Abstract
Leptospira immunoglobulin (Ig)-like (Lig) proteins are a novel family of surface-associated proteins in which the N-terminal 630 amino acids are conserved. In this study, we truncated the LigA conserved region into 7 fragments comprising the 1st to 3rd (LigACon1-3), 4th to 7.5th (LigACon4-7.5), 4th (LigACon4), 4.5th to 5.5th (LigACon4.5–5.5), 5.5th to 6.5th (LigACon5.5–6.5), 4th to 5th (LigACon4-5), and 6th to 7.5th (LigACon6-7.5) repeat domains. All 7 recombinant Lig proteins were screened using a slot-shaped dot blot assay for the diagnosis of equine leptospirosis. Our results showed that LigACon4-7.5 is the best candidate diagnostic antigen in a slot-shaped dot blot assay. LigACon4-7.5 was further evaluated as an indirect enzyme-linked immunosorbent assay (ELISA) antigen for the detection of Leptospira antibodies in equine sera. This assay was evaluated with equine sera (n = 60) that were microscopic agglutination test (MAT) negative and sera (n = 220) that were MAT positive to the 5 serovars that most commonly cause equine leptospirosis. The indirect ELISA results showed that at a single serum dilution of 1:250, the sensitivity and specificity of ELISA were 80.0% and 87.2%, respectively, compared to those of MAT. In conclusion, an indirect ELISA was developed utilizing a recombinant LigA fragment comprising the 4th to 7.5th repeat domain (LigACon4-7.5) as a diagnostic antigen for equine leptospirosis. This ELISA was found to be sensitive and specific, and it yielded results that concurred with those of the standard MAT.
INTRODUCTION
Leptospirosis is a worldwide bacterial disease caused by spirochetes of the genus Leptospira.It has been identified as one of the emerging infectious diseases in the United States (1). The disease affects humans, domestic animals, and wildlife, including reptiles and amphibians (1). In horses the disease has most often been associated with abortion and equine recurrent uveitis; sporadic cases of renal and hepatic disease have also been reported (2–4). More recently, Broux et al. reported acute respiratory failure caused by Leptospira spp. in 5 foals (5). Some studies have shown that horses were more likely to be seropositive than other domestic animal species (6); however, clinical signs are nonspecific, which is a major obstacle for the clinical diagnosis of equine leptospirosis (7). Establishing a definitive diagnosis of equine leptospirosis can be difficult. Due to the fastidious and slow-growing nature of Leptospira and difficulty in observing the organism in body fluid, diagnosis of leptospirosis often relies on serology, which can be difficult to interpret due to the high seroprevalence in the equine population (8). Currently, the standard reference method for serologic diagnosis of leptospirosis is the microscopic agglutination test (MAT), in which sera are reacted with live antigen suspensions of various Leptospira serovars (7). However, MAT can be a challenging assay to implement because it requires considerable expertise to perform and interpret and necessitates the continual maintenance of a panel of live strains of all serogroups plus locally isolated serovars, which is technically demanding and biohazardous. Therefore, MAT is usually restricted to reference laboratories (9). The current interpretive criteria indicative of active infection for the Leptospira MAT require a 4-fold rise in titer between the acute- and convalescent-phase sera (7). Although it is well recognized that seroconversion or increasing antibody titers in paired serum specimens provide strong evidence for true infection, the samples need to be taken 2 to 3 weeks apart in order to see changes in titer (7), which is not practical in the clinical setting. The complexities associated with MAT highlight the need to develop a simple and rapid screening test to detect Leptospira antibodies. Previously, several research groups have attempted to utilize different recombinant proteins from Leptospira spp. as antigens in an indirect enzyme-linked immunosorbent assay (ELISA) as a serodiagnostic test (10–19). However, as far as we know, no one has applied them to equine leptospirosis diagnosis. Leptospira immunoglobulin (Ig)-like (Lig) proteins are a novel family of surface-associated proteins that bind to extracellular matrices (20–25). Further, Lig proteins are expressed exclusively by pathogenic and not saprophytic Leptospira species (26, 27). Lig proteins are expressed during host infection and induce strong antibody responses in infected animals (27). In the present research, in order to increase the sensitivity and specificity of the serologic test, we truncated and expressed the conserved region of the Lig protein into 7 fragments comprising the 1st to 3rd (LigACon1-3), 4th to 7.5th (LigACon4-7.5), 4th (LigACon4), 4.5th to 5.5th (LigACon4.5-5.5), 5.5th to 6.5th (LigACon5.5-6.5), 4th to 5th (LigACon4-5), and 6th to 7.5th (LigACon6-7.5) bacterial immunoglobulin-like (Big) repeat domains of LigA from the L. interrogans serovar Pomona (Fig. 1). We compared the antigenicity of these recombinant proteins and evaluated their diagnostic potential in equine leptospirosis with an indirect ELISA using one of these recombinant proteins.
Fig 1.
