Abstract
In this study, we assessed in humans the immunogenicity and safety of one dose (7.5 or 15 μg of hemagglutinin [HA]) of a whole-virion inactivated prepandemic influenza vaccine adjuvanted with aluminum hydroxide. The vaccine strain was made by reverse genetics from the highly pathogenic avian A/Chicken/Astana/6/05 (H5N1) clade 2.2 strain isolated from a dead bird in Kazakhstan. The humoral immune response was evaluated after a single vaccination by hemagglutination inhibition (HI) and microneutralization (MN) assays. The vaccine was safe and immunogenic, inducing seroconversion in 55% of the evaluated patients, with a geometric mean titer (GMT) of 17.1 and a geometric mean increase (GMI) of 3.42 after a dose of 7.5 μg in the HI test against the vaccine strain. The rate of seroconversion increased up to 70% when the dose of 15 μg was used. The percentages of individuals achieving anti-HA titers of ≥1:40 were 52.5% and 57.5% for the 7.5- and 15-μg dose groups, respectively. Similar results were obtained when antibodies were analyzed in an MN test. Substantial cross-neutralization titers (seroconversion in 35% and 52.5% of subjects in the two dose groups, respectively) were detected against heterologous clade 1 strain NIBRG14 (H5N1). Thus, one dose of this whole-virion prepandemic vaccine adjuvanted with aluminum has the potential to be effective against H5N1 viruses of different clades.
INTRODUCTION
Since 2003, highly pathogenic avian influenza viruses (HPAIV) of the H5N1 subtype have continued to evolve and to rarely and sporadically infect humans. As of March 2013, the World Health Organization (WHO) reported 622 cases of avian influenza in humans, with 371 fatal outcomes. By sequencing the hemagglutinin (HA), the main influenza virus antigen, all H5N1 viruses were classified into 10 antigenically different clades (1). High mortality in humans, exceeding 60%, and the capability to acquire aerosol transmission in mammals heighten the threat of a global pandemic (2, 3). Vaccination remains the primary strategy for the prevention and control of an influenza pandemic (4, 5). In order to protect the population against a pandemic, the vaccine virus must match the newly emerged strain. In the case of an inactivated vaccine, two vaccinations are likely to be required to provide prime and boost immunizations for effective protection (6). Avian viruses A/Vietnam/1203/04 and A/Vietnam/1194/04 isolated in 2004 from infected humans and classified as H5N1 clade 1 were used for the development of the first human pandemic vaccines. Five inactivated vaccines produced in embryonated chicken eggs have already received marketing authorization from corresponding local regulatory agencies (7). These preparations were shown to meet the immunological criteria of the EU Committee for Medicinal Products for Human Use (CHMP) for pandemic vaccines when two doses supplemented with the adjuvant were administered. The existing manufacturers of seasonal influenza vaccine could provide the pandemic preparation for approximately 18% of the world population (8). Therefore, in the case of a H5N1 pandemic, it is unlikely that a vaccine will be available in time in adequate quantity. The WHO released an action plan to expand the global manufacturing capacity in order to produce the prepandemic vaccine, which can be used at an early pandemic stage, even if it poorly matches the pandemic strain. This decision was based on the observation that one dose of the prepandemic vaccine would mitigate the morbidity and mortality of a pandemic virus (9). In concordance with this plan, the National Authorities of Kazakhstan issued an order to develop the prepandemic vaccine from the domestic influenza H5N1 strain isolated in Kazakhstan. The genetic analysis of viruses circulating in Kazakhstan in 2005 revealed their similarities to the viruses inducing a large outbreak of H5 HPAIV in Qinghai Lake in western China. Qinghai Lake is known as a breeding place for migratory birds on the way to southern Asia, western Russia, and Australia (10). After this outbreak, highly pathogenic H5N1 viruses continued to spread to Europe, Russia, Mongolia, and Kazakhstan. Antigenic characterization of these viruses revealed that they underwent the genetic drift from the viruses of 2003 (clade 1) and were classified as clade 2.2. Virus A/Chicken/Astana/6/05 (H5N1), isolated from a dead bird in Kazakhstan, was selected for the generation of the vaccine strain by reverse genetics according to a procedure developed by the National Institute of Biological Standards and Controls (NIBSC; United Kingdom) (11). The objective of the performed clinical trial was to identify the immunogenicity and safety parameters of the inactivated whole-virus vaccine made from the A/Chicken/Astana/6/05 (H5N1) strain as a potential prepandemic vaccine.
MATERIALS AND METHODS
Cells.
A WHO-certified Vero cell line was obtained from the European Collection of Cell Cultures. After two consecutive passages, cells were adapted to a serum-free Opti-pro medium (Invitrogen). Further cultivation was done in this medium at 37°C and 5% CO2.
Virus generation and vaccine formulation.
