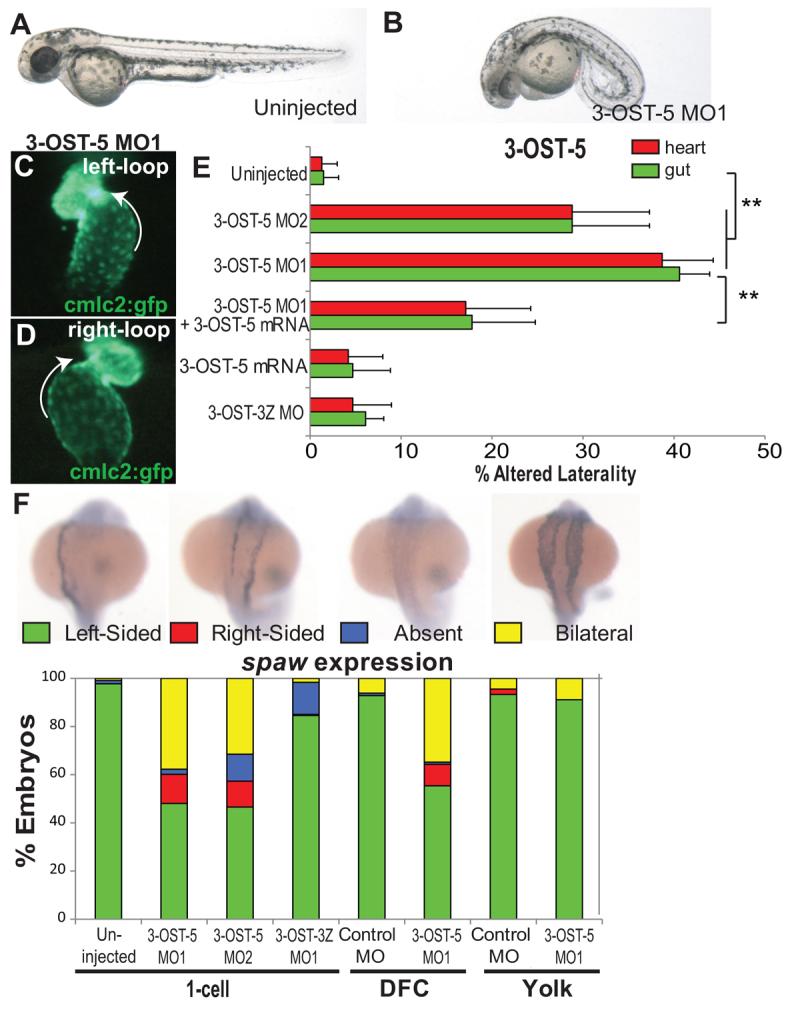
3-OST-5 is required in whole embryos and cell autonomously in Kupffer’s vesicle for LR development. (A,B) Differential interference contrast images of 48 hpf embryos. Uninjected (A) and 3-OST-5 MO-injected (B) embryos at 48 hpf showing dose-dependent curly down tail phenotype. Both 3-OST-5 MOs showed a dose-dependent increase in both heart and gut reversals. (C,D) cmlc2-gfp transgenic fish (expressing GFP in cardiomyocytes) were used to visualize normal left looping (C) and reversed right looping (D) in 3-OST-5 MO1 embryos at 48 hpf. (E) Percentages of reversed heart and gut looping in 3-OST-5 morphants. Injection of 3-OST-5 MO1 (n=120; **P<0.001) or 3-OST-5 MO2 (n=121; **P<0.001) resulted in significant heart and gut looping compared with uninjected embryos (n=361). Co-injection of 3-OST-5 MO1 and 3-OST-5 ‘rescue’ mRNA resulted in a significant rescue of heart and gut orientation (n=137; **P<0.001). Injection of 3-OST-5 ‘rescue’ mRNA alone (n=171) or 3-OST-3Z MO (n=106) did not alter normal heart and gut looping. Error bars represent s.d. for all heart and gut looping treatments. (F) Representative images of normal (left-sided), reversed (right-sided), absent and bilateral spaw expression in 3-OST-5 morphants. Percentages of normal (left-sided), reversed (right-sided), absent, and bilateral spaw expression in the LPM of 3-OST-5 morphants (MO1, n=183; MO2, n=206), 3-OST-3Z morphants (n=240) or uninjected control embryos (n=220). 3-OST-5 morphants had a preponderance of bilateral expression, in comparison with predominantly left-sided expression in uninjected and 3-OST-3Z MO1-injected embryos. Embryos injected for DFC/KV-targeted knockdown, DFC3-OST-5 MO1 (n=112), showed a high rate of bilateral spaw expression. This is in contrast to DFCcontrol MO (n=97), Yolk3-OST-5 MO1 (n=61) and Yolkcontrol MO (n=90) injected embryos showing left-sided spaw expression.
To test MO specificity, we performed rescue experiments by co-injecting 3-OST-5 MO1 with a 3-OST-5 MO-resistant ‘rescue’ mRNA, specifically engineered to prevent 3-OST-5 MO1 from binding injected mRNA. Co-injection of 3-OST-5 MO-resistant ‘rescue’ mRNA resulted in a statistically significant (P<0.001) rescue of 3-OST-5 MO1 induced heart (17%, n=137) and gut (18%, n=137) looping defects (Fig. 1E). Injection of 3-OST-5 MO-resistant ‘rescue’ mRNA alone did not alter heart and gut orientation (4% and 5%, respectively, n=171; Fig. 1E). The similar phenotype seen with two distinct morpholinos and the ability to rescue the phenotype indicate that these effects are specific for 3-OST-5, revealing its function in zebrafish LR development.
As an important control for functional specificity within the 3-OST family, we knocked down three other family members and found that none had roles in LR development. 3-OST-3Z (gene name: hs3st3b1b), a closely related family member concomitantly expressed with 3-OST-5 in early development (Cadwallader and Yost, 2006a), was targeted for knockdown by injection of a highly efficient splice-blocking MO resulting in reduction of 3-OST-3Z mRNA (supplementary material Fig. S1D) but normal LR patterning, with normal heart (95%, n=172) and gut (94%, n=172) orientation (Fig. 1E). This 3-OST-3Z MO was used as a parallel control morpholino throughout this study. Similarly, knockdown of another family member, 3-OST-7 (gene name: hs3st1l1), although expressed in overlapping patterns with 3-OST-5, resulted in normal LR development (100%, n=24; compared with 96% uninjected embryos, n=46). Although this 3-OST-7 MO did not alter LR patterning, it resulted in a distinct phenotype, cardiac ventricular non-contraction at 48 hpf, that could be partially rescued by transgene expression (S. C. Samson, T. Ferrer, C. J. Jou, F. B. Sachse, S. S. Shankaran, R. M. Shaw, N. C. Chi, M. Tristani-Firouzi and H.J.Y., unpublished)), indicating the efficacy and specificity of this MO. Together, these results indicate that the LR patterning defects seen in embryos injected with 3-OST-5 MO are not observed with reduction of at least two other 3-OST family members.
To assess where 3-OST-5 functions in the LR pathway, we analyzed the expression of southpaw (spaw; a zebrafish nodal homolog) by in situ hybridization. spaw is expressed in the left LPM directly downstream of the molecular cascade that initiates LR axis formation (Fig. 1F) (Long et al., 2003). In 3-OST-5 morphants, spaw expression was altered, resulting in high incidence of bilateral spaw expression (Fig. 1F). Control 3-OST-3Z morphants had normal spaw expression (Fig. 1F), consistent with normal organ LR orientation (Fig. 1E).
Two transient embryonic structures, the embryonic midline (floor plate and notochord) and KV, are required for normal asymmetric gene expression in the LPM (Danos and Yost, 1996; Amack and Yost, 2004; Essner et al., 2005). 3-OST-5 is expressed throughout the embryo at 70-90% epiboly, including midline cells and precursors of KV cells, dorsal forerunner cells (DFCs; supplementary material Fig. S1E). At the 7-10 somite stage (SS), when signals are being transferred from KV to the LPM (Essner et al., 2005), 3-OST-5 is expressed in KV, LPM and intervening tailbud tissues (supplementary material Fig. S1F). Thus, the ubiquitous expression patterns do not help define the location of 3-OST-5 function in the LR pathway.
