Abstract
In conjunction with the 2012 Yosemite hantavirus outbreak, the number of sera our facility tested for hantavirus antibodies increased. We tracked test results and used the data set to determine if a more efficient testing algorithm was possible. Sera were screened using laboratory-developed pan-hantavirus IgG and IgM enzyme immunoassays (EIAs), with an index of >1.10 defined as positive. Sera that were IgM positive by screening (screen IgM+) were tested for Sin Nombre virus (SNV)-specific IgM using a laboratory-developed EIA; screen IgM+ IgG+ sera were also tested for SNV IgG using a laboratory-developed immunoblot assay. SNV antibody-positive samples were sent to state public health laboratories (PHL) or the CDC for confirmation. Of 3,946 sera tested from July through December 2012, 205 were screen IgM+ IgG negative (IgG−); 7/205 were SNV IgM+, but only 1/5 sent to PHL/CDC was confirmed as SNV IgM+. Of 61 screen IgM+ IgG+ sera, 16 were SNV antibody positive; 13/16 sera (from 11 patients) went to PHL/CDC, where SNV infection was confirmed for all patients. Of 12 confirmed patients, 7 had been exposed at Yosemite. A modified algorithm defining screen indices of ≥2.00 as positive identified 11/12 confirmed cases while reducing the number of sera requiring SNV-specific antibody testing by 65%; the patient missed was not tested until 3 months after the onset of symptoms. Hantavirus antibody testing at our facility identified 12 SNV-infected patients, including 7 exposed at Yosemite. Some screen IgM+ IgG− SNV IgM+ results were false positives, emphasizing the value of PHL/CDC confirmatory testing. We identified a modified algorithm requiring analysis of fewer specimens for SNV-specific antibodies without loss of sensitivity.
INTRODUCTION
The major hantavirus-associated illness in North America is hantavirus pulmonary syndrome (HPS) (1). HPS is caused by Sin Nombre virus (SNV), which is transmitted to humans via inhalation of aerosols of excreta from infected rodents, particularly deer mice (Peromyscus maniculatus) (2–5). HPS is characterized by fever, thrombocytopenia, bilateral pulmonary infiltrates, and hemoconcentration (3, 4, 6). Treatment is supportive, and approximately 35% of HPS patients do not survive (4).
On 16 August 2012, a California Department of Public Health press release announced the diagnosis of HPS in two California residents who had recently visited Yosemite National Park and advised visitors to take precautions to prevent exposure to SNV (7). Another press release issued 30 August 2012 announced four more cases of HPS among recent Yosemite visitors (8). The next day, the National Park Service recommended that individuals who had visited Yosemite National Park between 10 June and 24 August 2012 seek medical attention at the first sign of symptoms consistent with SNV infection (9).
Detection of SNV-specific IgM is the main laboratory tool for identifying acute SNV infection (10, 11). Our facility is one of only two reference laboratories in the United States to offer such testing, and here we document the marked increase in hantavirus serologic testing that occurred as a result of the 2012 Yosemite hantavirus outbreak. Further, we took advantage of the large data set generated to determine if the efficiency of our hantavirus antibody testing algorithm could be improved.
MATERIALS AND METHODS
Sera submitted for hantavirus antibody testing were screened for pan-hantavirus IgM and IgG as previously described (12) using enzyme immunoassays (EIAs) employing microtiter wells coated with a cocktail of recombinant Seoul virus and SNV nucleocapsid proteins (NPs). For each assay, a positive result was defined as an index of >1.10 (12). All sera that were IgM positive by screening (screen IgM+) were reflexed at our facility to a laboratory-developed SNV-specific IgM EIA; this assay is similar to the screening IgM EIA except that it utilizes microtiter wells coated with SNV NP only, and a positive result is defined as an index of ≥0.80. The SNV-specific IgM EIA was validated in 2008 using 69 well-characterized sera and exhibited 96% (27/28) sensitivity and 95% (39/41) specificity. Screen IgM+ sera that were also screen IgG+ were additionally tested for SNV-specific IgG as previously described (12) using an in-house immunoblot assay employing recombinant SNV NP and SNV glycoprotein n envelope peptide, each conjugated to bovine serum albumin (11); reactivity with both SNV NP and the envelope peptide was interpreted as positive. As previously reported (12), screen IgM-negative (IgM−) IgG+ sera were not tested for SNV IgG because the negative IgM screen result rules out acute SNV infection. Sera positive for SNV-specific IgM and/or IgG were sent to the appropriate state public health laboratory (PHL) or the Centers for Disease Control and Prevention (CDC) for confirmatory SNV IgM and IgG testing (13). PHL/CDC testing results and hantavirus exposure locales were supplied by public health personnel.