The cDNAs of HA and neuraminidase (NA) segments of A/Chicken/Astana/6/05 (H5N1) virus were synthesized (Geneart, Regensburg, Germany) based on the results obtained by the direct sequencing of viral genes (GenBank accession numbers FJ390028 for HA and FJ390029 for NA). The original HA polybasic cleavage site was replaced by the sequence TETR/GLF, which was shown to be genetically more stable for low-pathogenicity avian viruses (12). The remaining segments originated from the A/Puerto Rico/8/34 (PR8) virus. All synthesized segments were cloned into the bidirectional plasmid pHW2006 (analog of pHW2000) containing a polymerase I (Pol I) and Pol II for bidirectional transcription (13). The Vero cells were transfected with plasmids as described by Kittel et al. (14). The rescued virus was propagated in the allantoic cavities of 10-day-old embryonated hen eggs at 37°C for 48 h.
Virus-containing fluid was clarified by low-speed centrifugation, inactivated with 0.04% formalin, and further purified and concentrated by ultrafiltration under tangential flow (up to 1/10 of initial volume). Then, filtration with the use of Sepharose size exclusion chromatography was carried out, and the purified virus was mixed with 0.4% aluminum hydroxide in a 1:1 ratio. This purification process allowed for a rich ovalbumin content of <2 μg/ml (15).
Sequencing.
RNA was extracted using the RNeasy minikit (Qiagen, Germany). The reverse transcription of RNA was performed by using the Reverta-L kit (InterLabService, Russia). The amplification of cDNA was performed by a standard method (16) using the described specific primers. The amplified bands were purified with a Qiaex II gel extraction kit (Qiagen, Germany). The sequencing of influenza virus genome fragments was carried out on an ABI Prism 3100 Avant genetic analyzer (Applied Biosystems) with the BigDye Terminator cycle sequencing kit (v3.1; Applied Biosystems).
Phylogenetic tree generation.
The HA amino acid sequences used for virus comparison were obtained from the GenBank database and have the following HA accession numbers: AAC32099 (A/HongKong/483/1997), AAC40508 (A/HongKong/156/97), ABI84497 (A/duck/Potsdam/1402-6/1986), ACZ48584 (A/Duck/Singapore/F119/3/1997), ABI85106 (A/Chicken/Scotland/1959), ABJ96698 (A/Goose/Guiyang/337/2006), ACO07033 (A/Chicken/Vietnam/NCVD-016/2008), ACO07038 (A/Chicken/Vietnam/NCVD-03/2008), ABW06108 (A/Indonesia/5/2005), ACA47835 (A/Duck/Hunan/795/2002), AAW80717 (A/VietNam/1203/2004), ABP51976 (A/VietNam/1194/2004), ACI06178 (A/Cambodia/R0405050/2007), ABP51975 (A/HongKong/213/2003), ACJ26242 (A/Commonmagpie/HongKong/5052/2007), AEO89181 (A/Hubei/1/2010), ABG67978 (A/Duck/Laos/3295/2006), ADG59080 (A/Anhui/1/2005), ABJ96775 (A/Japanesewhiteeye/HongKong/1038/2006), ABP96850 (A/Egypt/2321-NAMRU3/2007), ADG21447 (A/Egypt/N03072/2010), ABQ45850 (A/Chicken/India/NIV33487/06), ACI87705 (A/Chicken/Astana/6/2005), ABW70133 (A/Domesticgoose/Pavlodar/1/2005), ABD92945 (A/Chicken/Kurgan/05/2005), ABQ58921 (A/Turkey/Turkey/1/2005), and ACZ36881 (A/Whooper-swan/Mongolia/244/2005). The HA sequence of A/Bar-headedgoose/Qinghai/1/2005 virus was obtained from the GISAID database (HA with accession number EPI227594). Multiple alignments were done with the Vector NTI Advanced package (Invitrogen). The software MEGA 5 was used for the phylogenetic analysis (17). The evolutionary history was inferred with the use of the neighbor-joining method (18). Calculations of the evolutionary distances were performed using the Poisson correction method (19).
Clinical study design.
This phase II clinical study was conducted at two centers in Russia from 23 May to 30 June 2011. Before the study was initiated and any study procedures were performed, the study protocol (VRK-I-00-04/2011 version 01 from 29 October 2010) and proposed informed consent form were reviewed and approved by the Federal Service for the Surveillance in Health Care and Social Development of Russian Federation (protocol 15-20-04-2011) and by the Local Ethics Committee (registration number FWA00012786 at the Federal Wide Assurance [FWA] for the Protection of Human Subjects; protocol 45-24-05-2011).
The study was conducted in accordance with good clinical practice (GCP) guidelines and the current version of the Helsinki Declaration. Written informed consents were obtained from all the subjects prior to conducting any study-related procedures.
