Abstract
The staphylococcal adhesin clumping factor A (ClfA) has a variant amino acid sequence, generating the potential for alterations in epitope structure and immunogenicity of this vaccine candidate. We demonstrated for two recombinant ClfA40–531 (a slightly truncated version of the fibrinogen-binding domain of ClfA containing amino acids 40 to 531) genetic variants that strain-specific epitopes are immunodominant. This work indicates that immune responses elicited by ClfA may, at least in part, be dependent on the strain of origin of the ClfA.
TEXT
Staphylococcus aureus is a normal colonizing species of the human anterior nares and causes a wide variety of diseases if it breaches the epithelial layer. Increasingly, S. aureus strains are methicillin resistant (MRSA) and as such are recalcitrant to clearance with many standard antibiotics (1). Prophylactic vaccination could be an effective means of decreasing the incidence of infection by this pathogen. Several vaccine candidates have shown various levels of success in animal models (for reviews, see references 2 and 3). Clumping factor A (ClfA), an adhesin that can adhere to both fibrinogen and fibronectin, demonstrated some level of protection in sepsis (4, 5), arthritis (5, 6), prosthetic device infection (7), and mastitis (8) models. The fibrinogen-binding domain of ClfA (amino acids 40 to 559) is responsible for ligand binding. The entire domain and a slightly truncated version of the domain (amino acids 40 to 531) have been shown to elicit protection in animal models (5, 7, 9, 10). A vaccine including a ClfA antigen based on the fibrinogen-binding domain fragment is currently in clinical trials (11, 12). Antibodies against the fibrinogen-binding domain of ClfA have been shown to provide passive protection against S. aureus disease in sepsis, arthritis, and mastitis animal models (6, 13, 14).
Up to 14% of the primary amino acid sequence of ClfA varies between sequenced isolates (15, 16), which could alter epitope composition and change ClfA antigenicity and immunogenicity between S. aureus strains. The crystal structure of the N2 and N3 subdomains (amino acids 221 to 559) of the fibrinogen-binding domain of ClfA (17, 18) was used by Murphy and colleagues (15) to map the sequence diversity in the N2 and N3 subdomains of 39 ClfA strain variants. They found that the large majority of the variant regions were surface exposed.
In this study, we examined antibodies elicited by two different ClfA fibrinogen-binding domain genetic variants to determine the ability of antibodies elicited by one genetic variant to bind to the ClfA fibrinogen-binding domain from another genetic variant in order to examine the strain specificity of the antibody response. We chose to study ClfA genetic variants of two representative S. aureus strains, N315 and NRS384 (strain N315 belongs to clonal complex 5, and strain NRS384, of the pulsed-field gel electrophoresis [PFGE] type USA300, belongs to clonal complex 8; http://www.narsa.net/content/home.jsp). When we aligned the two proteins from amino acids 40 to 531, we found a 10% amino acid variation between S. aureus N315 and NRS384 ClfA (see Fig. S1 in the supplemental material). While a crystal structure of neither the N315 nor NRS384 ClfA fibrinogen-binding domain is available, the crystal structure of the Newman ClfA N2N3 fibrinogen-binding domain, which is 99% identical to that of NRS384 (Fig. S1), is available. The single amino acid difference falls in the N1 domain, allowing us to use the published crystal structure as a surrogate for the NRS384 ClfA N2N3 fibrinogen-binding domain. In order to determine whether the altered amino acids are surface associated, we compared the amino acid sequence of N315 ClfA to the published three-dimensional structure of the ClfA N2N3 fibrinogen-binding domain from S. aureus strain Newman (18) using PyMOL software (Schrödinger LLC, New York City, NY). Of the 41 amino acid variations within the N2N3 region, 40 appear to be at least partially surface exposed (Fig. 1), with only L508 appearing to be internal. These results agree with those of Murphy et al., where structural mapping of 39 ClfA variants indicated that the majority of diverse sites were surface exposed (15). Because the crystal structure elaborates the N2 and N3 subdomains of the ClfA fibrinogen binding domain but lacks the N1 subdomain (amino acids 40 to 220), we could not examine the locations of amino acid variations in the N1 subdomain.
Fig 1.