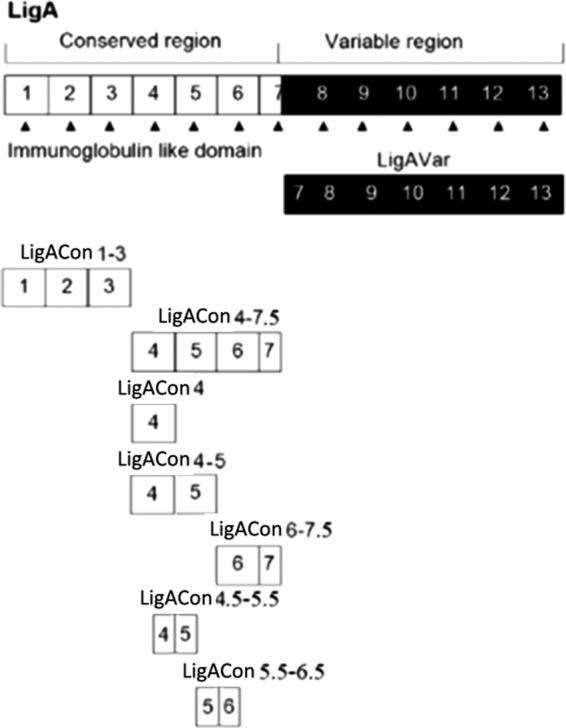
A schematic diagram showing the structure of LigA proteins and the truncated LigA proteins used in this study.
MATERIALS AND METHODS
Bacterial strain.
The L. interrogans serovar Pomona (NVSL 1427-35-093002) was used for this study (28). Leptospira isolates were maintained on Ellinghausen-McCullough-Johnson-Harris (EMJH) medium at 30°C. Growth of the Leptospira isolates was monitored using dark-field microscopy.
Sera.
All equine sera were collected from 2005 to 2009 by the New York State Animal Health Diagnostic Center (AHDC), Cornell University, Ithaca, NY. These serum samples were either positive or negative in MAT to the most common serovars causing equine leptospirosis, including L. interrogans serovar Pomona, L. kirschneri serovar Grippotyphosa, L. interrogans serovar Icterohaemorrhagiae, and L. interrogans serovar Bratislava.
Cloning, expression, and purification of proteins.
Primers for the gene segments encoding these truncated proteins are listed in Table 1. The underlined nucleotides indicate the restriction sites added to facilitate cloning. Genomic DNA from L. interrogans serovar Pomona was extracted using the QIAmp DNA blood minikit (Qiagen, Valencia, CA), following the manufacturer's instructions. The target genes were amplified using PCR with AccuPrime Taq polymerase (Invitrogen, CA), and the other reagents were added as outlined in the manufacturer's instructions (Invitrogen, CA). The amplified PCR products were cloned into the pCR2.1 vector (Invitrogen, CA) and confirmed by DNA sequencing with an automated 3730 DNA analyzer (Applied Biosystems, CA). After sequence confirmation, all the genes were subcloned into pGEX-4T-2 (GE, NJ). The expression and purification of glutathione S-transferase (GST) fusion protein were carried out as previously described (26). The GST was cut with thrombin (20 U/ml in phosphate-buffered saline [PBS] [pH 7.3]), while fusion protein was bound to the column by incubating at room temperature for 12 h. Following incubation, the eluate was collected, subjected to SDS-PAGE to check the purity, and stored at −80°C until used. The concentration of purified protein was then determined using the Bradford method and was used for ELISA.
Table 1.
Primers used to amplify and clone L. interrogans serovar-specific sequences
| Recombinant LigA fragment | Primer sequencea |
|---|---|
| LigACon1-3 | CGGTCGACTGGTAACTCTAATCCG (F) |
| CGGCGGCCGCAATAGAAACTAAGGC (R) | |
| LigACon4-7.5 | CGGTCGACTATCGTTACTCCAGCA (F) |
| CGGCGGCCGCAATATCCGTATTAGA (R) | |
| LigACon6-7.5 | CGGCATGCACTGTAGTTCCTGCG (F) |
| CGGTCGACAATATCCGTATTAGA (R) | |
| LigACon4.5–5.5 | CCGGAATTCTCTTCTAATACCGATATTCT (F) |
| AATCTCGAGATTCCAAGTGACTTGATCCGTAATATCCT (R) | |
| LigACon4-5 | CGGCATGCATCGTTACTCCAGCA (F) |
| CGGTCGACGAGAACCGCAGGAAC (R) | |
| LigACon5.5–6.5 | CCGGAATTCAATTCTTCTTCAGCAATCGT (F) |
| AATCTCGAGTGAGAACCACGTAACAGCGGAAGTGATGT (R) | |
| LigACon4 | CGGCATGCATCGTTACTCCAGCA (F) |
| CGGTCGACTAATACCTCTTGTGT (R) |
Underlined nucleotides indicate the restriction site added to facilitate cloning. F, forward; R, reverse.
MAT.
MAT was used as the reference method to determine the serum titers using live L. interrogans as the antigen as previously described (29). The 5 serovars that most commonly cause equine leptospirosis were tested, including L. interrogans serovar Pomona, L. interrogans serovar Hardjo, L. interrogans serovar Grippotyphosa, L. interrogans serovar Icterohaemorrhagiae/Copenhageni, and L. interrogans serovar Canicola. Briefly, serial 2-fold dilutions of the sera, starting with a dilution of 1:10, were mixed with an equal volume of viable Leptospira strains in a 96-well microtiter plate. After incubation at 30°C for 2 h, the samples were examined for agglutination by dark-field microscopy. Titers represented the highest serum dilution showing 50% agglutination of the Leptospira cells in the suspension.
Slot-shaped dot blot assay.