RESULTS
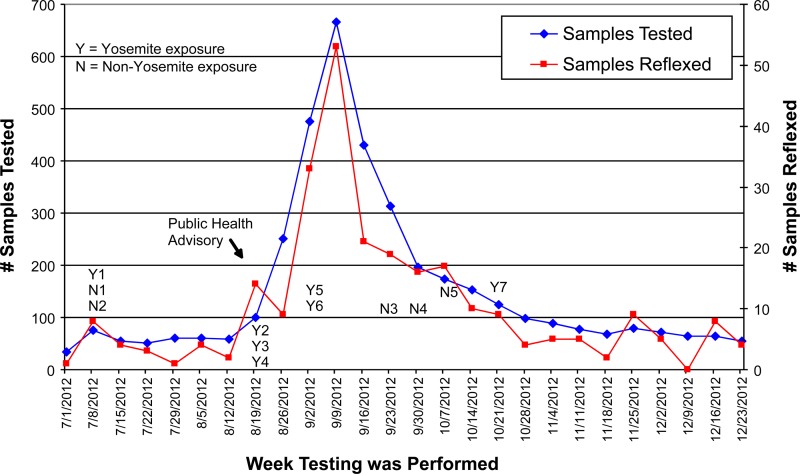
During the last half of 2012, 3,946 sera were submitted to Focus Diagnostics for hantavirus antibody testing. The number of submitted samples increased markedly between the weeks of 26 August and 30 September 2012, reaching a peak during the week of 9 September (Fig. 1). The number of screen-positive samples that were reflexed to SNV-specific IgM testing followed a nearly identical time trend (Fig. 1). Sera from 6 of the 12 patients with confirmed SNV infections were submitted before or during the week that the initial press release was issued by the California Department of Public Health (7).
Fig 1.
Hantavirus antibody testing timeline. Patient designations indicating location of exposure (e.g., Y1 or N1) for the 12 patients with confirmed SNV infection are placed at the week of specimen collection.
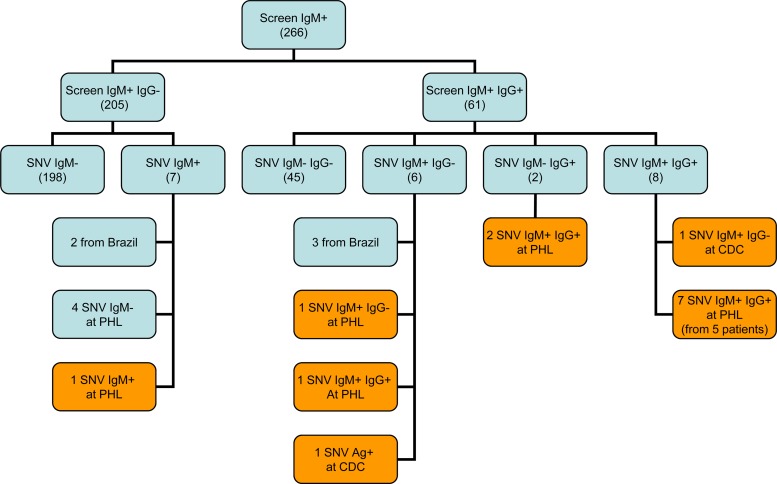
Of these 3,946 sera, 266 (6.7%) were reflexed to SNV-specific IgM testing; 205 were screen IgM+ IgG−, and 61 were screen IgM+ IgG+ (Fig. 2). Of the 205 screen IgM+ IgG− sera, only 7 (3%) were positive for SNV-specific IgM at our facility. Two of the seven SNV IgM+ sera were from Brazilian patients; the sera were presumed to represent Andes or Andes-like hantavirus infection (2, 14) and were excluded from further analysis. The five remaining SNV IgM+ sera were sent to PHL/CDC for confirmatory testing, where only one was confirmed to be SNV IgM+.
Fig 2.
Confirmatory SNV antibody results for hantavirus IgM screen-positive sera. Values in parentheses indicate the number of samples; orange boxes indicate samples with confirmed SNV infection.
Of the 61 screen IgM+ IgG+ sera, 16 (26%) were positive for SNV-specific IgM and/or IgG (Fig. 2) at our laboratory. Of these 16 sera, 3 were from Brazilian patients and were thus excluded from further testing; the 13 remaining sera (from 11 patients) were sent to PHL/CDC, where 12 sera (from 10 patients) were positive for SNV IgM and/or IgG. The one serum sample not tested at PHL was from a deceased patient whose SNV infection was confirmed by the detection of SNV antigen in lung tissue. Table 1 summarizes the Focus and PHL/CDC results and site of SNV exposure for the 12 patients (1 screen IgM+ IgG− sample plus 11 screen IgM+ IgG+ samples) with confirmed SNV infection.
