Abstract
Streptococcus zooepidemicus of Lancefield group C is a highly variable tonsillar and mucosal commensal that usually is associated with opportunistic infections of the respiratory tract of vertebrate hosts. More-virulent clones have caused epizootics of severe respiratory disease in dogs and horses. The virulence factors of these strains are poorly understood. The antiphagocytic protein SeM is a major virulence factor and protective antigen of Streptococcus equi, a clonal biovar of an ancestral S. zooepidemicus strain. Although the genome of S. zooepidemicus strain H70, an equine isolate, contains a partial homolog (szm) of sem, expression of the gene has not been documented. We have identified and characterized SzM from an encapsulated S. zooepidemicus strain from an epizootic of equine respiratory disease in New Caledonia. The SzM protein of strain NC78 (SzMNC78) has a predicted predominantly alpha-helical fibrillar structure with an LPSTG cell surface anchor motif and resistance to hot acid. A putative binding site for plasminogen is present in the B repeat region, the sequence of which shares homology with repeats of the plasminogen binding proteins of human group C and G streptococci. Equine plasminogen is activated in a dose-dependent manner by recombinant SzMNC78. Only 23.20 and 25.46% DNA homology is shared with SeM proteins of S. equi strains CF32 and 4047, respectively, and homology ranges from 19.60 to 54.70% for SzM proteins of other S. zooepidemicus strains. As expected, SzMNC78 reacted with convalescent-phase sera from horses with respiratory disease associated with strains of S. zooepidemicus. SzMNC78 resembles SeM in binding equine fibrinogen and eliciting strong protective antibody responses in mice. Sera of vaccinated mice opsonized S. zooepidemicus strains NC78 and W60, the SzM protein of which shared partial amino acid homology with SzMNC78. We conclude that SzM is a protective antigen of NC78; it was strongly reactive with serum antibodies from horses during recovery from S. zooepidemicus-associated respiratory disease.
INTRODUCTION
Streptococcus zooepidemicus (Streptococcus equi subsp. zooepidemicus) of Lancefield group C is a normal tonsillar and mucosal commensal of the upper respiratory tract of Equus spp. Although a variety of serovars are present in the tonsils of healthy horses, respiratory disease is associated with a single clone, which usually is present in large numbers in bronchial and nasopharyngeal secretions (1). Unlike its clonal derivative Streptococcus equi, comparisons of the genomic sequences of S. zooepidemicus in databases confirm genetic variability and extensive rearrangement/recombination, as suggested by early studies (2, 3). S. zooepidemicus opportunistically produces respiratory disease in situations involving viral infections, heat stress, or prolonged transportation (4). Select clones can be devastating pathogens in intensively housed dogs and guinea pigs and in humans following consumption of contaminated milk or cheese (5–7).
Few virulence factors of S. zooepidemicus have been recognized. SzP protein, an antiphagocytic, hypervariable, and protective M-like protein, is a mosaic of 2 variable N termini, at least 5 variable central regions, and a variable number of PEPK C-terminal repeats (8). Vaccination with recombinant SzP protein of S. zooepidemicus strain W60 protected mice against intraperitoneal homologous challenge (9). Intranasal administration of live attenuated Salmonella enterica serovar Typhimurium MGN707 expressing SzP from S. zooepidemicus serovar MB9 was effective in reducing the persistence of MB9 in utero (10). However, there is evidence that other protective antigens exist. A SzP deletion mutant from S. zooepidemicus strain ATCC 35246 protected mice against intramuscular challenge (11).