A total of 600 ng of purified protein in 200 μl PBS was transferred onto nitrocellulose membranes (Schleicher & Schuell Biosciences, Inc.) by using a Bio-Dot slot format (SF) microfiltration apparatus (Bio-Rad) according to the manufacturer's instructions. After washing with Tris-buffered saline (TBS), the membranes were removed and soaked in the blocking solution, Tris-buffered saline (20 mM Tris-HCl, 500 mM NaCl [pH 7.5]) containing 5% (wt/vol) nonfat powdered milk for 1 h at 37°C on a shaker to saturate the remaining protein-binding sites. The blocked membranes were then washed three times (15 min each) with Tris-buffered saline containing Tween (TTBS) and probed with MAT-positive equine serum at 4°C overnight. After washing, the membranes were transferred to the alkaline phosphatase-labeled goat anti-horse IgG (KPL, Inc., MD) solution and incubated for 1 h at 37°C. After this, the membranes were incubated in freshly prepared 5-bromo-4-chloro-3-indolylphosphate-nitroblue tetrazolium (BCIP-NBT) color development solution (Invitrogen) for 10 to 30 min and then rinsed in deionized water to stop the reaction.
Quantitative slot blot assay for LigA fragment interaction with equine anti-Leptospira-positive sera.
To determine the intensity of the interaction between the LigA fragments and equine anti-Leptospira antibodies, we used a quantitative slot assay as previously described (30). The membrane to be analyzed was scanned immediately following the blotting procedure. The scanned image of the membrane was analyzed by Phoretix 1D (TotalLab Ltd., England) image processing and analysis software, which can calculate peak height and area for each slot on the membrane. A line graph for each lane was plotted with pixel intensity units on the y axis and pixel location (position from left to right of the membrane in each lane) on the x axis. Thus, each peak in the graphs represented a slot containing detected recombinant protein interaction with antibodies.
ELISA.
Purified proteins were diluted in coating buffer (0.05 M NaHCO3-Na2CO3 buffer [pH 9.6]) at optimum concentration established by checkerboard titration. A total of 100 μl of the diluted antigen was used to coat 96-well microtiter plates (Corning, NY), incubated at 4°C overnight, and blocked with 1% bovine serum albumin in PBS. To determine the optimum dilution to test sera, 10 sera representing negative (MAT titer of <100) and positive (MAT titer of >800) serum samples were diluted 2-fold from 1:50 to 1:400 in 1% bovine serum albumin in PBS. Each dilution of serum was tested for Leptospira-specific antibodies. The dilution that provided a high optical density (OD) value while effectively differentiating between negative and positive sera was selected as the optimum dilution for ELISA. The optimum dilution of conjugates for the assay was also determined by checkerboard titration. The dilution that gave the highest OD was selected as the optimum dilution.
The test sera were diluted at optimum dilution in PBS containing 1% bovine serum albumin and 0.05% Tween 20 and then added to the wells for 1 h at 37°C. The IgG reactivity was detected with peroxidase-labeled anti-horse IgG (KPL, Inc., MD) and tetramethylbenzidine (TMB) 2-component microwell peroxidase substrate (KPL, Inc., MD). The plates were read at an optical density of 450 mm (OD450) on a microtiter plate reader (BioTek, VT) after the addition of the same volume of TMB stop solution (KPL, Inc., MD).
Western blot analysis.
Sera that were MAT negative and ELISA positive were further tested by Western blotting. Purified recombinant LigACon4-7.5 (rLigACon4-7.5) was transferred from the SDS-PAGE separation gel to a nitrocellulose membrane (Schleicher & Schuell Biosciences, Inc.) using a Mini Trans-Blot system (Bio-Rad, CA) according to the instruction manual. After washing with TBS, the membranes were removed and soaked in the blocking solution, Tris-buffered saline (20 mM Tris-HCl, 500 mM NaCl [pH 7.5]) containing 5% (wt/vol) nonfat powdered milk, for 1 h at 37°C on a shaker to saturate the remaining protein-binding sites. The blocked membranes were then washed 3 times for 15 min each with Tris-buffered saline containing Tween (TTBS). After washing, the membranes were subjected to assay using the equine serum to be tested as the primary antibody and l:3,000-diluted, alkaline phosphatase-labeled goat anti-horse IgG (KPL, Inc., MD) as the secondary antibody. After this, the membranes were incubated in freshly prepared BCIP-NBT color development solution (Invitrogen) for 10 to 30 min and then rinsed in deionized water to stop the reaction. A serum sample that was both MAT and ELISA negative was used as the negative control and an experimental positive serum was used as a positive control.
Additional samples from experimentally infected horses.
A total of 39 serum samples from 3 experimentally infected horses were included. These serum samples were from a horse trial that experimentally challenged the horses with Leptospira interrogans serovar Kennewicki (31). The serum samples were collected from horses at the start of the project and at 1, 2, 3, 4, 5, 6, 14, 21, 28, 35, 42, and 49 days postchallenge.
Statistical analysis.
The ELISA performance was evaluated using MAT as the reference method. The relative sensitivity, specificity, and accuracy of ELISA for the detection of anti-Leptospira antibodies in horse sera were determined in comparison to the MAT as follows: sensitivity = a/(a + b) × 100; specificity = d/(c + d) × 100; accuracy = [(a + d)/(a + b + c + d) × 100, where a is the number of samples positive by both ELISA and MAT, b is the number of samples positive by MAT but negative by ELISA, c is the number of samples negative by MAT but positive by ELISA, and d is the number of samples negative by both MAT and ELISA.