Considering that the embryonic midline is essential for LR development (Danos and Yost, 1995; Danos and Yost, 1996; Chen et al., 1997; Meno et al., 1998; Bisgrove et al., 1999; Yamamoto et al., 2003), we assessed whether 3-OST-5 morphants had intact midline structures. At 16 SS, in situ RNA expression of lefty1, ntl and shh in 3-OST-5 MO1-injected embryos showed normal midline and floor plate development (lefty1, present in 83%, n=82; ntl, present in 100%, n=122; shh, present in 100%, n=122; supplementary material Fig. S1G-J). These results indicate that LR patterning defects in 3-OST-5 morphants are not due to gross perturbations of midline development.
DFCs appeared normal in 3-OST-5 MO1-injected embryos, as assessed by whole-mount in situ analysis of sox17 expression (present in 95%, n=136). KV was analyzed in live embryos using stereo microscopy at 6 SS and appeared to form normally in 3-OST-5 MO1-injected embryos (present in 88%, n=94). Together, these results suggest that LR defects in 3-OST-5 morphants are not due to defects in midline structure, DFC specification or KV morphogenesis.
3-OST-5 is required cell-autonomously in KV cells for LR development
To test directly whether 3-OST-5 functions cell-autonomously in the DFC/KV cell lineage, we utilized a technique we developed to target MOs to DFC/KV cells by injection into the yolk cell at the 500-cell stage (Amack and Yost, 2004). This creates chimeric embryos in which targeted gene function is knocked down in DFC/KV cells, but remains wild type in all other embryonic cells. In embryos with targeted knockdown of 3-OST-5 in DFC/KV cells (DFC3-OST-5MO1), aberrant spaw expression was predominantly bilateral (Fig. 1F). This phenotype is similar to the spaw expression profile seen in global knockdown of 3-OST-5 in morphants (Fig. 1F). Asymmetric spaw expression was normal in DFCcontrol MO embryos (Fig. 1F). To examine whether these spaw defects were due to knockdown of 3-OST-5 in DFC/KV cells and not in the yolk cell, we injected 3-OST-5 MO1 into yolk at dome stage (Yolk3-OST-5MO1). At this stage, DFC precursors are no longer connected to the yolk cell and therefore do not receive MO (Amack and Yost, 2004). Yolk3-OST-5MO and YolkControl MO had normal left-sided spaw expression (Fig. 1E). Together, these results indicate that 3-OST-5 functions cell-autonomously in DFC/KV cells to regulate LR patterning.
3-OST-5 regulates KV cilia length
Motile KV cilia are essential for asymmetric fluid flow and normal LR development (Essner et al., 2005). In contrast to many other LR mutants or morphants that have perturbed KV shape (Amack et al., 2007; Hatler et al., 2009; Aamar and Dawid, 2010), 3-OST-5 morphants displayed normal KV morphogenesis and differentiation as assessed by confocal microscopy with the apical cell marker atypical Protein Kinase C (aPKC; Fig. 2A,B). No significant difference was detected in cilia number at both 4 SS (WT, 54.5±15.8, n=6; 3-OST-5 MO, 41.86±14.3, n=7; P<0.1587) and 8 SS when comparing 3-OST-5 morphants (49.27±14.90, n=28, P<0.67) and uninjected control embryos (51.07±16.54, n=27). However, cilia length was significantly shorter in 3-OST-5 morphants, compared with uninjected embryos (P<6.19e-10; Fig. 2C).
Fig. 2.
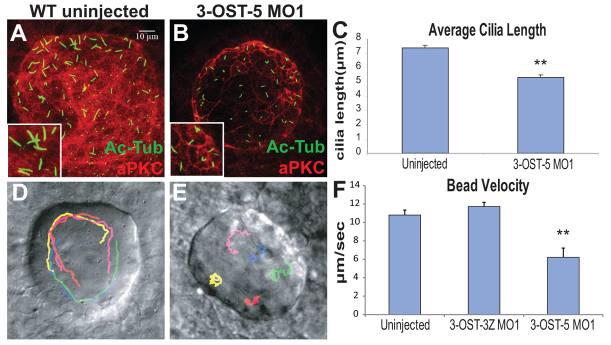
3-OST-5 is required for KV cilia length. (A,B) KV in uninjected (A) and 3-OST-5 MO1-injected (B) embryos at 8 SS. Cilia were labeled with anti-acetylated tubulin (green) and cells labeled by a marker of polarized epithelia anti-aPKC (red). (C) Quantification of cilia length. 3-OST-5 MO1 injected embryos (1429 cilia, 29 embryos) had shorter cilia at 8 SS compared with uninjected control embryos (1379 cilia, 27 embryos; *P<6.19e-10). (D,E) Beads tracks showing KV morphology and bead flow. 3-OST-5 MO1 morphants (E) lacked the coordinated movement seen in uninjected embryos (D) or 3-OST-3Z morphants (supplementary material Movies 1 and 2). Each color represents a single tracked bead. (F) Average velocity of fluorescent beads was significantly slower in 3-OST-5 MO1 morphants (five embryos, five beads/embryo; *P<0.001) compared with either uninjected (eight embryos, five beads/embryo) or 3-OST-3Z MO1 (three embryos, five beads/embryo) controls. Error bars represent s.e.m.
To determine whether knockdown of 3-OST-5 affected cilia-driven fluid flow in KV, fluorescent beads were injected into the KV of 3-OST-5 morphants. The majority of uninjected embryos and control 3-OST-3Z morphants showed a characteristic counterclockwise flow of beads (Essner et al., 2005) (Fig. 2D; supplementary material Fig. S1K,L; Movies 1, 2). By contrast, directional KV flow was perturbed in 3-OST-5 morphants (Fig. 2E; supplementary material Fig. S1M; Movie 3). In addition to perturbed directionality, the velocity of bead movement was significantly decreased in 3-OST-5 morphants compared with uninjected and 3-OST-3Z morphants (Fig. 2F; P<0.001).
Foxj1a and Rfx2 ciliogenic transcription factors are dependent on 3-OST-5
We next focused on several markers known to be involved in cilia formation and function. In 3-OST-5 morphants, expression of dnah9 mRNA, a dynein subunit, was expressed normally in DFCs in >89% of embryos (Fig. 3A,B; Fig. 5H). This concurred with our sox17 results (see above), indicating that the DFCs appear to be migrating and differentiating normally. Two transcription factor genes, foxj1a and rfx2, implicated in zebrafish KV ciliogenesis and downstream LR patterning (Yu et al., 2008; Neugebauer et al., 2009; Tian et al., 2009; Bisgrove et al., 2012), were diminished in 3-OST-5 morphants compared with controls (both P<0.001; Fig. 3C-F; Fig. 5H). The 3-OST-5 phenotypes of shorter cilia, loss of ciliogenic transcription factor expression, and altered LR patterning are similar to phenotypes seen in Fgfr1 signaling pathway manipulations (Hong and Dawid, 2009; Neugebauer et al., 2009). sef, a downstream FGF signaling response gene (Tsang et al., 2002), is downregulated specifically in the DFCs of 3-OST-5 morphants (P<0.001 compared with controls; Fig. 3G,H; Fig. 5H). Together, these results indicate that the mechanism by which 3-OST-5 controls cilia length includes regulation of the ciliogenic transcription factor genes foxj1a and rfx2, perhaps by modulating the FGF signaling pathway in KV cells.
Fig. 3.