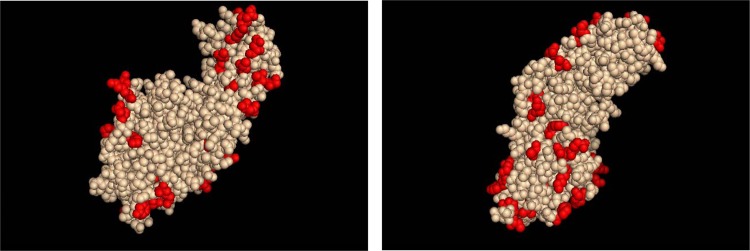
Locations of variant amino acids on ClfA crystal structure. Amino acids that are altered in S. aureus N315 ClfA N2N3 subdomains compared to S. aureus Newman/NRS384 ClfA N2N3 subdomains are highlighted in red. The left and right panels are 180° views.
We next cloned DNA encoding ClfA40–531 (a slightly truncated version of the fibrinogen-binding domain of ClfA containing amino acids 40 to 531) from S. aureus N315 and NRS384 in Escherichia coli and expressed and purified the resultant recombinant proteins. We immunized BALB/c mice with either ClfA40–531 variant (6-week-old females; 20 μg per dose, adsorbed to 200 μg Alhydrogel with 15 μg CpG, at days 0 and 14) and obtained immune sera 2 weeks following the second immunization. We then analyzed the ability of antibodies generated against each recombinant to bind to both the homologous and heterologous ClfA40–531 variants using enzyme-linked immunosorbent assay (ELISA). As shown in Fig. 2, both antisera bound to both recombinants, illustrating that the ClfA40–531 variants are cross-immunogenic. However, we noted that the ELISA curves were slightly shifted when heterologous antibodies were used, suggesting that antibody binding to the two variants may differ.
Fig 2.
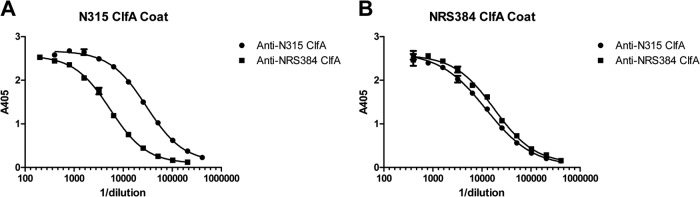
Antigen binding by heterologous ClfA antibodies in direct ELISA. (A and B) Serially diluted anti-N315 ClfA40–531 sera and serially diluted anti-NRS384 ClfA40–531 sera were applied to an S. aureus N315 ClfA40–531-coated plate (A) or an S. aureus NRS384 ClfA40–531-coated plate (B). The graphs were generated from data from duplicate plates with the mean ± standard error of the mean (SEM) shown for each point. The figure shows data from one experiment that is representative of two independent experiments.
In order to further investigate differences in antibody binding, we evaluated the ability of each ClfA40–531 variant to bind to antibodies generated against either itself or the heterologous variant using a competitive ELISA. In this assay, soluble recombinant competitor protein (either S. aureus N315 or NRS384 ClfA40–531) was titrated starting at 600 nM, mixed with a constant dilution of antisera (either anti-N315 ClfA40–531 or anti-NRS384 ClfA40–531), and incubated for 1 h. The antiserum/competitor mixture was then added to a blocked ELISA plate that was coated with the homologous recombinant ClfA40–531 (i.e., N315 ClfA40–531 for anti-N315 ClfA40–531 serum and NRS384 ClfA40–531 for anti-NRS384 ClfA40–531 serum). Anti-mouse IgG conjugated to horseradish peroxidase was used as the secondary antibody (KPL Inc., Gaithersburg, MD). The ability of the soluble competitor to bind to anti-ClfA40–531 antibodies and prevent those antibodies from binding to the antigen coated on the plate (resulting in a lower absorbance reading) reflects the affinity of those antibodies for the competitor—the higher the affinity of the antibody for the competitor, the lower the absorbance reading would be. The percent competition at 600 nM competitor protein was analyzed using a two-tailed Student's t test. A difference in the abilities of the homologous versus heterologous ClfA40–531 variants to compete for antibody binding would indicate differences in antibody affinity.