For each patient, a clinical examination, including medical history review, physical examination, testing for serological evidence of chronic viral infections (with human immunodeficiency virus or hepatitis B or C viruses), electrocardiography, routine biochemical and hematological blood tests, and urinalysis, was conducted. Only clinically healthy subjects were included in the study. Women were obliged to have a negative pregnancy test.
Healthy adult subjects, including 39 men and 41 women with an H5 hemagglutination inhibition (HI) titer of ≤10 and who were seronegative to human immunodeficiency and hepatitis B/C viruses, were selected. The age of the subjects ranged from 19 to 58 years, with a mean ± standard deviation of 35.3 ± 11.4 years. The low-dose group (7.5 μg) included 23 men (mean age, 33.8 ± 12 years) and 17 women (mean age, 35.1 ± 9.8 years), and the high-dose group (15 μg) included 16 men (mean age, 33.1 ± 11 years) and 24 women (mean age, 38.6 ± 12.2 years). The mean weight was 73.9 kg in the 7.5-μg dose group and 70.3 kg in the 15-μg dose group. The mean height of the subjects was 170.3 cm in the low-dose group and 174.8 cm in the high-dose group. All the patients were of the Caucasian race. The study did not include a placebo group.
Each participant was enrolled for approximately 5 weeks (14 screening days and 21 ± 2 study days). Subjects were randomly assigned to receive either 7.5 or 15 μg of H5N1 influenza vaccine. Assignments were enclosed in identical, numbered, opaque, and sealed envelopes. There were no differences in the distribution of baseline demographic characteristics between the two study vaccine groups. The vaccine dose (0.5-ml volume) was administered by deep intramuscular injection with a 1-inch needle.
The subjects were observed for immediate local and systemic reactions for 20 min and at 2 h after the injection. The symptoms observed over the next 7 days were recorded by volunteers in self-completed diaries. Solicited local (pain, swelling, bruising, induration, and erythema) and systemic (chills, malaise, muscle aches, nausea, and headache) adverse events (AEs) were assessed according to a three-point grading system. All AEs were recorded in the volunteer's medical record and in the case report form (CRF). The onset and end dates, severity, consequence, and relationship to study medication were recorded for each AE. All AEs, including the clinical laboratory test results, were assessed by a study clinician to quantify the severity, based on a protocol-defined grading system, as follows: 0, none, lack of a sign or symptom; 1, mild, events requiring minimal or no treatment and that did not interfere with the subject's daily activities; 2, moderate, events that resulted in a low level of inconvenience or concern to the subject with therapeutic measures or might have caused some interference with functioning; 3, severe, the event interrupted the subject's functioning and may have required systemic drug therapy or other treatment (usually incapacitating).
Injection site erythema, swelling, and induration were graded according to the measured diameter: 0, absent/none; 1, mild, erythema, swelling of <50 mm or induration of <25 mm; 2, moderate, erythema, swelling of >50 mm or induration of 26 to 50 mm; 3, severe, erythema, swelling of >50 mm and induration of >50 mm.
Furthermore, the daily axillary temperature and any unsolicited AEs, including serious AEs, were recorded throughout the duration of the study.
Serum samples were collected on the day of immunization (day zero) and 21 days postimmunization. Serum samples with antibody titers of <1:10 were assigned values of 5 for the purposes of mean titer calculations.
Immunogenicity evaluations.
Sera were analyzed for the presence of antibodies by HI and microneutralization (MN) assays according to methods described elsewhere (20) with some modifications. For the HI assay, sera diluted 1:4 with a receptor-destroying enzyme (RDE; Denka, Japan) were incubated at 37°C overnight. Then, sera were inactivated at 56°C for 30 min. An HI assay was performed with 1% horse erythrocytes.
For the detection of neutralizing antibody titers, serial 2-fold dilutions of heated sera (range, 1/10 to 1/1,280) were made in 60 μl of diluent (Dulbecco's modified Eagle's medium containing 100 U/ml penicillin, 100 μg/ml streptomycin, 2 μg/ml tosylsulfonyl phenylalanyl chloromethyl keton-treated trypsin) in round-bottom immunoassay plates. An equal volume of virus containing 100 50% tissue culture infectious doses/50 μl was added to serum samples. The plates were incubated for 2 h at 37°C and 5% CO2. Then, 100 μl of virus-serum mixture and 50 μl of diluent were transferred to wells with an 80 to 90% confluent monolayer of MDCK cells (Nunc plates, Denmark) previously washed twice with phosphate-buffered saline (PBS).
Control samples were included on each plate: control cells (CC) were distributed in 4 wells with diluent but without virus or serum, and virus dose control cells (CV) were distributed in 4 wells with virus.