3-OST-5 function is required for FGF signaling to regulate cilia length. (A,B) Motor molecule dnah9 mRNA was equivalently expressed in the DFC (white arrows) of tailbud stage in uninjected embryos (A; n=99) and 3-OST-5 MO1-injected embryos (B; n=62). (C,D) Ciliogenic transcription factor gene foxj1a expression in uninjected tailbud stage embryos (C, white arrow; n=111) was diminished or absent in 36% of 3-OST-5 MO1-injected embryos (D, red arrow; n=75). (E,F) Similarly, ciliogenic transcription factor gene rfx2 expression in uninjected 90% epiboly embryos (E, white circle; n=175) was diminished in 3-OST-5 MO1 DFCs (F, red circle; n=92). (G,H) FGF pathway response gene sef expression in DFCs of 90% epiboly uninjected embryos (G, white arrow; n=85) was diminished in 3-OST-5 MO1-injected embryos (H, red arrow; n=44). See Fig. 5 for quantification of in situ hybridization. (I) Schematic of experiments to test interactions between FGF signaling and 3-OST-5 function. Clutches of embryos from heterozygous crosses of fgf8 hypomorphic mutants were divided into two groups: uninjected or injected with sub-threshold dose of 3-OST-5 MO embryos. At 10 SS, individual embryos were fixed, assayed for KV cilia length, and then individually genotyped for fgf8/ace alleles by HRMA. (J) Average cilia length in WT, heterozygous and homozygous fgf8/ace mutants that were not injected or had been injected with a sub-threshold dose of 3-OST-5 MO1. Homozygous fgf8 mutant embryos injected with sub-threshold dose of 3-OST-5 MO1 had statistically shorter cilia length (**P<0.001) compared with all other classes of embryos. Error bars represent s.e.m. (K) Schematic summarizing 3-OST-5 modulation of FGF signaling and ciliogenic transcription factors to control cilia length.
Fig. 5.
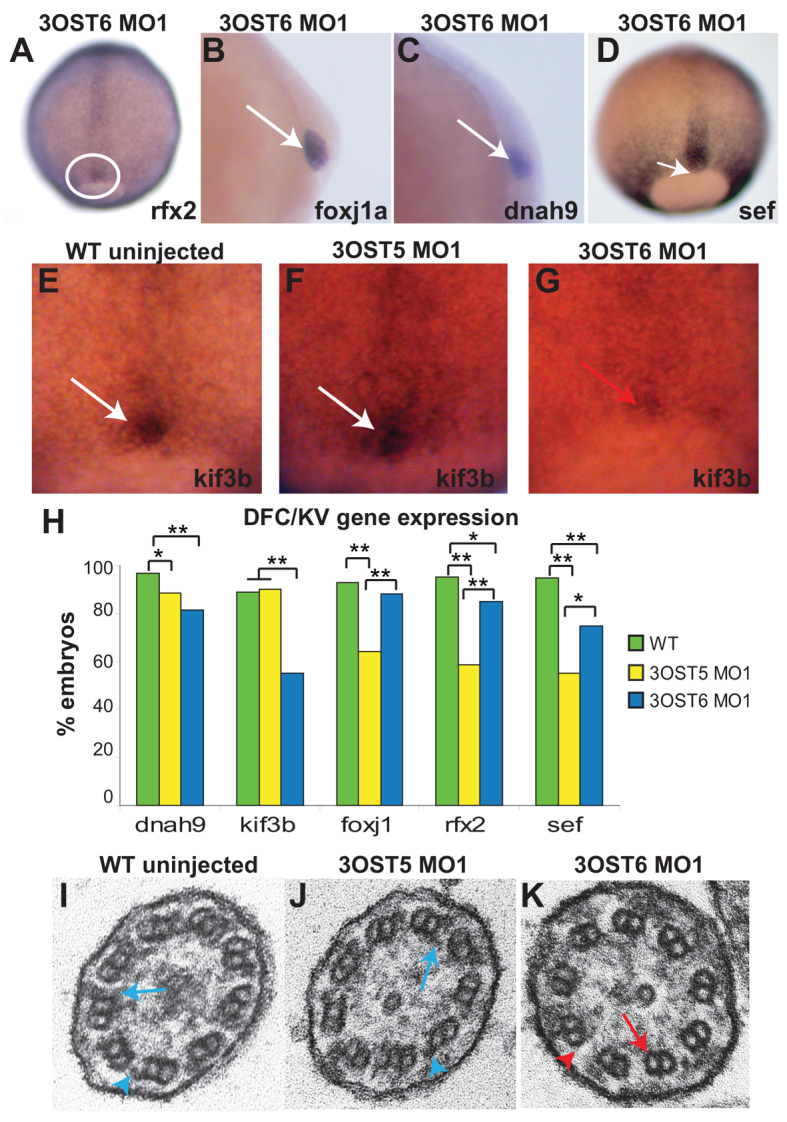
3-OST-6 regulates distinct cell signaling pathways to control cilia motility. (A-D) In contrast to diminished expression in 3-OST-5 MO-injected embryos (Fig. 3), 3-OST-6 MO1-injected embryos had normal expression of ciliogenic transcription factors rfx2 (A; n=74) and foxj1a (B; n=84). Motor molecule dnah9 (C; n=60) and the FGF-response gene sef (D; n=39) were also normally expressed in 3-OST-6 MO1-injected embryos. (E-G) Motor molecule kif3b mRNA normally expressed in the DFCs of 90% epiboly uninjected embryos (E, white arrow; n=79) and 3-OST-5 MO1-injected embryos (F, white arrow; n=48) was diminished in 3-OST-6 MO1-injected embryos (G, red arrow; n=38). (H) Percentages of embryos with normal levels of DFC expression of each marker gene in WT uninjected (green), 3-OST-5 MO1-injected (yellow) and 3-OST-6 MO1-injected (blue) embryos. *P<0.05, **P<0.001. The data represent a compilation of at least three independent experiments. (I-K) Electron micrographs showing cross-sections of cilia in the KV. Inner (blue arrows) and outer (blue arrowheads) dynein arms were visible in WT (I; n=3 embryos) and 3-OST-5 MO1-injected embryos embryos (J; n=4 embryos; P<1 compared with WT). By contrast, inner (red arrow) and outer (red arrowhead) dynein arms were largely absent in 3-OST-6 morphants (K; n=4 embryos; P<0.0018 compared with 3-OST-5 MO1 and WT).
3-OST-5 and FGF signaling interact to control normal length KV cilia
Our previous work has shown that two FGF ligands, Fgf8 and Fgf24, and the FGF receptor 1 (Fgfr1), regulate a ciliogenic transcription factor network controlling cilia length (Neugebauer et al., 2009). Interestingly, the Fgf8 hypomorphic mutant acerebellar (ace) does not have a shortened cilia phenotype. However, further reduction of FGF ligands by Fgf24 MO into ace mutants, or other combinations of Fgf8 and Fgf24 mutants and morphants, results in a shortened cilia phenotype (Neugebauer et al., 2009), indicating that cilia length determination is very sensitive to FGF ligand dosage. In order to determine whether 3-OST-5 works with the FGF pathway to control cilia length, we injected 3-OST-5 MO1 at a sub-threshold dose that does not cause a cilia length phenotype by itself, into embryos generated by mating ace heterozygotes (Fig. 3I). This produces embryos with sub-threshold knockdown of 3-OST-5 in the context of three distinct doses of Fgf8 ligand (homozygous wild type, heterozygous and homozygous mutant; Fig. 3I,J). If HS modifications incurred by 3-OST-5 gene function are required to modulate FGF signaling, then the partial loss of both Fgf8 and 3-OST-5 should be synergistic and lead to a shortened cilia phenotype. Embryos were individually assessed for KV structure, cilia length and cilia number, blinded to Fgf8 genotype. Each individual embryo was then genotyped using high resolution melt analysis (HRMA) (Parant et al., 2009), and identified as WT, ace heterozygous or ace mutant in both the 3-OST-5 MO1-injected and uninjected classes (Fig. 3I). Together, the results indicate (1) that the sub-threshold dose of 3-OST-5 MO1 did not produce a shortened cilia phenotype and (2) that combining sub-threshold reduction of 3-OST-5 and sub-threshold reduction of FGF ligand produced a shortened cilia phenotype (Fig. 3J). These results show that 3-OST-5 and FGF signaling work together to control cilia length through ciliogenic transcription factors (Fig. 3K).