RESULTS
Cloning, expression, and purification of recombinant proteins.
All the recombinant proteins were expressed as GST fusion proteins and the GST tags were excised by treatment with thrombin. SDS-PAGE and Coomassie blue staining of the purified recombinant proteins revealed the protein bands corresponding to the expected size of the proteins (Fig. 2). However, LigACon4-5 (lane 3) with an extra band is either a contaminant or its degraded product. All of these proteins were expressed in soluble form to retain their native conformation and allowed easy recovery and purification.
Fig 2.
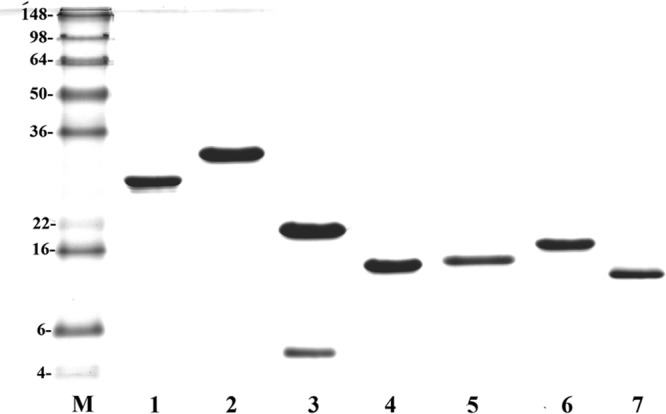
Expression of truncated LigA proteins. Shown are stained SDS-PAGE gels. Analysis of affinity chromatography-purified recombinant fragments by Coomassie brilliant blue-stained SDS-PAGE. Lane M, molecular mass (kDa)marker; lane 1, LigACon1-3; lane 2, LigACon4-7.5; lane 3, LigACon4-5; lane 4, LigACon4.5-5.5; lane 5, LigACon5.5-6.5; lane 6, LigACon6-7.5; lane 7, LigACon4.
MAT.
Serum with a titer equal to or higher than 1:100 against one or more serovars was considered MAT positive. As we reported before (31, 32), multiple titers against different serovars were detected in most seropositive cases.
Slot-shaped dot blot assay.
All recombinant proteins tested reacted with the Leptospira-positive serum; however, the level of reactivity varied (Fig. 3). LigACon4-7.5 showed the highest intensity when analyzed by the image analysis software. Thus, recombinant protein LigACon4-7.5 was chosen as the candidate antigen for the ELISA.
Fig 3.
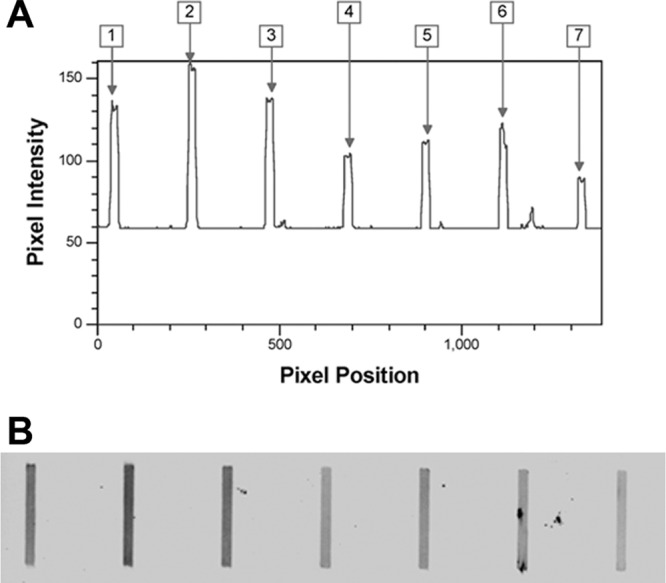
Reactivity of different truncated LigA proteins to Leptospira-positive serum in slot blot assays. (A) Analyzed results using Phoretix 1D software. Data are plotted as pixel intensity versus pixel location (the distance from left to right on the membrane). Lane 1, LigACon1-3; lane 2, LigACon4-7.5; lane 3, LigACon6-7.5; lane 4, LigACon4.5–5.5; lane 5, LigACon4-5; lane 6, LigACon5.5–6.5; lane 7, LigACon4. (B) Scanned slot blot assay image.
Optimization of reagents for ELISA.
A checkerboard titration technique was used to determine the optimum concentrations of reagents for ELISA. With the use of the equine serum with the high MAT titer, the optimum concentration of recombinant antigen yielding high specificity was obtained at 25 ng/well and the optimum dilutions of primary and secondary conjugated antibodies were found to be 1:250 and 1:500, respectively. The cutoff value of the ELISA was determined from 60 MAT-negative equine sera. The mean OD value was 232 with a standard deviation (SD) of 101. The cutoff value for estimation of positive and negative samples was 333 (mean +1 SD).
Detection of serum samples from experimentally infected horses.