The plates were incubated for 48 h at 37°C and 5% CO2. Postincubation, supernatant was removed and cells were fixed with 80% acetone for 15 min at −4°C. The presence of viral nucleoprotein (NP) in the cells was detected by enzyme-linked immunosorbent assay with peroxidase-labeled monoclonal antibody 4H1 (MAbs-Px; kindly provided by A. A. Sominina), which recognizes the NP of avian and human influenza viruses. The plates with fixed cells were washed two times with PBS. Then, 100 μl of MAbs-Px diluted in PBS containing 5% skim milk was added to each well. The plates were incubated at room temperature for 1 h and then washed four times with PBS. A 100-μl aliquot of substrate mixture consisting of 0.02% H2O2 and 0.1 mg/ml 3,3′,5,5′-tetramethylbenzidine (Sigma) in 0.1 M acetate-citrate buffer (pH 5.0) was added to each well. The reaction was stopped after 15 min by the addition of 50 μl 2 N H2SO4 to each well. The optical density at 450 nm (OD450) was measured by using an Anthos-2010 photometer (Austria). The neutralizing antibody endpoint titer was the last dilution with an OD450 below the threshold value (TV), calculated using the following formula: TV = [(OD450 of CV) − (OD450 of CC)]/2 + (OD450 of CC), where the OD450 CV was the average OD in control wells containing virus working dose and OD450 CC was the average OD in control wells with uninfected cells.
Statistics.
For both serum HI and MN antibody titers, the geometric mean titers (GMTs) of duplicate assays done in parallel in one test were calculated with 95% confidence intervals (CI). Comparisons between groups receiving 7.5 μg and 15 μg of HA were determined as the proportion of subjects with detected seroconversion or subjects with protective antibody titers (seroprotection) in the HI and MN assays by using Fisher's exact test.
RESULTS
Generation of H5N1 vaccine candidate.
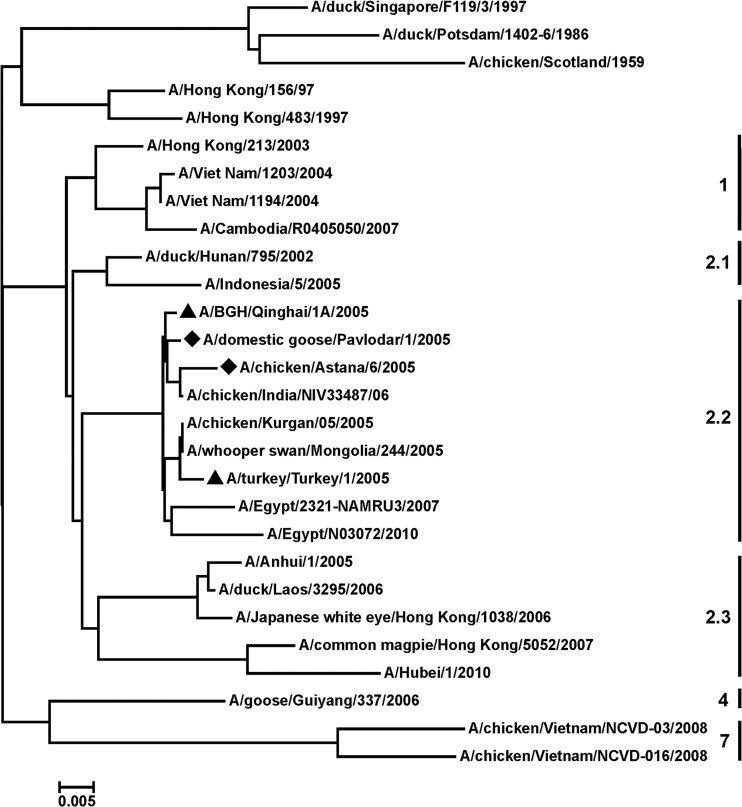
Two HPAI H5N1 viruses, A/Chicken/Astana/6/05 and A/Domestic goose/Pavlodar/1/05, were isolated from dead birds in Kazakhstan in 2005. The HA gene segments were sequenced, and the predicted amino acid sequences were subjected to phylogenetic analysis by using the neighbor-joining method. As shown in Fig. 1, the HAs of the studied viruses showed genetic relatedness to that of the H5N1 HPAIV circulating in Europe, Russia, and the Middle East since 2005 and belonging to clade 2.2, according to the unified H5N1 virus classification (98 to 99% sequence identity). Viruses A/BGH/Qinghai/1A/05 and A/Turkey/Turkey/1/05 of this clade 2.2 were recommended by the WHO for the generation of vaccine candidates and production of a prototype pandemic influenza vaccine. The HA and NA sequences of the selected virus in Kazakhstan A/Chicken/Astana/6/05 were used for the construction of the vaccine strain by use of reverse genetics. The remaining 6 segments originated from the PR8 strain, which is highly adapted to growth in embryonated chicken eggs. Vero cells were transfected with eight plasmids, and rescued reassortant virus was named A/Astana/6/05RG. All subsequent passages, including for vaccine bulk production, were performed solely in embryonated hen eggs. Virus A/Astana/6/05RG grew in embryonated hen eggs to an infectious titer of 8.5 to 9.0 log10 50% infectious dose/ml and a 1:512 HA titer. The vaccine was produced and characterized by the Research Institute for Biological Safety Problems (RIBSP; Kazakhstan) (15).