Table 1.
Sample collection details and results for 12 patients with confirmed Sin Nombre virus infection
| Patient no. | Exposure locale | Collection date (mo/day/yr) | Screen result (index) |
Subsequent test result |
|||
|---|---|---|---|---|---|---|---|
| IgM | IgG | Focus SNV IgM | Focus SNV IgG | Confirmatory lab SNV (location)a | |||
| Y1 | Yosemite | 7/6/2012 | 9.11 | 3.77 | Positive | Positive | IgM+ IgG+ (CA) |
| N1 | Oregon | 7/8/2012 | 10.36 | ≤1.10 | Positive | Not done | IgM+ IgG− (OR) |
| N2 | Colorado | 7/9/2012 | 10.74 | 1.52 | Positive | Negative | Antigen-positive lung tissue* |
| Y2 | Yosemite | 8/20/2012 | 5.11 | 6.26 | Positive | Positive | IgM+ IgG+ (CA) |
| Y3 | Yosemite | 8/20/2012 | 10.08 | 2.05 | Positive | Positive | IgM+ IgG+ (CA) |
| Y4 | Yosemite | 8/22/2012 | 8.66 | 1.11 | Positive | Negative | IgM+ IgG− (CA) |
| Y5 | Yosemite | 8/31/2012 | 3.66 | 8.16 | Positive | Positive | IgM+ IgG+ (CA) |
| Y6 | Yosemite | 9/4/2012 | 10.08 | 6.91 | Positive | Positive | IgM+ IgG+ (CA) |
| N3 | Washington | 9/21/2012 | 2.51 | 2.69 | Negative | Positive | IgM+ IgG+ (WA) |
| N4 | New York | 9/30/2012 | 5.45 | 1.31 | Positive | Positive | IgM+ IgG−* |
| N5 | Wyoming | 10/8/2012 | 6.00 | 2.11 | Positive | Positive | IgM+ IgG+ (MT) |
| Y7 | Yosemite | 10/20/2012 | 1.22 | 9.37 | Negative | Positive | IgM+ IgG+ (CA) |
Test was performed at the indicated state PHL except as noted otherwise. *, CDC.
Table 2 summarizes an analysis of an alternative reflex algorithm with a more stringent result cutoff (index of ≥2.00 instead of >1.10) for the screen IgM and IgG EIAs. The alternative algorithm would have reduced the number of samples that reflexed to SNV-specific IgM testing by 65% (from 266 to 93) and would still have identified 11 of 12 SNV-infected patients. The one patient who would have been missed using the alternative algorithm was not tested until 3 months after the onset of symptoms, a timeline not typical for identification of acute SNV infection. Of note, three samples from SNV-infected patients that were screen IgM+ IgG+ by the old algorithm would have been reclassified as screen IgM+ IgG− using the modified algorithm because they had screen IgG indices of >1.10 but <2.00; however, these patients would still have been tested for SNV-specific IgM because of screen IgM+ indices of ≥2.00.
Table 2.
Impact of a modified reflex algorithm based on different screen result cutoff values
| Parameter | Value for the parametera |
|
|---|---|---|
| Original algorithm | Modified algorithm | |
| No. of samples reflexed (screen IgM+) | 266 | 93 |
| No. of screen IgM+ IgG− samples | 205 | 79 |
| No. of SNV IgM+ samples (no. confirmed positive) | 7 (1) | 11 (4)b |
| No. of screen IgM+ IgG+ samples | 61 | 14 |
| No. of SNV IgM+ and/or IgG+ samples (no. confirmed positive) | 16 (13) | 11 (9) |
| No. of SNV-infected patients identified | 12 | 11c |
A positive screen was identified as an index of >1.10 with the original algorithm and of ≥2.00 with the modified algorithm.
Three confirmed positive patients (N2, Y4, and N4) with a screen IgG index of >1.10 but <2.00 were defined as screen IgM+ IgG+ using the original algorithm but were reclassified as screen IgM+ IgG− using the modified algorithm.
The serum from the patient who was not identified was collected 3 months after the onset of symptoms.