3-OST-6 controls LR development through a distinct KV cell-autonomous pathway
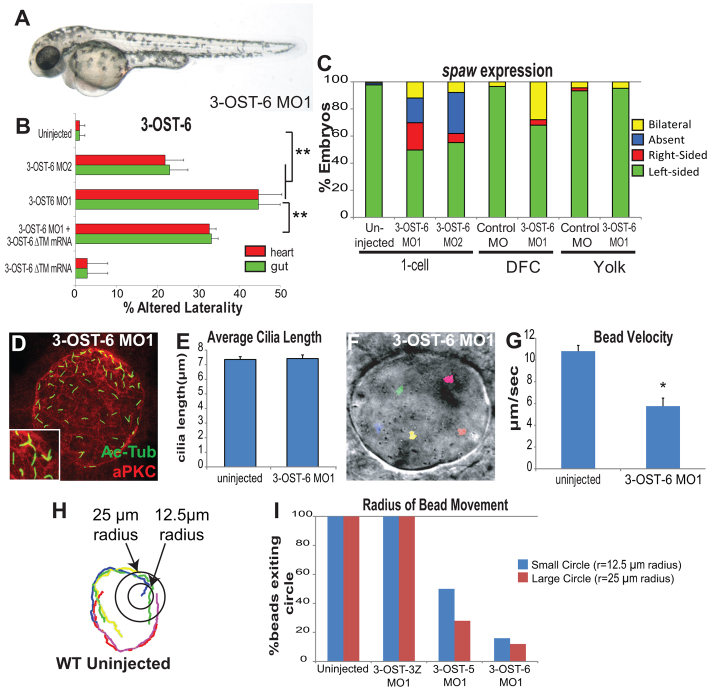
Substantiating the glycocode hypothesis that individual 3-OST family members have distinct functions (Cadwallader and Yost, 2013), we found that 3-OST-6 controls LR patterning through a mechanism that is strikingly different from that of 3-OST-5. In contrast to the curved body axis defects in 3-OST-5 morphants, the gross morphology of 3-OST-6 morphants appeared similar to uninjected control embryos (Fig. 4A). Embryos injected with either a translation-blocking MO targeting 3-OST-6 (3-OST-6 MO1) or a splice-blocking MO (3-OST-6 MO2) displayed a high frequency of reversed heart (MO1 45%, n=172; MO2 22%, n=89) and gut (MO1 45%, n=172; MO2 23%, n=89) orientations (Fig. 4B). The injection of 3-OST-6 MO2 effectively reduced accumulation of 3-OST-6 mRNA (supplementary material Fig. S2A,B). The 3-OST-6 MO1 was significantly rescued by injection of MO-resistant 3-OST-6 mRNA, which lacked the N-terminus and transmembrane coding regions (TM) and could not bind MO1 (3-OST-6 ΔTM; Fig. 4B). Expression of 3-OST-6 ΔTM alone did not alter heart (3% abnormal, n=140) or gut (3%, n=140) orientation (Fig. 4B). As with 3-OST-5, the observation that two distinct morpholinos share a similar LR phenotype, and that the phenotype can be rescued by mRNA co-injection, indicates that 3-OST-6 has a role in LR patterning.
Fig. 4.
3-OST-6 cell-autonomously controls KV cilia motility and LR patterning, but not cilia length. (A) 3-OST-6 morphant at 48 hpf. Except for heart and gut anomalies, 3-OST-6 morphants were indistinguishable from WT embryos at the gross level. (B) Percentages of reversed heart and gut looping in 3-OST-6 morphants (MO1: n=172, **P<0.001; MO2: n=89, **P<0.001) were increased compared with uninjected embryos (n=411). Co-injection of 3-OST-6 MO1 and MO-resistant 3-OST-6ΔTM ‘rescue’ mRNA resulted in a significant rescue of heart and gut orientation (n=184; **P<0.001). Overexpression of 3-OST-6ΔTM mRNA alone did not alter normal heart and gut looping (n=140). Error bars represent s.d. (C) Percentages of left-sided, right-sided, absent, and bilateral spaw in 3-OST-6 morphants (MO1, n=159; MO2, n=165; uninjected, n=220). DFC3-OST-6 MO1 embryos (n=150) showed a high level of bilateral spaw expression, statistically distinct (**P<0.001) from controls: DFCControl MO (n=145), Yolk3-OST-6 MO1 (n=79), and YolkControl MO (n=90). (D) 3-OST-6 morphants had normal KV cell differentiation (red) and normal cilia length (green) that was comparable to control embryos (see Fig. 2A). (E) 3-OST-6 MO (920 cilia, 18 embryos, 8 SS) had cilia lengths comparable to uninjected embryos (P<0.85). Error bars represent s.e.m. (F) 3-OST-6 MO1 morphants lacked directed movement of beads injected into KV. Each color represents a single tracked bead. (G) Average velocity of fluorescent beads injected into KV. Flow velocity was significantly decreased in 3-OST-6 MO1 (five embryos, five beads/embryo; **P<0.001) compared with controls (see Fig. 2D; supplementary material Movies 1 and 2). Error bars represent s.d. (H) Radius of bead movement assay. Concentric circles were placed at bead start. Beads were scored as exiting the inner circle with a radius of 12.5 μm and/or outer circle with a radius of 25 μm. Each color represents a single tracked bead. (I) Quantification of the radius of bead movement.
Upstream of organ laterality, we found that spaw expression was altered in 3-OST-6 morphants (Fig. 4C). Interestingly, the alteration in spaw expression pattern was different from that of 3-OST-5 morphants. 3-OST-5 morphants showed a high incidence of bilateral spaw expression (Fig. 1F), whereas 3-OST-6 morphants had a more randomized spaw expression profile (Fig. 4C). When comparing the classes of spaw, we found a statistically significant difference (Fisher’s Exact Test P<0.001) between 3-OST-5 MO2 morphants and 3-OST-6 MO2 morphants, with embryos derived from injections of sibling embryos. Similar to 3-OST-5, DFC3-OST-6 MO1 embryos had aberrant spaw expression that was predominantly bilateral (Fig. 1F; Fig. 4C). As with the controls in the 3-OST-5 analysis, DFCcontrol MO, YolkControl MO and Yolk3-OST-6 MO embryos did not alter left-sided spaw expression (Fig. 4C). Together, these results demonstrate that 3-OST-6 functions cell-autonomously in the DFC/KV cell lineage to regulate LR development.
Similar to 3-OST-5, 3-OST-6 is expressed ubiquitously in the early embryo, both at 70% epiboly and 7-10 SS (supplementary material Fig. S2C,D). In addition, it appears that early embryonic structures important for LR development are also intact in 3-OST-6 morphants, including normal DFC development and migration (98% normal sox17 whole-mount in situ expression patterns, n=138) and normal KV differentiation and morphogenesis (98% normal by live stereo microscopy, n=90; Fig. 4D, aPKC confocal imaging). At 16 SS, in situ RNA expression of lefty1, ntl and shh in 3-OST-6 MO1-injected embryos indicated normal floor plate and midline development (lefty1, expressed in 87.8%, n=49; ntl, expressed in 100%, n=108; shh, expressed in 100%, n=109; supplementary material Fig. S2E,F). These results indicate that LR patterning defects in 3-OST-6 morphants are not due to perturbations of DFC migration, KV formation or midline development.
Cilia motility, but not cilia length, is dependent on 3-OST-6
Despite having altered LR development, 3-OST-6 morphants had normal KV cilia length (Fig. 4D,E) and normal KV cilia number (51.11±10.47, n=18, P<0.99). This is in striking contrast to the short cilia phenotype in 3-OST-5 morphants (Fig. 2A-C). However, bead tracking in 3-OST-6 morphants showed very little bead movement, and average bead velocity was significantly reduced (Fig. 4F,G; supplementary material Fig. S2G; Movie 4), indicating that the normal length cilia were immotile in 3-OST-6 morphants. We further analyzed the images by drawing two concentric circles centered at the start of each bead track and scored whether each bead exited one or both circles. The inner circle had a 12.5-μm radius and the outer circle had a 25-μm radius (see example in Fig. 4H). In control and 3-OST-3Z morphants, beads exited both circles with 100% frequency (control, n=45 beads; 3-OST-3Z MO, n=15 beads). Consistent with reduced flow but in line with some residual directionality, beads exited the circles with reduced frequency in 3-OST-5 morphants, with only 50% of beads exiting the small circle and <20% exiting the large circle (n=32 beads; P<0.001 compared with controls). In 3-OST-6 morphants, only 12% of beads exited the small circle and <8% exited the large circle (n=25 beads; P<0.001 compared with controls), indicating that the normal length cilia in 3-OST-6 morphants were immotile. This further indicates a statistical difference between bead behaviors in 3-OST-6 morphants compared with 3-OST-5 morphants (P<0.01).