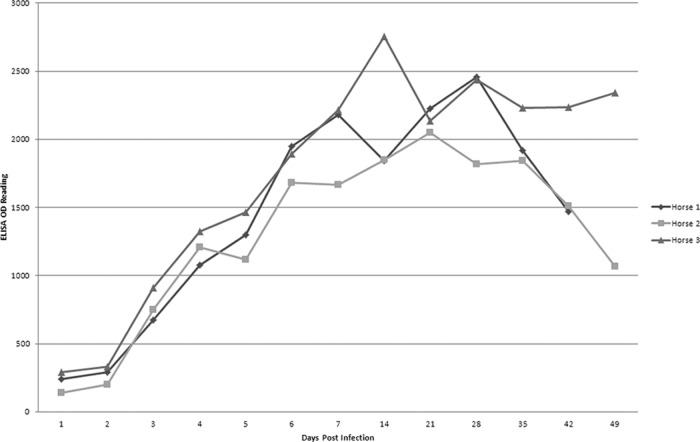
The earliest antibody response was detected at day 3 postchallenge with Leptospira. The OD reading of the ELISA increased as days postinfection increased (Fig. 4) and peaked at 14 days postinfection, which is consistent with the MAT results. However, the increasing of IgG as early as 3 days postinfection was unexpected. The early antibody response suggests that all three horses had been exposed to Leptospira before challenge, even though the MAT was negative before challenge.
Fig 4.
Kinetic curves of horse antibodies to rLigACon4-7.5 tested by ELISA at different days postinfection. The graph shows the OD readings of rLigACon4-7.5 ELISA using sera from three horses experimentally infected with Leptospira interrogans serovar Kennewicki at different days postinfection. Antibody was detected as early as day 3 after infection, increased over time, and reached a peak after day 14.
Evaluation of ELISA in comparison with MAT.
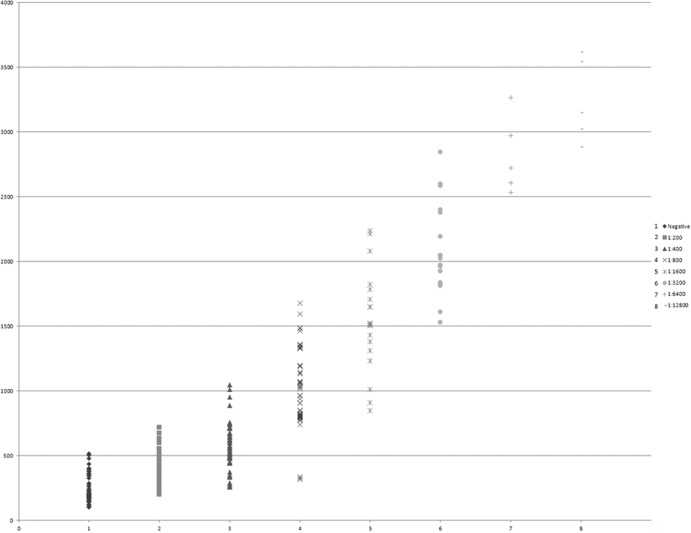
All together a total of 280 horse serum samples were studied with both ELISA and MAT. Sixty of the samples were negative and 220 positive by MAT; 76 were negative and 204 positive by ELISA. ELISA had a sensitivity of 87.2%, a specificity of 80.0%, and an accuracy of 85.7%, based on the assumption that the MAT test, considered the gold standard for Leptospira antibody detection, is 100% accurate (Fig. 5).
Fig 5.
Graph of the ELISA samples showing the IgG ELISA reactivity of 220 equine sera. The x axis indicates the MAT titers of the tested sera. The y axis indicates the ELISA reading of OD450.
Western blot analysis.
Of the sera that were negative in MAT but positive in ELISA, 10 recognized the LigACon4-7.5 fragment in the Western blot (WB) assay (Fig. 6). This indicated that some of these horses may have been infected before and that antibody levels declined below the detection threshold of MAT. However, these low antibody levels were detected by ELISA and confirmed by Western blotting when the LigACon4-7.5 antigen was used.
Fig 6.
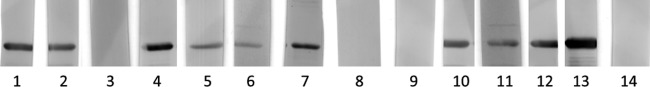
Western blot results of sera that were MAT negative but ELISA positive. Lanes 1 through 12, sera that are MAT negative and ELISA positive; lane 13, MAT positive and ELISA positive (positive control); lane 14, MAT negative and ELISA negative (negative control).
DISCUSSION
Although MAT is an imperfect reference test (33), it is still considered the gold standard for leptospirosis diagnosis in both humans and animals (2, 32). Most state animal diagnostic laboratories in the United States use MAT for the serologic diagnosis of animal leptospirosis. Because of the serious drawbacks of the MAT assay, numerous attempts have been made to develop a serodiagnostic ELISA (10–18) or to develop a dual-path platform (DPP) assay (19). We previously used the LigACon protein for diagnosis of equine and canine leptospirosis (26, 27). The full length of LigA was poorly expressed in Escherichia coli, as previously described (27). Therefore, we were not able to use the full length of LigA in this study. The N-terminal 630 amino acids of LigA and LigB are conserved. However, the N-terminal conserved region is very large (62 kDa) (20). LigACon was expressed well in E. coli; however, it was in an insoluble form (inclusion body) (26, 34). Therefore, we hypothesized that one of several truncated LigA conserved regions would be expressed well and in a soluble form that could be useful as a source of antigens in a serologic-testing format for animal leptospirosis. We truncated the LigA conserved region into 7 fragments and performed a quantitative slot blot assay to assess the interaction of the LigA fragments with equine anti-Leptospira-positive sera. The rationale to use a quantitative slot blot assay was that the test proteins were not in denatured condition and thus retained their native conformation to a large extent. So, at least in principle, most of the antibodies from infected animals should interact with these nondenatured proteins (35). This is especially important for detection of antibodies that bind to higher-order protein structures. In this study, we found that LigACon4-7.5 gave the strongest response and thus it was chosen for further ELISA evaluation.