DISCUSSION
The highly publicized 2012 hantavirus outbreak among visitors to Yosemite National Park led to a marked increase in the number of serum specimens submitted to our laboratory for hantavirus antibody testing. Of the 10 HPS cases identified as part of the Yosemite outbreak (15), 7 cases (all California residents) were identified through investigations triggered by positive SNV-specific antibody results generated by our facility. During the same time period as the Yosemite outbreak, we also aided in the identification of five additional HPS patients whose SNV infections occurred in other geographic areas.
Our algorithm for identifying patients with acute hantavirus infection begins with pan-hantavirus IgM and IgG screening assays, designed to identify patients potentially infected with Old World (e.g., Seoul, Hantaan, or Puumala) as well as New World (SNV or Andes) hantaviruses. This approach capitalizes on the observation that antibodies induced by infection with one hantavirus species exhibit various levels of cross-reactivity with NPs of other hantaviruses (14, 16); due to the distant relationship between Seoul hantavirus and SNV (16), an assay employing their NPs should detect antibodies induced by all known hantaviruses. Serum samples positive in the IgM screening assay are then tested in an IgM EIA employing SNV NP only as the antigenic target in order to identify samples representing possible SNV infection; SNV IgM+ samples then require confirmatory testing by a public health laboratory. Screen IgM+ samples that are also screen IgG+ are also tested for SNV-specific IgG using a peptide from the SNV G1 envelope protein as the antigenic target. Because antibodies recognizing hantavirus envelope proteins do not exhibit the broad reactivity associated with NP antibodies (10, 11), a positive IgG result in SNV IgM+ patients provides additional support for SNV infection, whereas a negative IgG result in SNV IgM+ patients may indicate infection with a hantavirus other than SNV (e.g., Andes or Andes-like hantavirus). As demonstrated in an earlier publication from our group (12), screen IgM− IgG+ serum samples are not tested for SNV IgG since the negative IgM screen result rules out acute hantavirus infection.
Although 6.7% of sera submitted to our laboratory for hantavirus antibody testing were positive in the screening IgM assay, >90% of these were negative for SNV-specific IgM, most likely indicating nonspecific reactivity in the screen IgM assay rather than recent exposure to a related hantavirus. Further analyses showed that all screen IgM+ sera that were positive in our SNV IgM assay had screen IgM index values of >2.00. We thus assessed the impact of a modified testing algorithm in which the cutoff index for defining screen IgM+ results was increased from 1.10 to 2.00. This assessment demonstrated that the number of screen IgM+ samples (thus requiring SNV-specific IgM testing) would have been reduced by 65% without sacrificing sensitivity. Nonetheless, the modified algorithm still identified 79 screen IgM+ IgG− samples that were negative in our SNV IgM assay. Although this subset of samples represents only 2% of all samples tested, further studies are needed to identify the mechanism(s) responsible for this nonspecific reactivity, which in turn should lead to a modified IgM screening assay with improved specificity.
Another limitation of our hantavirus antibody testing system was the rare occurrence of screen IgM+ IgG− sera exhibiting false-positive SNV IgM reactivity. All four samples demonstrating this pattern had screen IgM index values of >2.00 and would thus still have been considered positive at our facility using the modified reflex testing algorithm. The explanation for the SNV IgM false-positive reactivity remains unclear and is under investigation; one possibility is our use of recombinant SNV nucleocapsid protein, in contrast to the PHL/CDC's use of a whole-virus extract prepared from SNV grown in cell culture (13). Whatever the resolution, this type of assay discordance emphasizes the importance of a collaborative relationship between reference laboratories and public health agencies for identifying acute SNV infections.
In addition to evaluating the effect of increasing the cutoff for defining a screen IgM+ result, we also assessed the impact of using a cutoff index of ≥2.00 for defining a screen IgG+ result. This modification, when combined with a screen IgM cutoff of ≥2.00, would have reduced the number of samples requiring SNV IgG immunoblot testing from 61 sera to only 14 sera (a reduction of 77%) without compromising the identification of SNV-infected patients.
Based on these findings and data from additional collaborative studies with the CDC, our laboratory now routinely uses the modified hantavirus reflex algorithm, which employs a ≥2.00 positive cutoff value for both the screen IgM and screen IgG EIAs. While reflexing 65% fewer samples to confirmatory testing, this algorithm successfully detected 11 of the 12 SNV-infected patients identified with the original algorithm during the last half of 2012. The sole patient missed by the modified algorithm (due to a screen IgM index of <2.00) was tested at 3 months after the onset of symptoms in response to the press releases about the Yosemite outbreak. This timeline is atypical for detecting acute hantavirus infection and would not be expected in a routine clinical setting.