Fig 1.
Phylogenetic tree of the HA amino acid sequences of H5N1 viruses. The HA amino acid sequences of H5 viruses published in GenBank and GISAID databases were subjected to phylogenetic analysis by the neighbor-joining method. ▲, virus classified as clade 2.2; ◆, virus isolated in Kazakhstan. The virus clade is indicated on the right side. The phylogenetic tree was constructed with the use of Vector NTI and the MEGA 5 software package.
Vaccine safety evaluation.
A total of 80 subjects with the age range of 19 to 58 years were enrolled. Forty subjects were vaccinated with the 7.5-μg dose and 40 received the 15-μg dose. Clinical laboratory evaluations and physical examinations did not reveal any significant abnormalities or safety concerns for the study medication. The AE profile was comparable for both vaccine dose groups (Table 1). No serious AEs and no unexpected events were reported. Adverse reactions, which were observed for 25 of 80 (31%) volunteers, were mild and largely local (pain at the site of injection) and lasted no more than 2 days. No erythema or infiltrates were registered. Swelling for 1 week (of grade 1) was observed in the 15-μg group. One systemic reaction (rhinitis and sore throat) was noticed in the 7.5-μg vaccine group, and two reactions (headache and sore throat) were recorded in the 15-μg dose group.
Table 1.
Local and systemic reactions after the vaccination
| Reaction | No. of responders in dose group |
|
|---|---|---|
| 7.5 μg | 15 μg | |
| Total no. of subjects | 40 | 40 |
| No. with any AE | 14 | 19 |
| Local reactions | ||
| Pain | 8 | 10 |
| Tenderness | 4 | 3 |
| Swelling | 0 | 1 |
| Induration | 0 | 0 |
| Erythema | 0 | 0 |
| Infiltrate | 0 | 0 |
| Systemic reactions | ||
| Headache | 0 | 1 |
| Sore throat | 1 | 2 |
| Rhinitis | 1 | 1 |
| Malaise | 0 | 1 |
Vaccine immunogenicity evaluation.
In a preliminary phase I study in adults, we found that the vaccine induced seroconversion and seroprotection in 90.9% of the vaccinated individuals in response to a 2-dose schedule with the 15-μg vaccine dose (O. S. Konshina et al., unpublished data). Therefore, in the phase II study, we were interested in determining whether a one-dose regimen with 7.5 or 15 μg of the vaccine would be immunogenic enough.
Antibody responses were measured based on serum HI antibody titers and MN antibody titers against the vaccine virus A/Astana/6/05RG. The results are presented in Table 2. The vaccine dose of 7.5 μg induced seroconversion in 55% of subjects in the HI test, with a GMT of 17.1 and geometric mean increase (GMI) of 3.42. The rate of seroconversion increased to 70% when the dose of 15 μg was used. The GMT and GMI for the higher dose group reached 23.38 and 4.68, respectively. The percentages of individuals with HI titers of ≥1:40 were 52.5% and 57.5% for the 7.5- and 15-μg dose groups, respectively. Similar results were obtained when antibodies were analyzed in an MN test, showing seroconversion in 55% of subjects in the 7.5-μg dose group and in 67.5% in the 15-μg group. Statistical analysis with Fisher's exact test revealed no significant differences between the dose groups in the percentages of seroconversion (P = 0.25 for HI and P = 0.36 for MN) or seroprotection (P = 0.82 for HI and P = 0.26 for MN). Substantial immunogenicity (seroconversion in 35% and 52.5% of subjects in the low- and high-dose groups, respectively) was demonstrated in the MN test against clade 1 strain NIBRG14 (reassortant of A/Vietnam/1194/04 H5N1 and PR8) (Table 3), although the cross-neutralization titers measured against a heterologous strain were lower than those against homologous virus.
Table 2.
HI and MN antibody responses against the homologous A/Astana/6/05RG strain
| Dose level (n) | Response to vaccine based on: |
|||||
|---|---|---|---|---|---|---|
| Seroconversiona |
Geometric mean increaseb |
Seroprotectionc |
||||
| HI | MN | HI | MN | HI | MN | |
| 7.5 μg (40) | 22 (55.0; 38.5–70.7) | 22 (55.0; 38.5–70.7) | 3.5 (2.4–5.2) | 4.37 (2.88–6.6) | 21 (52.5; 36.1–68.5) | 16 (40.0; 24.9–56.7) |
| 15 μg (40) | 28 (70.0; 53.5–83.4) | 27 (67.5; 50.9–81.4) | 4.7 (3.3–6.7) | 7.46 (4.79–11.6) | 23 (57.5; 40.9–73) | 22 (55.0; 38.5–70.7) |
Seroconversion was defined as a ≥4-fold rise in antibody titer compared to the day zero titer. Data are the number of responders, with the percentage and 95% CI provided in parentheses.