Results from the Animal Health Diagnostic Center indicate five serovars that occur commonly in New York State and are used routinely in our MAT for equine leptospirosis. We collected 220 MAT-positive and 60 MAT-negative equine sera from the 2005-2009 period, as well as equine serum samples from our previous challenge study (31) for further ELISA evaluation. Since the MAT targeted both IgM and IgG but skewed toward IgG (2, 32), we used a LigACon4-7.5 fragment as the coating antigen to establish an ELISA for improved detection of specific IgG in sera from equine patients with positive MAT titers. In this study, we found that 95% of the equine serum samples with MAT titers of ≥400 were also positive by ELISA. This is not surprising, as a titer of 1:100 or 1:200 is considered low positive and may require other clinical data, follow-up serological testing, or a direct demonstration of the presence of Leptospira in samples, such as a positive blood culture, to help establish the accuracy of the low-titer MAT results. This information was not available, so we cannot prove that these serum samples with low MAT titers are truly positive for leptospirosis.
MAT has some limitations for the diagnosis of leptospirosis. Currently, a 4-fold rise in titer or seroconversion is the most definitive criterion for diagnosis of active leptospirosis. Therefore, a second sample is required, which is often not practical in the clinical setting. In such circumstances, a single high titer in MAT may be taken as evidence of active infection. For human cases, laboratory case definitions have been developed for single (unpaired) MAT results. The WHO Leptospirosis Burden Epidemiology Reference Group recently defined a MAT titer of 1:400 in a single serum specimen as evidence supporting laboratory confirmation (http://www.who.int/zoonoses/diseases/lerg/en/). A similar definition has been made by the U.S. Centers for Disease Control and Prevention (CDC) (http://wwwn.cdc.gov/NNDSS/script/casedef.aspx?CondYrID=907&DatePub=1/1/2013%2012:00:00%20AM). However, to our knowledge, no such titer has been defined for animal leptospirosis diagnosis. A defined positive titer is also needed in horses. Based on the results from this study, a definition similar to those of the WHO and CDC may be applied to equine leptospirosis, i.e., a 1:400 MAT titer in a single serum specimen.
The sensitivity and specificity of this ELISA are 80.0% and 87.2%, respectively. These values are based on the assumption that the MAT is 100% accurate. If we had additional defined leptospira-positive serum samples for this study, the sensitivity and specificity of this ELISA could be higher. In our experimental challenge horses, we performed the MAT before challenge, and the MAT titers of these horses were all below 1:100. However, all three horses developed anti-LigACon4-7.5 IgG antibodies 3 days after challenge. It has been reported that 7 days after the onset of clinical symptoms in human patients, 62% had anti-LigB IgG antibodies as detected by WB analysis (13). Similar results with IgG antibody responses to other leptospiral recombinant proteins have also been reported (12, 36–38). This suggests that these horses were previously infected with Leptospira and developed a memory response upon re-exposure to Leptospira. However, another possible explanation for the robust IgG response to LigACon4-7.5 or other leptospiral antigens may be a rapid antibody class switch during the incubation period, as previously described (13, 32). This phenomenon has also been found in Lyme disease and syphilis patients (39–41).
Interestingly, 12 sera were negative by MAT but positive in our ELISA. We subjected these sera to WB analysis, and to our surprise, 10 out of 12 sera with a negative MAT titer were positive by WB analysis. This suggests that these horses were infected previously but the MAT antibody titers to Leptospira lipopolysaccharide antigens declined below the detection threshold (<1:100). Another possibility is that these horses were infected with other pathogens and developed cross-reacting antibody to LigACon4-7.5. Further study is needed to clarify this issue.
In conclusion, LigACon4-7.5 is a good candidate antigen to develop an ELISA to detect leptospirosis in horses. This ELISA may replace or supplement the current equine MAT for the diagnosis of equine leptospirosis in the near future after further validation with additional defined equine serum samples.
ACKNOWLEDGMENTS
This study was partially supported by the Biotechnology Research and Development Corporation (BRDC) and the New York State Science and Technology Foundation, Centers for Advanced Technology (CAT), to Y.F.C.