ACKNOWLEDGMENTS
We gratefully acknowledge the collaborative assistance of Barbara Knust from the CDC hantavirus laboratory and David Cottam from the California Department of Public Health.
Footnotes
Published ahead of print 5 June 2013
REFERENCES
- 1. Peters CJ, Simpson GL, Levy H. 1999. Spectrum of hantavirus infection: hemorrhagic fever with renal syndrome and hantavirus pulmonary syndrome. Annu. Rev. Med. 50: 531– 545 [DOI] [PubMed] [Google Scholar]
- 2. Khaiboullina SF, St Jeor SC. 2002. Hantavirus immunology. Viral Immunol. 15: 609– 625 [DOI] [PubMed] [Google Scholar]
- 3. Lednicky JA. 2003. Hantaviruses. Arch. Pathol. Lab. Med. 127: 30– 35 [DOI] [PubMed] [Google Scholar]
- 4. Maes P, Clement J, Gavrilovskaya I, Van Ranst M. 2004. Hantaviruses: immunology, treatment, and prevention. Viral Immunol. 17: 481– 497 [DOI] [PubMed] [Google Scholar]
- 5. Childs JE, Ksiazek TG, Spiropoulou CF, Krebs JW, Morzunov S, Maupin GO, Gage KL, Rollin PE, Sarisky J, Enscore RE, Frey JK, Peters CJ, Nichol ST. 1994. Serologic and genetic identification of Peromyscus maniculatus as the primary rodent reservoir for a new hantavirus in the southwestern United States. J. Infect. Dis. 169: 1271– 1280 [DOI] [PubMed] [Google Scholar]
- 6. Schmidt J, Meisel H, Hjelle B, Kruger DH, Ulrich R. 2005. Development and evaluation of serological assays for detection of human hantavirus infections caused by Sin Nombre virus. J. Clin. Virol. 33: 247– 253 [DOI] [PubMed] [Google Scholar]
- 7. California Department of Public Health 2012. Hantavirus pulmonary syndrome found in two California residents. http://www.cdph.ca.gov/Pages/NR12-040.aspx
- 8. California Department of Public Health 2012. Hantavirus found in four more visitors to Yosemite National Park. http://www.cdph.ca.gov/Pages/NR12-049.aspx
- 9. National Park Service 2012. Hantavirus pulmonary syndrome response continues at Yosemite National Park (August 31, 2012). http://www.nps.gov/yose/parknews/hanta812b.htm
- 10. Bharadwaj M, Nofchissey R, Goade D, Koster F, Hjelle B. 2000. Humoral immune responses in the hantavirus cardiopulmonary syndrome. J. Infect. Dis. 182: 43– 48 [DOI] [PubMed] [Google Scholar]
- 11. Hjelle B, Jenison S, Torrez-Martinez N, Herring B, Quan S, Polito A, Pichuantes S, Yamada T, Morris C, Elgh F, Lee HW, Artsob H, Dinello R. 1997. Rapid and specific detection of Sin Nombre virus antibodies in patients with hantavirus pulmonary syndrome by a strip immunoblot assay suitable for field diagnosis. J. Clin. Microbiol. 35: 600– 608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Prince HE, Su X, Hogrefe WR. 2007. Utilization of hantavirus antibody results generated over a five-year period to develop an improved serologic algorithm for detecting acute Sin Nombre hantavirus infection. J. Clin. Lab. Anal. 21: 7– 13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. MacNeil A, Comer JA, Ksiazek TG, Rollin PE. 2010. Sin Nombre virus-specific immunoglobulin M and G kinetics in hantavirus pulmonary syndrome and the role played by serologic responses in predicting disease outcome. J. Infect. Dis. 202: 242– 246 [DOI] [PubMed] [Google Scholar]
- 14. Schmidt J, Meisel H, Capria SG, Petraityte R, Lundkvist A, Hjelle B, Vial PA, Padula P, Kruger DH, Ulrich R. 2006. Serological assays for the detection of human Andes hantavirus infections based on its yeast-expressed nucleocapsid protein. Intervirology 49: 173– 184 [DOI] [PubMed] [Google Scholar]
- 15. Centers for Disease Control and Prevention 2012. Outbreak of hantavirus infection in Yosemite National Park. http://www.cdc.gov/hantavirus/outbreaks/yosemite-national-park-2012.html
- 16. Elgh F, Linderholm M, Wadell G, Tarvnik A, Juto P. 1998. Development of humoral cross-reactivity to the nucleocapsid protein of heterologous hantaviruses in nephropathia epidemica. FEMS Immunol. Med. Microbiol. 22: 309– 315 [DOI] [PubMed] [Google Scholar]