The geometric mean increase (with 95% CI in parentheses) reflects the increase in the GMT.
Seroprotection is the number of volunteers whose titer was ≥1:40 (with the percentage of responders and the 95% CI shown in parentheses).
Table 3.
MN antibody response against heterologous strain NIBRG14
| HA dose | No. of subjects | MN antibody response based on: |
||
|---|---|---|---|---|
| Seroconversiona | Geometric mean increaseb | Seroprotectionc | ||
| 7.5 μg | 40 | 14 (35; 22.1–50.5) | 2.8 (1.9–4.2) | 11 (27.5; 14.6–43.9) |
| 15 μg | 40 | 21 (52.5; (37.5–67.1) | 4.1 (2.8–6.3) | 16 (40; 24.9–56.7) |
Seroconversion was defined as a ≥4-fold rise in antibody titer compared to the day zero titer. Data are the number of responders, with the percentage and 95% CI provided in parentheses.
The geometric mean increase (with 95% CI in parentheses) reflects the increase in the GMT.
Seroprotection is the number of volunteers whose titer was ≥1:40 (with the percentage of responders and the 95% CI shown in parentheses).
DISCUSSION
We have described herein the phase II clinical trial results of an inactivated whole-virion vaccine made from the Kazakhstan domestic influenza strain A/Chicken/Astana/6/05 of H5N1 subtype clade 2.2. The vaccine, formulated with an aluminum hydroxide adjuvant, demonstrated good safety and tolerability in terms of local and systemic reactions and appeared to be immunogenic. The single immunization of adults with 7.5 μg of HA antigen resulted in seroconversion in 55% of the evaluated patients and a GMI of 3.42. In addition, cross-reactive neutralizing antibodies against the clade 1 virus NIBRG-14 were demonstrated. These parameters exceeded two licensing criteria of the CHMP for pandemic influenza vaccine. With respect to seroprotection, lower rates (52.5% for the 7.5-μg group and 57.5% for the 15-μg group) were obtained, compared to those indicated in the CHMP guidelines.
Numerous clinical trials performed with different formulations of inactivated H5N1 vaccine have shown relatively low rates of seroconversion and seroprotection, especially when antibodies were measured by HI assay. The enhanced dose of HA antigen and a two-dose vaccination regimen heightened the vaccine's immunogenicity to a level consistent with the CHMP criteria for an interpandemic influenza vaccine (21–23).
The addition of an adjuvant to the H5N1 vaccine enabled the reduction of the HA dose in order to elicit an adequate immune response. Thus, the best-known prepandemic vaccines today (split formulations) have demonstrated immunogenicity rates of ≥70% when administered two times with an oil-in-water adjuvant (MF59 or ASO3) at a dose of 3.8 to 7.5 μg of HA antigen (24, 25). However, these adjuvants are prone to induce adverse events, such as site pain, muscle aches, headache, and fatigue.
The whole-virion influenza vaccines are less safe but potentially more immunogenic than split or subvirion formulations. The stimulation of innate immunity by viral RNA via Toll-like receptor 7 is the most likely reason for this effect (26). Supplementation of the whole-virion influenza vaccine with aluminum adjuvant enabled the enhancement of the immunogenicity to the CHMP-required level after just a single immunization with some preparations. Thus, one whole-virion vaccine made from clade 1 NIBRG-14 virus fulfilled all three CHMP criteria (seroconversion in 90.9% and seroprotection in 79.5% of the evaluated patients) after a single dose of 6 μg HA administered with aluminum phosphate (27, 28). However, the same NIBRG-14 strain supplemented with aluminum hydroxide induced seroconversion and seroprotection in only 39% of patients when it was administered once at a dose of 10 μg in another study (29). A similar level of immunogenicity (seroconversion and seroprotection in 35.7% of the evaluated patients) was achieved with a whole-virion Vero cell-derived vaccine made from the clade 1 wild-type A/Vietnam/1203/04 H5N1 strain administered with aluminum adjuvant once at a dose of 7.5 μg (30).
Therefore, the data regarding the sufficiency of a single dose for the prepandemic H5N1 vaccine are not conclusive, and more comparative studies are needed. The immunogenicity of the clade 2.2 A/Astana/6/05RG aluminum-adjuvanted vaccine studied here is higher than that shown for one dose of whole-virion inactivated vaccines in the previous, above-mentioned studies (29, 30). High immunogenicity rates and demonstrated cross-reactive immunity enable us to conclude that this vaccine is capable of priming the immune system against the vaccine virus and has the potential to be effective against H5N1 viruses of different clades.
ACKNOWLEDGMENTS
This study was funded by the Research Institute for Biological Safety Problems (RIBSP), Gvardeiskiy, Kazakhstan.