3-OST-6 is required for kinesin motor molecule expression and ciliary dynein arms
Consistent with normal length cilia, and in striking contrast to 3-OST-5 morphants, the ciliogenic transcription factor genes rfx2 and foxj1a had normal expression patterns in a majority of 3-OST-6 morphants (P<0.05 and P<0.325, respectively, compared with controls; Fig. 5A,B). In addition, in 3-OST-6 morphants the number of embryos expressing dnah9 and sef was only slightly decreased compared with controls (P<0.05 and P<0.001, respectively; Fig. 5C,D). To explore further the cause of cilia immotility, we assessed other motor molecules. Kinesin 3b, kif3b, is expressed in DFCs from 30% epiboly to tailbud stages. Kif3b was reduced in the DFCs of 3-OST-6 morphants but not in the DFCs of 3-OST-5 morphants or controls (P<0.001 3-OST-6 MO1 compared with WT and 3-OST5 MO1; Fig. 5E-H). This suggests that the loss of cilia motility in 3-OST-6 morphants is due to a novel regulatory pathway that controls kinesin expression in KV ciliated cells.
Electron microscopy analysis of KV cilia revealed another important distinction between 3-OST-5 and 3-OST-6 morphants. Cilia were scored for the presence or absence of dynein arms (inner and outer); if a single dynein arm was present in a cilium, the entire cilium was scored as having dynein arms. Under this criterion, most cilia in WT uninjected and 3-OST-5 MO embryos had dynein arms (seven out of ten cilia from three WT embryos had detectable dynein arms; nine out of 12 cilia from four 3-OST-5 MO1 embryos had detectable dynein arms; Fig. 5I,J). However, in 3-OST-6 MO1 embryos, only 10% of cilia had detectable dynein arms (one out of 11 cilia from four 3-OST-6 MO embryos had detectable dynein arms; Fig. 5K), which was significantly different (P<0.0018) from WT and 3-OST-5 MO-injected embryos. This suggests that the non-motile cilia, loss of KV fluid flow and LR patterning defects in 3-OST-6 morphants are due to an absence of dynein arms in KV cilia.
DISCUSSION
Here, we present the first in vivo evidence in vertebrates, to our knowledge, that two closely related enzymes in the HS biosynthetic pathway, 3-OST-5 and 3-OST-6, have distinct cellular and developmental functions in the same cells. Significantly, at least two other members, 3-OST-3Z and 3-OST-7, are expressed in these cells, but are unable to compensate for these functions, and knockdown of these two 3-OSTs did not result in LR patterning defects. It is striking that knockdown of two members of the 3-OST family result in a shared developmental end-point phenotype, altered organ LR asymmetry, through cell-autonomous functions in ciliated KV cells. However, knockdown of each of these two 3-OSTs generates this convergent LR phenotype by apparently discrete cellular and molecular mechanisms (Fig. 6): 3-OST-5 regulates cilia length through FGF-dependent ciliogenic transcription factor expression, and 3-OST-6 regulates cilia motility through kinesin expression and cilia dynein arm morphology.
Fig. 6.
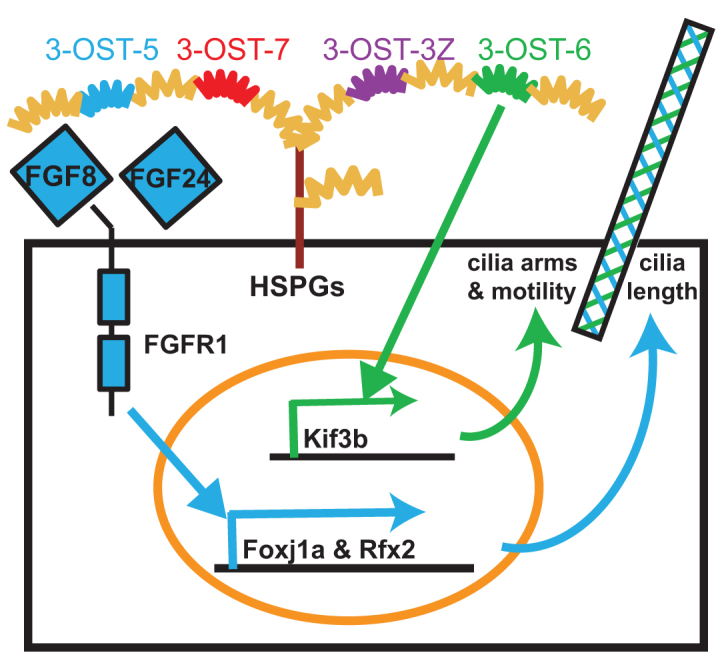
Glycocode model: 3-OST-5 and 3-OST-6 regulate distinct cell signaling pathways for ciliogenesis and cilia motility. Schematic of glycocode regulation of cilia length and cilia motility. 3-OST-5 and FGF pathway perturbations have the same downstream molecular and short cilia phenotypes, and display dose-dependent genetic interactions. Therefore, we propose that 3-OST-5 generates a glycocode (blue) on HS chains on proteoglycans (indicated by HSPGs) that modulates interactions of FGF ligands (for example, Fgf8 and Fgf24) and Fgfr1 at the cell surface. It is important to note that the modifications can be on the same HSPG core protein (as shown HSPG for simplicity) or multiple different HSPGs in the same cell. This interaction activates downstream ciliogenic transcription factors including foxj1a and rfx2 for normal length KV cilia. By contrast, 3-OST-6 generates a different glycocode (green) that modulates a distinct pathway regulating kif3b expression, dynein arms and cilia motility.
3-OST-5 controls cilia length through a pathway that controls the expression of two ciliogenic transcription factor genes in KV, foxj1a and rfx2, and the FGF response gene sef. Fgfr1 and 3-OST-5 have similar functions: they regulate DFC/KV expression of foxj1a and rfx2, cilia length and asymmetric fluid flow in KV, and are cell-autonomously required in KV cells for LR development (Neugebauer et al., 2009). In addition, they have similar phenotypes, such as curved body axis, that are common in morphants or mutants of ciliogenic genes such as polaris (ift88) and foxj1a (Bisgrove et al., 2005; Yu et al., 2008; Tian et al., 2009; Hellman et al., 2010). Thus, it is likely that 3-OST-5 and Fgfr1 signaling function in the same ciliogenic pathway to regulate LR development; this is further substantiated by dosage experiments with 3-OST-5 and the Fgf8 ligand (Fig. 3). We propose that 3-OST-5 in DFC/KV cells generates a glycocode necessary for FGF signaling at the cell surface, which then maintains the downstream ciliogenic pathway to control cilia length (Fig. 6). Interestingly, involvement of 3-OST-5 in FGF signaling appears to be tissue specific. For example, 3-OST-5 has otolith defects, but not the pronephric duct defects observed in fgfr1 morphants (Neugebauer et al., 2009) (data not shown). Downregulation of FGF pathway targets in 3-OST-5 morphants is unique to the DFCs during early embryonic development (Fig. 3G,H), suggesting that other glycocode combinations participate in FGF signaling in other tissues. Preliminary examination of 3-OST-5 putative mutants did not reveal an embryonic phenotype (data not shown). Given the maternally contributed stores of several family members in embryos (Cadwallader and Yost, 2006a), further analysis of maternal-zygotic mutants of this and other family members, perhaps in different strain backgrounds, will be required to fully understand the functional relationships within the 3-OST family, and will allow analysis of later phenotypes that are beyond the reach of morpholino technology.