The 58-kDa antiphagocytic SeM protein is a major virulence factor and protective antigen in S. equi, a host-specific clonal biovar of an ancestral S. zooepidemicus strain that causes equine strangles. SeM binds fibrinogen, which reduces deposition of C3b on the bacterial surface and phagocytosis by neutrophils (12). SeM elicits strong serum IgG and mucosal IgA responses following infection (13), and vaccines rich in SeM reduce disease severity and morbidity (14). Although the N-terminal sequence of SeM varies, different isolates are uniformly susceptible to the opsonobactericidal effect of a single opsonic serum, suggesting that some opsonogenic epitopes are invariant (15–17). Whole-genome annotation of S. zooepidemicus strain H70 has revealed a partial sem homolog designated szm (18). Expression of SzM by S. zooepidemicus and stimulation of an antibody response and protective efficacy have not been documented. The aims of this study were to clone and to express SzM from S. zooepidemicus strain NC78 (SzMNC78) from a clonal epizootic of equine respiratory disease, to compare its amino acid sequence with that of SeM, to determine its fibrinogen binding ability, opsonogenicity, and reactivity with convalescent-phase sera, and to evaluate its protective efficacy in mouse immunization and challenge experiments.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
S. zooepidemicus isolates from different cases and outbreaks of equine respiratory disease are listed in Table 1. Isolates from a case of peritonitis in a pony and one isolate from an outbreak of canine hemorrhagic pneumonia also are included. NC78 was a representative isolate from an epizootic of equine respiratory disease in New Caledonia in 1997 to 1998. The epizootic persisted for 10 months and involved weanling and adult horses at at least 13 riding premises in different parts of New Caledonia. Clinical signs included coughing and purulent nasal discharge. A specific clone of mucoid S. zooepidemicus (ST-307) was isolated as a pure culture from transtracheal aspirates from some affected animals and as heavy growths from the majority of nasal swabs (n = 56). Only 4% of swabs from unaffected horses were positive for S. zooepidemicus. Viral culture combined with early/late serum antibody screening for influenza virus, equine herpesvirus 1, adenovirus, and rhinovirus failed to indicate a viral etiology. The szp gene of mucoid strains of S. zooepidemicus isolated from stables in the epizootic expressed a protein with N1 N-terminal and HV4 hypervariable domains (GenBank accession numbers HM565772, HM565773, and HM565774). This isolate was subsequently cultured overnight at 37°C in Todd-Hewitt broth (THB) with 0.2% yeast extract.
Table 1.
Isolates of Streptococcus zooepidemicus
| Isolate | Year of outbreak | Location of outbreak | Comments |
|---|---|---|---|
| NC78 | 1997 | New Caledonia | Mucoid S. zooepidemicus from nasal swab |
| NC32 | 1997 | New Caledonia | Mucoid S. zooepidemicus from nasal swab |
| NC88 | 1998 | New Caledonia | Mucoid S. zooepidemicus from nasal swab |
| W60 | 1976 | New York | Nonmucoid S. zooepidemicus from mandibular lymph node abscess |
| RT | 2009 | Indiana | Nonmucoid S. zooepidemicus from nasal discharge |
| NH55426 | 2011 | Maryland | Nonmucoid S. zooepidemicus from nasal swab |
| NH38 | 2011 | Maryland | Nonmucoid S. zooepidemicus from nasal swab |
| NH182 | 2011 | Maryland | Nonmucoid S. zooepidemicus from nasal swab |
| 631 | 1979 | New York | Nonmucoid S. zooepidemicus from case of peritonitis |
| UK30 | 2009 | Kentucky | Nonmucoid S. zooepidemicus from mandibular lymph node abscess of foal |
| 7e | 1993 | Kentucky | Mucoid S. zooepidemicus from pneumonic donkey |
| 007 | 2006 | Kansas | Nonmucoid S. zooepidemicus from canine pneumonia |
| E69 | 2008 | Washington | Nonmucoid S. zooepidemicus from nasal swab |
pET-15b and Escherichia coli strains NovaBlue and BL21 were obtained from Novagen (Madison, WI). pBluescript phagemid, Lambda ZAP II predigested vector, ExAssist helper phage, and E. coli strains XL1-Blue MRF′ and SOLR were from Stratagene (La Jolla, CA). All E. coli strains were grown at 37°C in LB medium, supplemented with ampicillin (100 μg/ml) when necessary.
Convalescent-phase and hyperimmune sera.
Equine convalescent-phase sera were from the Gluck Equine Research Center collection. All samples screened at a 1:200 dilution were from weanling or adult horses with clinical evidence of respiratory disease (nasal discharge, cough, fever, and lung consolidation) and large numbers of S. zooepidemicus organisms detected in cultures of nasal swabs and nasopharyngeal lavage fluid specimens. Polyclonal antisera were raised in yearling goats by subcutaneous administration of 150 μg of purified recombinant SzM (rSzM), with QuilA (10 mg/ml) as adjuvant. Booster injections contained 100 μg of recombinant protein and were administered 14 and 28 days after the primary immunization. Sera were obtained 2 weeks after the final booster. Antibody titers in sera collected on days 0, 28, and 42 were determined by enzyme-linked immunosorbent assay (ELISA) using rSzM as the antigen.