We have no conflicts of interest.
Footnotes
Published ahead of print 26 June 2013
REFERENCES
- 1. Obenauer JC, Denson J, Mehta PK, Su X, Mukatira S, Finkelstein DB, Xu X, Wang J, Ma J, Fan Y, Rakestraw KM, Webster RG, Hoffmann E, Krauss S, Zheng J, Zhang Z, Naeve CW. 2006. Large-scale sequence analysis of avian influenza isolates. Science 311:1576–1580 [DOI] [PubMed] [Google Scholar]
- 2. Palese P, Wang TT. 2012. H5N1 influenza viruses: facts, not fear. Proc. Natl. Acad. Sci. U. S. A. 109:2211–2213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Imai M, Watanabe T, Hatta M, Das SC, Ozawa M, Shinya K, Zhong G, Hanson A, Katsura H, Watanabe S, Li C, Kawakami E, Yamada S, Kiso M, Suzuki Y, Maher EA, Neumann G, Kawaoka Y. 2012. Experimental adaptation of an influenza H5 HA confers respiratory droplet transmission to a reassortant H5 HA/H1N1 virus in ferrets. Nature 486:420–428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Nichol KL, Treanor JJ. 2006. Vaccines for seasonal and pandemic influenza. J. Infect. Dis. 194(Suppl 2):S111–S118 [DOI] [PubMed] [Google Scholar]
- 5. Monto AS. 2006. Vaccines and antiviral drugs in pandemic preparedness. Emerg. Infect. Dis. 12:55–60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Schwartz B, Wortley P. 2006. Mass vaccination for annual and pandemic influenza. Curr. Top. Microbiol. Immunol. 304:131–152 [DOI] [PubMed] [Google Scholar]
- 7. Prieto-Lara E, Llanos-Mendez A. 2010. Safety and immunogenicity of prepandemic H5N1 influenza vaccines: a systematic review of the literature. Vaccine 28:4328–4334 [DOI] [PubMed] [Google Scholar]
- 8. Jennings LC, Monto AS, Chan PK, Szucs TD, Nicholson KG. 2008. Stockpiling prepandemic influenza vaccines: a new cornerstone of pandemic preparedness plans. Lancet Infect. Dis. 8:650–658 [DOI] [PubMed] [Google Scholar]
- 9. Germann TC, Kadau K, Longini IM, Jr, Macken CA. 2006. Mitigation strategies for pandemic influenza in the United States. Proc. Natl. Acad. Sci. U. S. A. 103:5935–5940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Chen H, Li Y, Li Z, Shi J, Shinya K, Deng G, Qi Q, Tian G, Fan S, Zhao H, Sun Y, Kawaoka Y. 2006. Properties and dissemination of H5N1 viruses isolated during an influenza outbreak in migratory waterfowl in western China. J. Virol. 80:5976–5983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Robertson JS, Engelhardt OG. 2010. Developing vaccines to combat pandemic influenza. Viruses 2:532–546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Horimoto T, Takada A, Fujii K, Goto H, Hatta M, Watanabe S, Iwatsuki-Horimoto K, Ito M, Tagawa-Sakai Y, Yamada S, Ito H, Ito T, Imai M, Itamura S, Odagiri T, Tashiro M, Lim W, Guan Y, Peiris M, Kawaoka Y. 2006. The development and characterization of H5 influenza virus vaccines derived from a 2003 human isolate. Vaccine 24:3669–3676 [DOI] [PubMed] [Google Scholar]
- 13. Hoffmann E, Neumann G, Kawaoka Y, Hobom G, Webster RG. 2000. A DNA transfection system for generation of influenza A virus from eight plasmids. Proc. Natl. Acad. Sci. U. S. A. 97:6108–6113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Kittel C, Sereinig S, Ferko B, Stasakova J, Romanova J, Wolkerstorfer A, Katinger H, Egorov A. 2004. Rescue of influenza virus expressing GFP from the NS1 reading frame. Virology 324:67–73 [DOI] [PubMed] [Google Scholar]
- 15. Mamadaliyev MM, Sandybayev NT, Kydyrbayev ZK, Khairullin BM, Zaitsev VL, Mambetaliyev Kassenov MMM, Chervyakova OV, Ryskeldinova SZ, Volgin YN, Nurpeissova AS, Bogdanov NV, Sarsenbayeva GZ, Kisselyov OI, Tsybalova LM, Grudinin MP, Migunov AI, Stukova MA. 2011. Basic results of development of a production technology and control of a pandemic influenza A/H5N1 vaccine. Influenza Other Respi. Viruses 5(Suppl 1):1 [Google Scholar]
- 16. Hoffmann E, Stech J, Guan Y, Webster RG, Perez DR. 2001. Universal primer set for the full-length amplification of all influenza A viruses. Arch. Virol. 146:2275–2289 [DOI] [PubMed] [Google Scholar]