In both fgfr1 MO- and 3-OST-5 MO-treated embryos, it was somewhat surprising that foxj1a is diminished but dnah9 expression is relatively normal (Neugebauer et al., 2009), in contrast to other reports linking dnah9 expression to foxj1a (Yu et al., 2008; Hellman et al., 2010; Caron et al., 2012). foxj1a expression turns on early, during 30% epiboly (Aamar and Dawid, 2008), and is initiated by the Wnt/β-catenin pathway (Caron et al., 2012). We suggest that FGF signaling, and now 3-OST-5, are required to maintain foxj1a expression. Loss of this pathway results in a reduction of foxj1a mRNA expression, but perhaps a sufficient amount of early Foxj1a protein is available to initiate dnah9 expression. It is important to note that in both 3-OST-5 and Fgfr1 knockdown, cilia are motile even in the face of diminished foxj1a expression.
In contrast to 3-OST-5, 3-OST-6 controls cilia motility without affecting cilia length, through an apparently separate pathway that is necessary for normal kif3b expression and normal LR axis formation (Fig. 6). Thus, 3-OST-6 is a novel and specific cell regulator of cilia motility that controls the organization or maintenance of dynein arms within cilia by generating a distinct glycocode from that generated by 3-OST-5. 3-OST-6 morphants have immotile but normal length cilia, consistent with their relatively normal expression of foxj1a and rfx2 ciliogenic transcription factor genes. Other non-motile cilia mutants have similar phenotypes to 3-OST-6 morphants, such as the LR defects exhibited by seahorse mutants (Serluca et al., 2009), and the axonemal dynein arm defects, exhibited by ktu mutants (Omran et al., 2008; Mitchison et al., 2012). The observation that knockdown of 3-OST-6 in whole embryos and knockdown in DFC/KV have different spaw expression patterns suggests that, in addition to its cell-autonomous role in DFC/KV, 3-OST-6 might also have roles in LR patterning in other cell lineages. It will be interesting to pursue the possibility that 3-OST-6 modulates a distinct signaling pathway to control kif3b transcription and/or assembly of dynein arms in cilia through cargo sorting. In addition, although 3-OST6 morphants have a very distinct phenotype from modulations of the Fgf8/Fgf24/Fgfr1 pathway, we have not excluded the possibility that 3-OST-6 modulates other components of FGF signaling in DFC/KV or other tissues.
We have demonstrated that distinct cell signaling pathways are regulated by distinct 3-OST functions in ciliated cells. These findings suggest that each member of the 3-OST family generates a distinct component of the HSPG glycocode at the cell surface of ciliated cells, perhaps by placing sulfation in the specific context of other regional modifications on the HS chain (Cadwallader and Yost, 2013). Previous reports have demonstrated that different members of the 3-OST family can generate the same sulfated disaccharides in vitro (Shworak et al., 1999; Xia et al., 2002; Xu et al., 2005). Unfortunately, current HS sequencing technology has a limit of ∼6-14 disaccharides from a homogeneous in vitro source (Stringer et al., 2003; Zaia and Costello, 2003; Thanawiroon et al., 2004; Saad and Leary, 2005; Volpi and Linhardt, 2010). In vivo, HS chains range in size from 40 to 160 disaccharides with a heterogeneous composition (Esko and Selleck, 2002). Technical advances in HS structural analysis will be necessary to determine experimentally the sequences of distinct glycocodes, and ultimately the mechanisms by which different members of the 3-OST family generate functionally distinct HS glycocodes. We predict that each cell type will utilize different combinations of OST family members to build distinct combinations of glycocodes at the cell surface in order to discriminate among the myriad of cell-cell signaling pathways available during vertebrate development.
Supplementary Material
Acknowledgments
We thank Bradley Demarest, Todd Townsend, Nick Trede, Manju Karthikeyan, Megan Smith and Elaine Martini for technical assistance; Maureen Condic, Jonathon Hill and Brad Olwin for comments on the manuscript; M. Condic and Yukio Sajioh for blind scoring of cilia TEMs; and Nancy Chandler for outstanding assistance with electron microscopy.
Footnotes
Funding
This work was supported by grants from the National Heart, Lung, and Blood Institute [HL075472 to H.J.Y.]; and the Primary Children’s Medical Foundation. Deposited in PMC for release after 12 months.
Competing interests statement
The authors declare no competing financial interests.
Author contributions
Morpholino design, laterality characterization and rescue experiments were performed by A.B.C. Cilia length and bead tracking experiments were carried out by J.D.A. Kif3b was cloned by B.W.B. D.F.C. and K.V. carried out gene expression, electron microscopy and Fgf8/ace epistasis experiments; radius of bead movement analysis was conducted by J.M.N. Figures and statistics were prepared by J.M.N. The manuscript was written by A.B.C., J.M.N. and H.J.Y. with input from B.W.B. and J.D.A.
Supplementary material
Supplementary material available online at http://dev.biologists.org/lookup/suppl/doi:10.1242/dev.096388/-/DC1
References
- Aamar E., Dawid I. B. (2008). Isolation and expression analysis of foxj1 and foxj1.2 in zebrafish embryos. Int. J. Dev. Biol. 52, 985–991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aamar E., Dawid I. B. (2010). Sox17 and chordin are required for formation of Kupffer’s vesicle and left-right asymmetry determination in zebrafish. Dev. Dyn. 239, 2980–2988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adams D. S., Robinson K. R., Fukumoto T., Yuan S., Albertson R. C., Yelick P., Kuo L., McSweeney M., Levin M. (2006). Early, H+-V-ATPase-dependent proton flux is necessary for consistent left-right patterning of non-mammalian vertebrates. Development 133, 1657–1671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albertson R. C., Yelick P. C. (2005). Roles for fgf8 signaling in left-right patterning of the visceral organs and craniofacial skeleton. Dev. Biol. 283, 310–321 [DOI] [PubMed] [Google Scholar]
- Alexander C. M., Reichsman F., Hinkes M. T., Lincecum J., Becker K. A., Cumberledge S., Bernfield M. (2000). Syndecan-1 is required for Wnt-1-induced mammary tumorigenesis in mice. Nat. Genet. 25, 329–332 [DOI] [PubMed] [Google Scholar]
- Amack J. D., Yost H. J. (2004). The T box transcription factor no tail in ciliated cells controls zebrafish left-right asymmetry. Curr. Biol. 14, 685–690 [DOI] [PubMed] [Google Scholar]
- Amack J. D., Wang X., Yost H. J. (2007). Two T-box genes play independent and cooperative roles to regulate morphogenesis of ciliated Kupffer’s vesicle in zebrafish. Dev. Biol. 310, 196–210 [DOI] [PubMed] [Google Scholar]
- Belenkaya T. Y., Han C., Yan D., Opoka R. J., Khodoun M., Liu H., Lin X. (2004). Drosophila Dpp morphogen movement is independent of dynamin-mediated endocytosis but regulated by the glypican members of heparan sulfate proteoglycans. Cell 119, 231–244 [DOI] [PubMed] [Google Scholar]
- Bellaiche Y., The I., Perrimon N. (1998). Tout-velu is a Drosophila homologue of the putative tumour suppressor EXT-1 and is needed for Hh diffusion. Nature 394, 85–88 [DOI] [PubMed] [Google Scholar]
- Bisgrove B. W., Essner J. J., Yost H. J. (1999). Regulation of midline development by antagonism of lefty and nodal signaling. Development 126, 3253–3262 [DOI] [PubMed] [Google Scholar]