Genomic DNA library.
A genomic DNA library for strain NC78 was constructed as described previously (19).
Library screening.
The library was screened with a pool of 5 convalescent-phase sera (diluted 1:200) from horses from the respiratory disease epizootic. Bound antibody was detected with horseradish peroxidase (HRP)-labeled protein G (Zymed, San Francisco, CA) diluted 1:1,000, followed by 4-chloro-1-naphthol. Positive plaques on agar plugs were allowed to elute overnight at 4°C in 500 μl of sodium-magnesium buffer. Reactive plaques were rescreened until clonal. Plasmids containing inserts of S. zooepidemicus DNA from selected reactive phages were generated by using ExAssist helper phage and E. coli SOLR, according to the manufacturer's protocol. Following SDS-PAGE, proteins in lysates of each reactive phage were transferred to nitrocellulose and blotted using the equine convalescent-phase serum pool, and the molecular masses of the proteins represented by reactive bands were calculated.
DNA sequencing and analysis.
Plasmid DNA was isolated using a Zyppy plasmid miniprep kit (Zymo Research, Irvine, CA) and sequenced at a commercial sequencing facility (Eurofins MWG Operon, Huntsville, AL), using standard T3 and T7 primers. The complete nucleotide sequences from inserts were then compared with the H70 sequence (http://www.sanger.ac.uk/cgi-bin/blast/submitblast/s_zooepidemicus). Putative coding sequences were predicted using the Sequence Manipulation Suite, PSORT, and SignalP. Protein secondary structures and transmembrane domains were predicted using SABLE (http://www.expasy.ch/tools).
Sequencing of SzM proteins from different S. zooepidemicus isolates.
The open reading frames (ORFs) of the szm genes from S. zooepidemicus strains NC78, RT, NH55426, NH38, NH182, and W60 were amplified by PCR using chromosomal DNA as the template and szm-specific primers (SzM forward, 5′-ATA AAG AAG TTC CTG TCA TTA-3′, and SzM reverse, 5′-CAA CAG ACA GGA GAC TGT TGC-3′). The PCR protocol consisted of 30 cycles each of 94°C for 30 s, 55°C for 30 s, and 72°C for 2 min. Amplicons were purified using a GeneJET PCR purification kit (Fermentas), and sequences were obtained (Eurofins MWG Operon) using primers from the initial amplification. Sequences were analyzed as described above.
Recombinant SzMNC78.
The open reading frame of szm without the signal sequence was amplified by PCR using NC78 chromosomal DNA as the template and szm-specific primers (SzMNC78 forward, 5′-TTG CTC GAG GAG GAT TTT AAT GGC GCT AAT TCT-3′, and SzMNC78 reverse, 5′-CAT GGA TCC TTA ACC TGC TTT AGG TGC TG-3′). The amplicon thus generated was digested using BamHI and XhoI and was ligated into predigested pET-15b with polyhistidine residues. The ligate was then transformed into E. coli NovaBlue to increase the plasmid copy number. Positive clones were identified by colony PCR, and recombinant plasmids were transformed into E. coli BL21. High-level expression of polyhistidine-tagged rSzM was achieved by overnight growth in Overnight Express instant terrific broth (TB) medium (Novagen, Madison, WI). Recombinant protein was extracted using Talon Superflow metal affinity resin (Clontech Laboratories, Inc.), in buffer containing 8 M urea, according to the manufacturer's recommendations. rSzM was dialyzed against 20 mM Tris buffer containing 50 mM NaCl (pH 7.5), and purity was checked by SDS-PAGE.
Gel electrophoresis and immunoblotting.