- 17. Tamura K, Dudley J, Nei M, Kumar S. 2007. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol. Biol. Evol. 24:1596–1599 [DOI] [PubMed] [Google Scholar]
- 18. Saitou N, Nei M. 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4:406–425 [DOI] [PubMed] [Google Scholar]
- 19. Zuckerkandl E, Pauling L. 1965. Molecules as documents of evolutionary history. J. Theor. Biol. 8:357–366 [DOI] [PubMed] [Google Scholar]
- 20. Rowe T, Abernathy RA, Hu-Primmer J, Thompson WW, Lu X, Lim W, Fukuda K, Cox NJ, Katz JM. 1999. Detection of antibody to avian influenza A (H5N1) virus in human serum by using a combination of serologic assays. J. Clin. Microbiol. 37:937–943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Nicholson KG, Colegate AE, Podda A, Stephenson I, Wood J, Ypma E, Zambon MC. 2001. Safety and antigenicity of non-adjuvanted and MF59-adjuvanted influenza A/Duck/Singapore/97 (H5N3) vaccine: a randomised trial of two potential vaccines against H5N1 influenza. Lancet 357:1937–1943 [DOI] [PubMed] [Google Scholar]
- 22. Treanor JJ, Campbell JD, Zangwill KM, Rowe T, Wolff M. 2006. Safety and immunogenicity of an inactivated subvirion influenza A (H5N1) vaccine. N. Engl. J. Med. 354:1343–1351 [DOI] [PubMed] [Google Scholar]
- 23. Treanor JJ, Wilkinson BE, Masseoud F, Hu-Primmer J, Battaglia R, O'Brien D, Wolff M, Rabinovich G, Blackwelder W, Katz JM. 2001. Safety and immunogenicity of a recombinant hemagglutinin vaccine for H5 influenza in humans. Vaccine 19:1732–1737 [DOI] [PubMed] [Google Scholar]
- 24. Manzoli L, Schioppa F, Boccia A, Villari P. 2007. The efficacy of influenza vaccine for healthy children: a meta-analysis evaluating potential sources of variation in efficacy estimates, including study quality. Pediatr. Infect. Dis. J. 26:97–106 [DOI] [PubMed] [Google Scholar]
- 25. Langley JM, Risi G, Caldwell M, Gilderman L, Berwald B, Fogarty C, Poling T, Riff D, Baron M, Frenette L, Sheldon E, Collins H, Shepard M, Dionne M, Brune D, Ferguson L, Vaughn D, Li P, Fries L. 2011. Dose-sparing H5N1 A/Indonesia/05/2005 pre-pandemic influenza vaccine in adults and elderly adults: a phase III, placebo-controlled, randomized study. J. Infect. Dis. 203:1729–1738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Geeraedts F, Goutagny N, Hornung V, Severa M, de Haan A, Pool J, Wilschut J, Fitzgerald KA, Huckriede A. 2008. Superior immunogenicity of inactivated whole virus H5N1 influenza vaccine is primarily controlled by Toll-like receptor signalling. PLoS Pathog. 4(8):e1000138. 10.1371/journal.ppat.1000138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Fazekas G, Martosne-Mendi R, Jankovics I, Szilvasy I, Vajo Z. 2009. Cross-reactive immunity to clade 2 strains of influenza virus A subtype H5N1 induced in adults and elderly patients by Fluval, a prototype pandemic influenza virus vaccine derived by reverse genetics, formulated with a phosphate adjuvant, and directed to clade 1 strains. Clin. Vaccine Immunol. 16:437–443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Vajo Z, Wood J, Kosa L, Szilvasy I, Paragh G, Pauliny Z, Bartha K, Visontay I, Kis A, Jankovics I. 2010. A single-dose influenza A (H5N1) vaccine safe and immunogenic in adult and elderly patients: an approach to pandemic vaccine development. J. Virol. 84:1237–1242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Lin J, Zhang J, Dong X, Fang H, Chen J, Su N, Gao Q, Zhang Z, Liu Y, Wang Z, Yang M, Sun R, Li C, Lin S, Ji M, Wang X, Wood J, Feng Z, Wang Y, Yin W. 2006. Safety and immunogenicity of an inactivated adjuvanted whole-virion influenza A (H5N1) vaccine: a phase I randomised controlled trial. Lancet 368:991–997 [DOI] [PubMed] [Google Scholar]
- 30. Ehrlich HJ, Muller M, Oh HM, Tambyah PA, Joukhadar C, Montomoli E, Fisher D, Berezuk G, Fritsch S, Low-Baselli A, Vartian N, Bobrovsky R, Pavlova BG, Pollabauer EM, Kistner O, Barrett PN. 2008. A clinical trial of a whole-virus H5N1 vaccine derived from cell culture. N. Engl. J. Med. 358:2573–2584 [DOI] [PubMed] [Google Scholar]