- Bisgrove B. W., Morelli S. H., Yost H. J. (2003). Genetics of human laterality disorders: insights from vertebrate model systems. Annu. Rev. Genomics Hum. Genet. 4, 1–32 [DOI] [PubMed] [Google Scholar]
- Bisgrove B. W., Snarr B. S., Emrazian A., Yost H. J. (2005). Polaris and Polycystin-2 in dorsal forerunner cells and Kupffer’s vesicle are required for specification of the zebrafish left-right axis. Dev. Biol. 287, 274–288 [DOI] [PubMed] [Google Scholar]
- Bisgrove B. W., Makova S., Yost H. J., Brueckner M. (2012). RFX2 is essential in the ciliated organ of asymmetry and an RFX2 transgene identifies a population of ciliated cells sufficient for fluid flow. Dev. Biol. 363, 166–178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cadwallader A. B., Yost H. J. (2006a). Combinatorial expression patterns of heparan sulfate sulfotransferases in zebrafish: I. The 3-O-sulfotransferase family. Dev. Dyn. 235, 3423–3431 [DOI] [PubMed] [Google Scholar]
- Cadwallader A. B., Yost H. J. (2006b). Combinatorial expression patterns of heparan sulfate sulfotransferases in zebrafish: II. The 6-O-sulfotransferase family. Dev. Dyn. 235, 3432–3437 [DOI] [PubMed] [Google Scholar]
- Cadwallader A. B., Yost H. J. (2013) The Glycocode: Translating Heparan Sulfate Fine Structure into Developmental Function, New York, NY: Springer; [Google Scholar]
- Caron A., Xu X., Lin X. (2012). Wnt/β-catenin signaling directly regulates Foxj1 expression and ciliogenesis in zebrafish Kupffer’s vesicle. Development 139, 514–524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J. N., van Eeden F. J., Warren K. S., Chin A., Nüsslein-Volhard C., Haffter P., Fishman M. C. (1997). Left-right pattern of cardiac BMP4 may drive asymmetry of the heart in zebrafish. Development 124, 4373–4382 [DOI] [PubMed] [Google Scholar]
- Colliec-Jouault S., Shworak N. W., Liu J., de Agostini A. I., Rosenberg R. D. (1994). Characterization of a cell mutant specifically defective in the synthesis of anticoagulantly active heparan sulfate. J. Biol. Chem. 269, 24953–24958 [PubMed] [Google Scholar]
- Danos M. C., Yost H. J. (1995). Linkage of cardiac left-right asymmetry and dorsal-anterior development in Xenopus. Development 121, 1467–1474 [DOI] [PubMed] [Google Scholar]
- Danos M. C., Yost H. J. (1996). Role of notochord in specification of cardiac left-right orientation in zebrafish and Xenopus. Dev. Biol. 177, 96–103 [DOI] [PubMed] [Google Scholar]
- Esko J. D., Selleck S. B. (2002). Order out of chaos: assembly of ligand binding sites in heparan sulfate. Annu. Rev. Biochem. 71, 435–471 [DOI] [PubMed] [Google Scholar]
- Essner J. J., Branford W. W., Zhang J., Yost H. J. (2000). Mesendoderm and left-right brain, heart and gut development are differentially regulated by pitx2 isoforms. Development 127, 1081–1093 [DOI] [PubMed] [Google Scholar]
- Essner J. J., Vogan K. J., Wagner M. K., Tabin C. J., Yost H. J., Brueckner M. (2002). Conserved function for embryonic nodal cilia. Nature 418, 37–38 [DOI] [PubMed] [Google Scholar]
- Essner J. J., Amack J. D., Nyholm M. K., Harris E. B., Yost H. J. (2005). Kupffer’s vesicle is a ciliated organ of asymmetry in the zebrafish embryo that initiates left-right development of the brain, heart and gut. Development 132, 1247–1260 [DOI] [PubMed] [Google Scholar]
- Habuchi H., Tanaka M., Habuchi O., Yoshida K., Suzuki H., Ban K., Kimata K. (2000). The occurrence of three isoforms of heparan sulfate 6-O-sulfotransferase having different specificities for hexuronic acid adjacent to the targeted N-sulfoglucosamine. J. Biol. Chem. 275, 2859–2868 [DOI] [PubMed] [Google Scholar]
- Häcker U., Nybakken K., Perrimon N. (2005). Heparan sulphate proteoglycans: the sweet side of development. Nat. Rev. Mol. Cell Biol. 6, 530–541 [DOI] [PubMed] [Google Scholar]
- HajMohammadi S., Enjyoji K., Princivalle M., Christi P., Lech M., Beeler D., Rayburn H., Schwartz J. J., Barzegar S., de Agostini A. I., et al. (2003). Normal levels of anticoagulant heparan sulfate are not essential for normal hemostasis. J. Clin. Invest. 111, 989–999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamada H., Meno C., Watanabe D., Saijoh Y. (2002). Establishment of vertebrate left-right asymmetry. Nat. Rev. Genet. 3, 103–113 [DOI] [PubMed] [Google Scholar]
- Hatler J. M., Essner J. J., Johnson R. G. (2009). A gap junction connexin is required in the vertebrate left-right organizer. Dev. Biol. 336, 183–191 [DOI] [PubMed] [Google Scholar]
- Hellman N. E., Liu Y., Merkel E., Austin C., Le Corre S., Beier D. R., Sun Z., Sharma N., Yoder B. K., Drummond I. A. (2010). The zebrafish foxj1a transcription factor regulates cilia function in response to injury and epithelial stretch. Proc. Natl. Acad. Sci. USA 107, 18499–18504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hojo M., Takashima S., Kobayashi D., Sumeragi A., Shimada A., Tsukahara T., Yokoi H., Narita T., Jindo T., Kage T., et al. (2007). Right-elevated expression of charon is regulated by fluid flow in medaka Kupffer’s vesicle. Dev. Growth Differ. 49, 395–405 [DOI] [PubMed] [Google Scholar]
- Hong S. K., Dawid I. B. (2009). FGF-dependent left-right asymmetry patterning in zebrafish is mediated by Ier2 and Fibp1. Proc. Natl. Acad. Sci. USA 106, 2230–2235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang C. J., Tu C. T., Hsiao C. D., Hsieh F. J., Tsai H. J. (2003). Germ-line transmission of a myocardium-specific GFP transgene reveals critical regulatory elements in the cardiac myosin light chain 2 promoter of zebrafish. Dev. Dyn. 228, 30–40 [DOI] [PubMed] [Google Scholar]
- Kamimura K., Rhodes J. M., Ueda R., McNeely M., Shukla D., Kimata K., Spear P. G., Shworak N. W., Nakato H. (2004). Regulation of Notch signaling by Drosophila heparan sulfate 3-O sulfotransferase. J. Cell Biol. 166, 1069–1079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kramer K. L., Yost H. J. (2002). Ectodermal syndecan-2 mediates left-right axis formation in migrating mesoderm as a cell-nonautonomous Vg1 cofactor. Dev. Cell 2, 115–124 [DOI] [PubMed] [Google Scholar]
- Kramer K. L., Barnette J. E., Yost H. J. (2002). PKCgamma regulates syndecan-2 inside-out signaling during xenopus left-right development. Cell 111, 981–990 [DOI] [PubMed] [Google Scholar]
- Kreiling J. A., Williams G., Creton R.; Prabhat (2007). Analysis of Kupffer’s vesicle in zebrafish embryos using a cave automated virtual environment. Dev. Dyn. 236, 1963–1969 [DOI] [PubMed] [Google Scholar]
- Levin M. (2005). Left-right asymmetry in embryonic development: a comprehensive review. Mech. Dev. 122, 3–25 [DOI] [PubMed] [Google Scholar]
- Long S., Ahmad N., Rebagliati M. (2003). The zebrafish nodal-related gene southpaw is required for visceral and diencephalic left-right asymmetry. Development 130, 2303–2316 [DOI] [PubMed] [Google Scholar]
- López-Gracia M. L., Ros M. A. (2007). Left-right asymmetry in vertebrate development. Adv. Anat. Embryol. Cell Biol. 188, 1–121 [PubMed] [Google Scholar]
- Marjoram L., Wright C. (2011). Rapid differential transport of Nodal and Lefty on sulfated proteoglycan-rich extracellular matrix regulates left-right asymmetry in Xenopus. Development 138, 475–485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meno C., Shimono A., Saijoh Y., Yashiro K., Mochida K., Ohishi S., Noji S., Kondoh H., Hamada H. (1998). lefty-1 is required for left-right determination as a regulator of lefty-2 and nodal. Cell 94, 287–297 [DOI] [PubMed] [Google Scholar]