SDS-PAGE was performed for 2 h at 100 V in an XCell SureLock minicell system (Invitrogen, Carlsbad, CA), in Tris-glycine running buffer (25 mM Tris, 250 mM glycine, 0.1% SDS [pH 8.8]). Samples were adjusted to equalize protein concentrations, mixed with equal volumes of 2× gel loading buffer (100 mM Tris-HCl [pH 6.8], 10% SDS, 50% glycerol, 500 mM dithiothreitol, 0.1% bromphenol blue), and boiled for 2 to 3 min before loading. Gels were rinsed twice in distilled water and stained with 0.3% Coomassie brilliant blue R-250 (Sigma). Separated proteins also were transferred electrophoretically to nitrocellulose membranes (0.2 μm; Schleicher & Schuell, Keene, NH) and blocked with 5% bovine serum albumin (BSA) in Tris-buffered saline (50 mM Tris, 150 mM NaCl, 0.05% Tween 20 [pH 7.5]). The membranes were then incubated with goat antiserum against recombinant SzMNC78 (rSzMNC78) (diluted 1:200), followed by protein G conjugated to horseradish peroxidase (diluted 1:1,000; Zymed, San Francisco, CA). Bound conjugate was detected by using 4-chloro-1-naphthol.
Hot acid extracts.
The procedure of Lancefield and Perlmann (20) was used to prepare hot acid extracts of S. zooepidemicus strain NC78.
ELISA.
Ninety-six-well, flat-bottomed polystyrene ELISA plates (Costar, Corning, NY) were coated overnight at 4°C with rSzM (1 μg/well) in 100 μl of coating buffer (0.1 M carbonate-bicarbonate [pH 9.2]). Optimal concentrations of antigen and antibody were determined by checkerboard titration. After washing in phosphate-buffered saline (PBS) with 0.05% Tween 20 (PBS-T), plates were blocked for 1 h at 37°C with 5% nonfat dry milk in PBS-T. Convalescent-phase sera diluted 1:200 were then added, and the plates were incubated for 2 h at 37°C. Bound IgG was detected by incubation with HRP-protein G (diluted 1:8,000; Zymed, San Francisco, CA) for 1 h at 37°C. Plates were then developed with 15 mg o-phenylenediamine (Sigma, St. Louis, MO) in 15 ml of 0.1 M citrate buffer and 20 μl H2O2, the reaction was terminated by the addition of 2 M H2SO4, and optical density (OD) values were read at 490 nm. Wells without coating antigen and serum served as negative controls for background reactivity.
Fibrinogen binding.
Wells of 96-well polystyrene ELISA plates were coated overnight with 100 μl rSzM solution, ranging in concentration from 0.005 to 10.0 μg per well. After washing and blocking with 5% (wt/vol) nonfat milk, equine fibrinogen (3.0 μg/well) was added to separate wells and incubated for 2 h at 37°C. After washing, 100 μl of rabbit antiserum specific for equine fibrinogen (diluted 1:100) was added and incubated for 2 h at 37°C. Controls consisted of wells from which fibrinogen or rabbit antiserum was omitted. Binding of fibrinogen-specific rabbit antibody was detected by incubation with HRP-conjugated goat anti-rabbit IgG. Plates were then developed as described in “ELISA.” The dose-response assay was repeated 3 times in duplicate.
Plasminogen activation.
Wells of 96-well polystyrene ELISA plates were coated overnight with 100 μl rSzM solution, ranging in concentration from 10.0 to 0.625 μg per well. After washing and blocking with 5% (wt/vol) nonfat milk, equine plasminogen (5.0 μg/well; Molecular Innovations, Inc., Novi, MI) was added to separate wells and incubated for 2 h at 37°C. After washing, 100 μl of recombinant streptokinase solution (6.0 μg/well) and equine fibrinogen (5.0 μg/well) were added to each well and incubated at 37°C. After 10 min of incubation, 50 μl of the plasmin substrate d-VLK-p-nitroanilide (pNA) (25 μg/ml; Molecular Innovations, Inc., Novi, MI) was added, the wells were incubated for 1 h at 37°C, and OD values were recorded at 405 nm. Controls consisted of wells from which plasminogen, rSzM, or streptokinase was omitted. The assay was repeated twice in duplicate.
Mouse immunization and challenge.
Two sets of 10 (5 male and 5 female) outbred, 8-week-old, Hsd:ICR(CD-1) mice (weight, 12 to 14 g) were immunized by subcutaneous injection of 40 μg of rSzM in 0.15 ml PBS with QuilA (25 μg/mice). Two booster doses were given (with a 2-week interval) by subcutaneous injection of 40 and 20 μg of protein in 0.15 ml of PBS. A set of 5 male and 5 female mice immunized with sterile PBS plus QuilA served as controls.