We found that while soluble homologous antigen effectively competed for antibody binding in the competitive ELISA, soluble heterologous antigen was largely unable to compete (Fig. 3). The level of competition achieved with 600 nM soluble competitor was significantly higher when homologous antigen was added than when heterologous antigen was used (P = 0.0005 for N315 ClfA40–531 competitor compared to NRS384 ClfA40–531 competitor using anti-N315 ClfA40–531 sera; P = 0.0001 for NRS384 ClfA40–531 compared to N315 ClfA40–531 competitor using anti-NRS384 ClfA40–531 sera). These findings suggest that both anti-N315 ClfA40–531 and anti-NRS384 ClfA40–531 antibodies have a higher affinity for the homologous variant than for the heterologous variant. Because the heterologous variant is ineffective at competing for antibodies in this assay, the data further suggest that the variant epitopes of ClfA40–531 are immunodominant.
Fig 3.
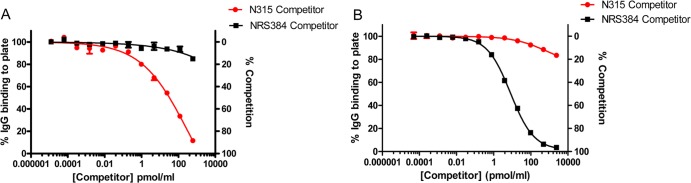
Limited antibody binding by heterologous ClfA in competitive ELISA. (A and B) Diluted anti-N315 ClfA40–531 sera (A) and diluted anti-NRS384 ClfA40–531 sera (B) were mixed with the indicated concentrations of either S. aureus N315 ClfA40–531 or S. aureus NRS384 ClfA40–531 prior to addition to an N315 ClfA40–531-coated plate (A) or an NRS384 ClfA40–531-coated plate (B). Percent competition was calculated by normalizing the data points to the highest A405 reading for each antibody/competitor sample (normalized to 100% IgG binding to plate or 0% competition). The graphs were generated from data from duplicate plates with the mean ± standard error of the mean (SEM) shown for each point. The figure shows data from one experiment that is representative of three independent experiments.
The data presented herein highlight the potential for small differences in protein composition across S. aureus strains to have large effects on antigenicity. Recombinant S. aureus N315 and NRS384 ClfA40–531 proteins exhibit only 10% variation in their amino acid sequences, and antibodies elicited by one genetic variant will bind to another. However, we demonstrated a large difference in the ability of each variant to bind to the homologous versus heterologous antisera in a competitive ELISA. This suggests that minor amino acid variations may affect the antibody repertoire elicited by the ClfA fibrinogen-binding domain in that the affinity of the antibodies generated is higher for the homologous variant than for the heterologous variant. We do not know what effect, if any, lower antibody affinity to heterologous ClfA might have on protection afforded by this antigen in vivo. However, these results provide the cautionary note that if ClfA is included in a vaccine, the potential strain specificity of protection afforded by the ClfA antigen should be considered and evaluated.
Supplementary Material
ACKNOWLEDGMENT
This work was supported by the Intramural Research Program of the Center for Biologics Evaluation and Research, Food and Drug Administration.
Footnotes
Published ahead of print 26 June 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/CVI.00275-13.