- Mitchison H. M., Schmidts M., Loges N. T., Freshour J., Dritsoula A., Hirst R. A., O’Callaghan C., Blau H., Al Dabbagh M., Olbrich H., et al. (2012). Mutations in axonemal dynein assembly factor DNAAF3 cause primary ciliary dyskinesia. Nat. Genet. 44, 381-389, S1-S2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nasevicius A., Ekker S. C. (2000). Effective targeted gene ‘knockdown’ in zebrafish. Nat. Genet. 26, 216–220 [DOI] [PubMed] [Google Scholar]
- Neugebauer J. M., Amack J. D., Peterson A. G., Bisgrove B. W., Yost H. J. (2009). FGF signalling during embryo development regulates cilia length in diverse epithelia. Nature 458, 651–654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nogami K., Suzuki H., Habuchi H., Ishiguro N., Iwata H., Kimata K. (2004). Distinctive expression patterns of heparan sulfate O-sulfotransferases and regional differences in heparan sulfate structure in chick limb buds. J. Biol. Chem. 279, 8219–8229 [DOI] [PubMed] [Google Scholar]
- Odenthal J., Nüsslein-Volhard C. (1998). fork head domain genes in zebrafish. Dev. Genes Evol. 208, 245–258 [DOI] [PubMed] [Google Scholar]
- Omran H., Kobayashi D., Olbrich H., Tsukahara T., Loges N. T., Hagiwara H., Zhang Q., Leblond G., O’Toole E., Hara C., et al. (2008). Ktu/PF13 is required for cytoplasmic pre-assembly of axonemal dyneins. Nature 456, 611–616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parant J. M., George S. A., Pryor R., Wittwer C. T., Yost H. J. (2009). A rapid and efficient method of genotyping zebrafish mutants. Dev. Dyn. 238, 3168–3174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rapraeger A. C., Krufka A., Olwin B. B. (1991). Requirement of heparan sulfate for bFGF-mediated fibroblast growth and myoblast differentiation. Science 252, 1705–1708 [DOI] [PubMed] [Google Scholar]
- Reifers F., Böhli H., Walsh E. C., Crossley P. H., Stainier D. Y., Brand M. (1998). Fgf8 is mutated in zebrafish acerebellar (ace) mutants and is required for maintenance of midbrain-hindbrain boundary development and somitogenesis. Development 125, 2381–2395 [DOI] [PubMed] [Google Scholar]
- Saad O. M., Leary J. A. (2005). Heparin sequencing using enzymatic digestion and ESI-MSn with HOST: a heparin/HS oligosaccharide sequencing tool. Anal. Chem. 77, 5902–5911 [DOI] [PubMed] [Google Scholar]
- Schweickert A., Weber T., Beyer T., Vick P., Bogusch S., Feistel K., Blum M. (2007). Cilia-driven leftward flow determines laterality in Xenopus. Curr. Biol. 17, 60–66 [DOI] [PubMed] [Google Scholar]
- Serluca F. C., Xu B., Okabe N., Baker K., Lin S. Y., Sullivan-Brown J., Konieczkowski D. J., Jaffe K. M., Bradner J. M., Fishman M. C., et al. (2009). Mutations in zebrafish leucine-rich repeat-containing six-like affect cilia motility and result in pronephric cysts, but have variable effects on left-right patterning. Development 136, 1621–1631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shworak N. W., Liu J., Petros L. M., Zhang L., Kobayashi M., Copeland N. G., Jenkins N. A., Rosenberg R. D. (1999). Multiple isoforms of heparan sulfate D-glucosaminyl 3-O-sulfotransferase. Isolation, characterization, and expression of human cdnas and identification of distinct genomic loci. J. Biol. Chem. 274, 5170–5184 [DOI] [PubMed] [Google Scholar]
- Soroldoni D., Bajoghli B., Aghaallaei N., Czerny T. (2007). Dynamic expression pattern of Nodal-related genes during left-right development in medaka. Gene Expr. Patterns 7, 93–101 [DOI] [PubMed] [Google Scholar]
- Stringer S. E., Kandola B. S., Pye D. A., Gallagher J. T. (2003). Heparin sequencing. Glycobiology 13, 97–107 [DOI] [PubMed] [Google Scholar]
- Tecle E., Diaz-Balzac C. A., Bülow H. E. (2013). Distinct 3-O-sulfated heparan sulfate modification patterns are required for kal-1-dependent neurite branching in a context-dependent manner in Caenorhabditis elegans. G3 (Bethesda) 3, 541–552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thanawiroon C., Rice K. G., Toida T., Linhardt R. J. (2004). Liquid chromatography/mass spectrometry sequencing approach for highly sulfated heparin-derived oligosaccharides. J. Biol. Chem. 279, 2608–2615 [DOI] [PubMed] [Google Scholar]
- Tian T., Zhao L., Zhang M., Zhao X., Meng A. (2009). Both foxj1a and foxj1b are implicated in left-right asymmetric development in zebrafish embryos. Biochem. Biophys. Res. Commun. 380, 537–542 [DOI] [PubMed] [Google Scholar]
- Topczewski J., Sepich D. S., Myers D. C., Walker C., Amores A., Lele Z., Hammerschmidt M., Postlethwait J., Solnica-Krezel L. (2001). The zebrafish glypican knypek controls cell polarity during gastrulation movements of convergent extension. Dev. Cell 1, 251–264 [DOI] [PubMed] [Google Scholar]
- Tsang M., Friesel R., Kudoh T., Dawid I. B. (2002). Identification of Sef, a novel modulator of FGF signalling. Nat. Cell Biol. 4, 165–169 [DOI] [PubMed] [Google Scholar]
- Tsuda M., Kamimura K., Nakato H., Archer M., Staatz W., Fox B., Humphrey M., Olson S., Futch T., Kaluza V., et al. (1999). The cell-surface proteoglycan Dally regulates Wingless signalling in Drosophila. Nature 400, 276–280 [DOI] [PubMed] [Google Scholar]
- Volpi N., Linhardt R. J. (2010). High-performance liquid chromatography-mass spectrometry for mapping and sequencing glycosaminoglycan-derived oligosaccharides. Nat. Protoc. 5, 993–1004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westerfield M. (2000) The Zebrafish Book. A Guide For The Laboratory Use Of Zebrafish (Danio Rerio). Eugene, OR: University of Oregon Press; [Google Scholar]
- Xia G., Chen J., Tiwari V., Ju W., Li J. P., Malmstrom A., Shukla D., Liu J. (2002). Heparan sulfate 3-O-sulfotransferase isoform 5 generates both an antithrombin-binding site and an entry receptor for herpes simplex virus, type 1. J. Biol. Chem. 277, 37912–37919 [DOI] [PubMed] [Google Scholar]
- Xu D., Tiwari V., Xia G., Clement C., Shukla D., Liu J. (2005). Characterization of heparan sulphate 3-O-sulphotransferase isoform 6 and its role in assisting the entry of herpes simplex virus type 1. Biochem. J. 385, 451–459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto M., Mine N., Mochida K., Sakai Y., Saijoh Y., Meno C., Hamada H. (2003). Nodal signaling induces the midline barrier by activating Nodal expression in the lateral plate. Development 130, 1795–1804 [DOI] [PubMed] [Google Scholar]
- Yayon A., Klagsbrun M., Esko J. D., Leder P., Ornitz D. M. (1991). Cell surface, heparin-like molecules are required for binding of basic fibroblast growth factor to its high affinity receptor. Cell 64, 841–848 [DOI] [PubMed] [Google Scholar]
- Yelon D., Horne S. A., Stainier D. Y. (1999). Restricted expression of cardiac myosin genes reveals regulated aspects of heart tube assembly in zebrafish. Dev. Biol. 214, 23–37 [DOI] [PubMed] [Google Scholar]
- Yost H. J. (2001). Establishment of left-right asymmetry. Int. Rev. Cytol. 203, 357–381 [DOI] [PubMed] [Google Scholar]
- Yu X., Ng C. P., Habacher H., Roy S. (2008). Foxj1 transcription factors are master regulators of the motile ciliogenic program. Nat. Genet. 40, 1445–1453 [DOI] [PubMed] [Google Scholar]
- Zaia J., Costello C. E. (2003). Tandem mass spectrometry of sulfated heparin-like glycosaminoglycan oligosaccharides. Anal. Chem. 75, 2445–2455 [DOI] [PubMed] [Google Scholar]
- Zhou X., Sasaki H., Lowe L., Hogan B. L., Kuehn M. R. (1993). Nodal is a novel TGF-beta-like gene expressed in the mouse node during gastrulation. Nature 361, 543–547 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.