Vaccinated and control groups of mice were challenged 2 weeks after the final vaccine booster. The challenge doses were 3 × 104 CFU of log-phase cultures of NC78 inoculated intraperitoneally into female mice and 3 × 103 CFU inoculated into male mice. In preliminary trials, these doses caused 100% mortality between 2 and 6 days after inoculation. A lower dose was used in male mice because a shorter time to death was noted when 3 × 104 CFU was inoculated, suggesting greater susceptibility of male mice. Mice were observed for 8 days after inoculation, at approximately 8-h intervals, for signs of illness (depression, rough coat, and lack of activity). Sick mice were immediately euthanized, and heart blood was collected for culture on colistin-nalidixic acid blood agar and assay of SzM-specific antibodies in serum. Heart blood also was cultured from mice that died in the intervals between observations. β-Hemolytic colonies were identified as S. zooepidemicus by fermentation of lactose and sorbitol.
Surviving mice were euthanized at 8 days, and heart blood was collected for culture and assay of serum antibodies. Cumulative morbidity curves were prepared for vaccinated and control mice. The χ2 test was used to test for significant differences in morbidities between groups of control and vaccinated mice. The immunization-challenge protocol with prompt euthanasia for humane reasons was required by the University of Kentucky Animal Care and Welfare Committee.
Opsonophagocytic assay.
Overnight cultures of NC78 in THB were diluted 1:10,000 in sterile PBS. One hundred microliters of this dilution was combined with 50 μl of pooled sera from 3 SzM-immunized mice, and the mixture was incubated for 30 min at 37°C. Opsonized bacteria were then added to 1.0 ml of fresh heparinized horse blood and mixed well. An aliquot of 575 μl was immediately removed and placed on ice (time 0 [T0] sample). The remaining suspension was rotated at 37°C for 90 min and placed on ice (time 90 [T90] sample). Pour plates prepared in triplicate contained 15 ml THB agar at 56°C, 500 μl heparinized horse blood, and 150 μl test sample. Colonies were counted after overnight incubation at 37°C, and percent reductions in counts from T0 to T90 were calculated. The donor horse was selected based on a low level of SzP-specific serum antibodies.
Statistical analysis.
P values for the significance of differences in morbidity rates were calculated using the χ2 test.
Nucleotide sequence accession numbers.
The nucleotide sequences of the szm genes of NC78, RT, NH38, NH55426, NH182, W60, and UK30 have been deposited in GenBank under accession numbers JX014303, KC146014, KC146015, KC146016, KC146017, KC146018, and KC146019, respectively.
RESULTS
Identification and analysis of szm.
Seventy immunoreactive plaques were identified during screening of the lambda library of NC78 with equine convalescent-phase sera from the New Caledonia epizootic. Plasmids rescued from these phages were sequenced and compared with the annotated genomic sequences of strains H70, ATCC 35246, and MGCS10565. Sequencing analysis coupled with immunoblotting patterns confirmed 33 plaques expressing different proteins. Thirty-three open reading frames (ORFs) were identified in the inserts, some of which overlapped. A subset of these ORFs was predicted to have surface exposure, based on LPXTG or LXYC sequences or a structure typical of a transmembrane domain. The sequence from one phagemid revealed an open reading frame encoding a protein, designated SzM (GenBank accession number JX014303), of 591 