Footnotes
Published ahead of print 29 May 2013
REFERENCES
- 1. Ko AI, Goarant C, Picardeau M. 2009. Leptospira: the dawn of the molecular genetics era for an emerging zoonotic pathogen. Nat. Rev. Microbiol. 7:736–747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Palaniappan RU, Ramanujam S, Chang YF. 2007. Leptospirosis: pathogenesis, immunity, and diagnosis. Curr. Opin. Infect. Dis. 20:284–292 [DOI] [PubMed] [Google Scholar]
- 3. Divers TJ, Byars TD, Shin SJ. 1992. Renal dysfunction associated with infection of Leptospira interrogans in a horse. J. Am. Vet. Med. Assoc. 201:1391–1392 [PubMed] [Google Scholar]
- 4. Faisal SM, McDonough SP, Chang YF. 2012. Leptospira: invasion, pathogenesis and persistence, p 143–172 In Embers ME. (ed), The pathogenic spirochetes: strategies for evasion of host immunity and persistence. Springer Science, New York, NY [Google Scholar]
- 5. Broux B, Torfs S, Wegge B, Deprez P, van Loon G. 2012. Acute respiratory failure caused by Leptospira spp. in 5 foals. J. Vet. Intern. Med. 26:684–687 [DOI] [PubMed] [Google Scholar]
- 6. Cerri D, Ebani VV, Fratini F, Pinzauti P, Andreani E. 2003. Epidemiology of leptospirosis: observations on serological data obtained by a “diagnostic laboratory for leptospirosis” from 1995 to 2001. New Microbiol. 26:383–389 [PubMed] [Google Scholar]
- 7. Levett PN. 2001. Leptospirosis. Clin. Microbiol. Rev. 14:296–326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Toyokawa T, Ohnishi M, Koizumi N. 2011. Diagnosis of acute leptospirosis. Expert Rev. Anti Infect. Ther. 9:111–121 [DOI] [PubMed] [Google Scholar]
- 9. Woodward MJ, Swallow C, Kitching A, Dalley C, Sayers AR. 1997. Leptospira Hardjo serodiagnosis: a comparison of MAT, ELISA and Immunocomb. Vet. Rec. 141:603–604 [PubMed] [Google Scholar]
- 10. Bomfim MR, Ko A, Koury MC. 2005. Evaluation of the recombinant LipL32 in enzyme-linked immunosorbent assay for the serodiagnosis of bovine leptospirosis. Vet. Microbiol. 109:89–94 [DOI] [PubMed] [Google Scholar]
- 11. Chalayon P, Chanket P, Boonchawalit T, Chattanadee S, Srimanote P, Kalambaheti T. 2011. Leptospirosis serodiagnosis by ELISA based on recombinant outer membrane protein. Trans. R. Soc. Trop. Med. Hyg. 105:289–297 [DOI] [PubMed] [Google Scholar]
- 12. Flannery B, Costa D, Carvalho FP, Guerreiro H, Matsunaga J, Da Silva ED, Ferreira AG, Riley LW, Reis MG, Haake DA, Ko AI. 2001. Evaluation of recombinant Leptospira antigen-based enzyme-linked immunosorbent assays for the serodiagnosis of leptospirosis. J. Clin. Microbiol. 39:3303–3310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Croda J, Ramos JG, Matsunaga J, Queiroz A, Homma A, Riley LW, Haake DA, Reis MG, Ko AI. 2007. Leptospira immunoglobulin-like proteins as a serodiagnostic marker for acute leptospirosis. J. Clin. Microbiol. 45:1528–1534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Hartleben CP, Leal FM, Monte LG, Hartwig DD, Seixas FK, Vasconcellos SA, Brihuega B, Dellagostin OA. 2013. Serological analysis by enzyme-linked immunosorbent assay using recombinant antigen LipL32 for the diagnosis of swine leptospirosis. Curr. Microbiol. 66:106–109 [DOI] [PubMed] [Google Scholar]
- 15. Joseph S, Thomas N, Thangapandian E, Singh VP, Verma R, Srivastava SK. 2012. Evaluation and comparison of native and recombinant LipL21 protein-based ELISAs for diagnosis of bovine leptospirosis. J. Vet. Sci. 13:99–101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Oliveira TR, Longhi MT, de Morais ZM, Romero EC, Blanco RM, Kirchgatter K, Vasconcellos SA, Nascimento AL. 2008. Evaluation of leptospiral recombinant antigens MPL17 and MPL21 for serological diagnosis of leptospirosis by enzyme-linked immunosorbent assays. Clin. Vaccine Immunol. 15:1715–1722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Sankar S, Harshan HM, Somarajan SR, Srivastava SK. 2010. Evaluation of a recombinant LigB protein of Leptospira interrogans serovar Canicola in an enzyme-linked immunosorbent assay for the serodiagnosis of bovine leptospirosis. Res. Vet. Sci. 88:375–378 [DOI] [PubMed] [Google Scholar]
- 18. Srimanote P, Wongdeethai N, Jieanampunkul P, Samonkiert S, Leepiyasakulchai C, Kalambaheti T, Prachayasittikul V. 2008. Recombinant ligA for leptospirosis diagnosis and ligA among the Leptospira spp. clinical isolates. J. Microbiol. Methods 72:73–81 [DOI] [PubMed] [Google Scholar]
- 19. Nabity SA, Ribeiro GS, Aquino CL, Takahashi D, Damiao AO, Goncalves AH, Miranda-Filho DB, Greenwald R, Esfandiari J, Lyashchenko KP, Reis MG, Medeiros MA, Ko AI. 2012. Accuracy of a dual path platform (DPP) assay for the rapid point-of-care diagnosis of human leptospirosis. PLoS Negl. Trop. Dis. 6:e1878. 10.1371/journal.pntd.0001878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Lin YP, McDonough SP, Sharma Y, Chang YF. 2011. Leptospira immunoglobulin-like protein B (LigB) binding to the C-terminal fibrinogen αC domain inhibits fibrin clot formation, platelet adhesion and aggregation. Mol. Microbiol. 79:1063–1076 [DOI] [PubMed] [Google Scholar]