REFERENCES
- 1.DeLeo FR, Otto M, Kreiswirth BN, Chambers HF. 2010. Community-associated meticillin-resistant Staphylococcus aureus. Lancet 375:1557–1568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Daum RS, Spellberg B. 2012. Progress toward a Staphylococcus aureus vaccine. Clin. Infect. Dis. 54:560–567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Verkaik NJ, van Wamel WJ, van Belkum A. 2011. Immunotherapeutic approaches against Staphylococcus aureus. Immunotherapy 3:1063–1073 [DOI] [PubMed] [Google Scholar]
- 4.Hu DL, Narita K, Hyodo M, Hayakawa Y, Nakane A, Karaolis DK. 2009. c-di-GMP as a vaccine adjuvant enhances protection against systemic methicillin-resistant Staphylococcus aureus (MRSA) infection. Vaccine 27:4867–4873 [DOI] [PubMed] [Google Scholar]
- 5.Josefsson E, Higgins J, Foster TJ, Tarkowski A. 2008. Fibrinogen binding sites P336 and Y338 of clumping factor A are crucial for Staphylococcus aureus virulence. PLoS One 3:e2206. 10.1371/journal.pone.0002206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Josefsson E, Hartford O, O'Brien L, Patti JM, Foster T. 2001. Protection against experimental Staphylococcus aureus arthritis by vaccination with clumping factor A, a novel virulence determinant. J. Infect. Dis. 184:1572–1580 [DOI] [PubMed] [Google Scholar]
- 7.Arrecubieta C, Matsunaga I, Asai T, Naka Y, Deng MC, Lowy FD. 2008. Vaccination with clumping factor A and fibronectin binding protein A to prevent Staphylococcus aureus infection of an aortic patch in mice. J. Infect. Dis. 198:571–575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gong R, Hu C, Xu H, Guo A, Chen H, Zhang G, Shi L. 2010. Evaluation of clumping factor A binding region A in a subunit vaccine against Staphylococcus aureus-induced mastitis in mice. Clin. Vaccine Immunol. 17:1746–1752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.McDevitt D, Francois P, Vaudaux P, Foster TJ. 1995. Identification of the ligand-binding domain of the surface-located fibrinogen receptor (clumping factor) of Staphylococcus aureus. Mol. Microbiol. 16:895–907 [DOI] [PubMed] [Google Scholar]
- 10.Stranger-Jones YK, Bae T, Schneewind O. 2006. Vaccine assembly from surface proteins of Staphylococcus aureus. Proc. Natl. Acad. Sci. U. S. A. 103:16942–16947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Anderson AS, Scully IL, Timofeyeva Y, Murphy E, McNeil LK, Mininni T, Nunez L, Carriere M, Singer C, Dilts DA, Jansen KU. 2012. Staphylococcus aureus manganese transport protein C is a highly conserved cell surface protein that elicits protective immunity against S. aureus and Staphylococcus epidermidis. J. Infect. Dis. 205:1688–1696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hawkins J, Kodali S, Matsuka YV, McNeil LK, Mininni T, Scully IL, Vernachio JH, Severina E, Girgenti D, Jansen KU, Anderson AS, Donald RG. 2012. A recombinant clumping factor A-containing vaccine induces functional antibodies to Staphylococcus aureus that are not observed after natural exposure. Clin. Vaccine Immunol. 19:1641–1650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hall AE, Domanski PJ, Patel PR, Vernachio JH, Syribeys PJ, Gorovits EL, Johnson MA, Ross JM, Hutchins JT, Patti JM. 2003. Characterization of a protective monoclonal antibody recognizing Staphylococcus aureus MSCRAMM protein clumping factor A. Infect. Immun. 71:6864–6870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tuchscherr LP, Buzzola FR, Alvarez LP, Lee JC, Sordelli DO. 2008. Antibodies to capsular polysaccharide and clumping factor A prevent mastitis and the emergence of unencapsulated and small-colony variants of Staphylococcus aureus in mice. Infect. Immun. 76:5738–5744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Murphy E, Lin SL, Nunez L, Andrew L, Fink PS, Dilts DA, Hoiseth SK, Jansen KU, Anderson AS. 2011. Challenges for the evaluation of Staphylococcus aureus protein based vaccines: monitoring antigenic diversity. Hum. Vaccin. 7(Suppl):51–59 [DOI] [PubMed] [Google Scholar]
- 16.McCarthy AJ, Lindsay JA. 2010. Genetic variation in Staphylococcus aureus surface and immune evasion genes is lineage associated: implications for vaccine design and host-pathogen interactions. BMC Microbiol. 10:173. 10.1186/1471-2180-10-173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Deivanayagam CC, Wann ER, Chen W, Carson M, Rajashankar KR, Hook M, Narayana SV. 2002. A novel variant of the immunoglobulin fold in surface adhesins of Staphylococcus aureus: crystal structure of the fibrinogen-binding MSCRAMM, clumping factor A. EMBO J. 21:6660–6672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ganesh VK, Rivera JJ, Smeds E, Ko YP, Bowden MG, Wann ER, Gurusiddappa S, Fitzgerald JR, Hook M. 2008. A structural model of the Staphylococcus aureus ClfA-fibrinogen interaction opens new avenues for the design of anti-staphylococcal therapeutics. PLoS Pathog. 4:e1000226. 10.1371/journal.ppat.1000226 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