- 21. Lin YP, Lee DW, McDonough SP, Nicholson LK, Sharma Y, Chang YF. 2009. Repeated domains of Leptospira immunoglobulin-like proteins interact with elastin and tropoelastin. J. Biol. Chem. 284:19380–19391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Lin YP, Greenwood A, Nicholson LK, Sharma Y, McDonough SP, Chang YF. 2009. Fibronectin binds to and induces conformational change in a disordered region of leptospiral immunoglobulin-like protein B. J. Biol. Chem. 284:23547–23557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Lin YP, Raman R, Sharma Y, Chang YF. 2008. Calcium binds to leptospiral immunoglobulin-like protein, LigB, and modulates fibronectin binding. J. Biol. Chem. 283:25140–25149 [DOI] [PubMed] [Google Scholar]
- 24. Lin YP, Chang YF. 2007. A domain of the Leptospira LigB contributes to high affinity binding of fibronectin. Biochem. Biophys. Res. Commun. 362:443–448 [DOI] [PubMed] [Google Scholar]
- 25. Choy HA, Kelley MM, Chen TL, Moller AK, Matsunaga J, Haake DA. 2007. Physiological osmotic induction of Leptospira interrogans adhesion: LigA and LigB bind extracellular matrix proteins and fibrinogen. Infect. Immun. 75:2441–2450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Palaniappan RU, Chang YF, Hassan F, McDonough SP, Pough M, Barr SC, Simpson KW, Mohammed HO, Shin S, McDonough P, Zuerner RL, Qu J, Roe B. 2004. Expression of leptospiral immunoglobulin-like protein by Leptospira interrogans and evaluation of its diagnostic potential in a kinetic ELISA. J. Med. Microbiol. 53:975–984 [DOI] [PubMed] [Google Scholar]
- 27. Palaniappan RU, Chang YF, Jusuf SS, Artiushin S, Timoney JF, McDonough SP, Barr SC, Divers TJ, Simpson KW, McDonough PL, Mohammed HO. 2002. Cloning and molecular characterization of an immunogenic LigA protein of Leptospira interrogans. Infect. Immun. 70:5924–5930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Palaniappan RU, McDonough SP, Divers TJ, Chen CS, Pan MJ, Matsumoto M, Chang YF. 2006. Immunoprotection of recombinant leptospiral immunoglobulin-like protein A against Leptospira interrogans serovar Pomona infection. Infect. Immun. 74:1745–1750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Cole JR, Jr, Sulzer CR, Pursell AR. 1973. Improved microtechnique for the leptospiral microscopic agglutination test. Appl. Microbiol. 25:976–980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Zhu D, Saul AJ, Miles AP. 2005. A quantitative slot blot assay for host cell protein impurities in recombinant proteins expressed in E. coli. J. Immunol. Methods 306:40–50 [DOI] [PubMed] [Google Scholar]
- 31. Yan W, Faisal SM, Divers T, McDonough SP, Akey B, Chang YF. 2010. Experimental Leptospira interrogans serovar Kennewicki infection of horses. J. Vet. Intern. Med. 24:912–917 [DOI] [PubMed] [Google Scholar]
- 32. Faine S, Adher B, Bloin C, Perolat P. 1999. Leptospira and leptospirosis, 2nd ed MedSci, Melbourne, Australia [Google Scholar]
- 33. Limmathurotsakul D, Turner EL, Wuthiekanun V, Thaipadungpanit J, Suputtamongkol Y, Chierakul W, Smythe LD, Day NP, Cooper B, Peacock SJ. 2012. Fool's gold: why imperfect reference tests are undermining the evaluation of novel diagnostics: a reevaluation of 5 diagnostic tests for leptospirosis. Clin. Infect. Dis. 55:322–331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Matsunaga J, Barocchi MA, Croda J, Young TA, Sanchez Y, Siqueira I, Bolin CA, Reis MG, Riley LW, Haake DA, Ko AI. 2003. Pathogenic Leptospira species express surface-exposed proteins belonging to the bacterial immunoglobulin superfamily. Mol. Microbiol. 49:929–945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Burke E, Barik S. 1998. Specific detection of native multi-subunit proteins by slot-blot assay. Biotechniques 25:574–576, 578, 582 [DOI] [PubMed] [Google Scholar]
- 36. Chapman AJ, Adler B, Faine S. 1988. Antigens recognised by the human immune response to infection with Leptospira interrogans serovar Hardjo. J. Med. Microbiol. 25:269–278 [DOI] [PubMed] [Google Scholar]
- 37. Chapman AJ, Everard CO, Faine S, Adler B. 1991. Antigens recognized by the human immune response to severe leptospirosis in Barbados. Epidemiol. Infect. 107:143–155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Guerreiro H, Croda J, Flannery B, Mazel M, Matsunaga J, Galvao Reis M, Levett PN, Ko AI, Haake DA. 2001. Leptospiral proteins recognized during the humoral immune response to leptospirosis in humans. Infect. Immun. 69:4958–4968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Engstrom SM, Shoop E, Johnson RC. 1995. Immunoblot interpretation criteria for serodiagnosis of early Lyme disease. J. Clin. Microbiol. 33:419–427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Magnarelli LA, Ijdo JW, Padula SJ, Flavell RA, Fikrig E. 2000. Serologic diagnosis of Lyme borreliosis by using enzyme-linked immunosorbent assays with recombinant antigens. J. Clin. Microbiol. 38:1735–1739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Schmidt BL, Edjlalipour M, Luger A. 2000. Comparative evaluation of nine different enzyme-linked immunosorbent assays for determination of antibodies against Treponema pallidum in patients with primary syphilis. J. Clin. Microbiol. 38:1279–1282 [DOI] [PMC free article] [PubMed] [Google Scholar]