amino acids (predicted molecular mass, 66 kDa) that shared 23.20 and 25.46% homology with the SeM proteins of S. equi strains CF32 and 4047, respectively. SzMNC78 shared 19.66, 22.30, 23.30, 24.05, 24.17, 43.52, and 54.66% homology with the SzM proteins of S. zooepidemicus strains H70, NH55426, UK30, NH38, RT, W60, and NH182, respectively. The N and C termini of SzM carried a 42-amino acid signal sequence and a specific sortase recognition sequence (LPSTG), respectively. SzMNC78 is hot acid resistant (Fig. 1c), with a secondary structure predicted to contain an extensive region of alpha-helix extending from residue 48 to residue 511. The secondary structure prediction shows loops in the vicinity of residues 1 to 48 and residues 511 to 590, with a small β-strand at the C terminus (data not shown). Comparisons of the amino acid sequence of SzMNC78 with those of SeM and SzM proteins from different strains of S. equi and S. zooepidemicus revealed 2 clades (Fig. 2). Clade 1 was composed of S. zooepidemicus strains RT, NH55426, NH38, UK30, H70, and ATCC 35246 and S. equi strains CF32 and 4047, and clade 2 was composed of S. zooepidemicus strains NC78, NH182, and W60. SzMNC78 showed two tandem repeats (repeat A, amino acids 308 to 332 and 336 to 360; repeat B, amino acids 374 to 404 and 416 to 446) and 9 or 10 proline-rich repeats at the C terminus. Proline-rich repeats appeared to be conserved and unique to clade 2 strains of S. zooepidemicus; they were absent from SeM and from all clade 1 S. zooepidemicus strains. Comparison of repeat sequences in SeM with the sequences of the SzM proteins from the 9 strains of S. zooepidemicus revealed some interesting features (Table 2). The clade 1 S. zooepidemicus strains all had B repeats similar to the B repeats of S. equi. However, 3 different A repeats (A1, A2, and A3) were present in clade 1 strains. The A3 (A3a, A3b, and A3c) repeats in both S. equi strains and four S. zooepidemicus strains probably represent degenerate versions of each other (Table 2). The B repeat of SzMNC78 is homologous to the C2, C3, and C4 repeats of the plasminogen binding proteins (MLC36 and MLG72) of human group C and G streptococci and also the C repeats of the Arp4 protein of Streptococcus pyogenes (Fig. 3).
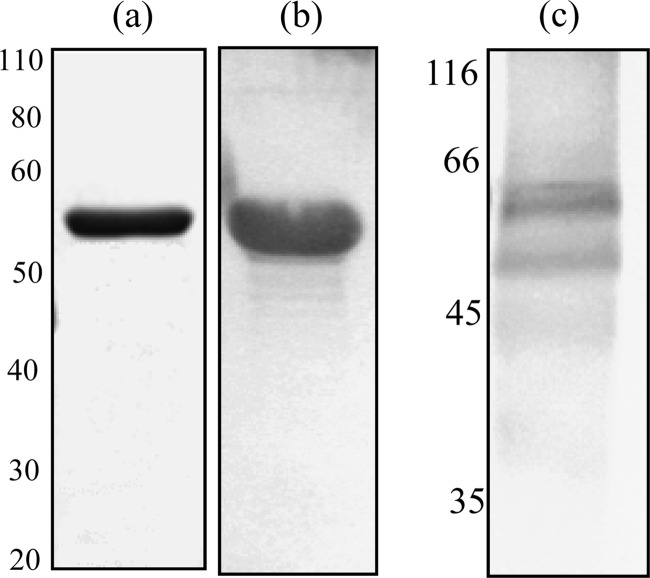
Fig 1.
Immunoblot showing rSzM and hot acid extracts of S. zooepidemicus NC78. (a) Coomassie-stained rSzM resolved by SDS-PAGE (12% gel); (b and c) reactivity of rSzM (b) and a hot acid extract (c) with rSzM-specific goat hyperimmune serum. Numbers, molecular mass markers (in kDa).
Fig 2.
Phylogenetic analyses of SzM and SeM in isolates of S. zooepidemicus and S. equi. The phylogenies were generated by neighborhood joining with 400 bootstrap replicates, rooted at the midpoint. The scale bar represents the measure of phylogenetic distance, and numbers indicate the bootstrap values.
Table 2.
Tandem repeats in SeM and SzM proteins of S. equi and S. zooepidemicus
| Repeat | Tandem repeat sequences | Presence of tandem repeats in indicated isolate |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Clade 1 |
Clade 2, S. zooepidemicus |
|||||||||||
|
S. equi |
S. zooepidemicus |
|||||||||||
| CF32 | 4047 | RT | NH38 | NH55426 | H70 | ATCC 35246 | UK30 | NC78 | NH182 | W60 | ||
| A1 | KDLDKFNRNLLGNAKLDLEKLGKEN | − | − | − | − | − | − | − | − | + | + | − |
| KDLDRINRNLLGNAKGELDKLSAKN | ||||||||||||
| ****::********* :*:**. :* | ||||||||||||
| A2 | KEKEKAAKMTKELADKLS | − | − | + | + | − | − | − | − | − | − | − |
| KDKDRAIQITKELADKLS | ||||||||||||
| *:*::* ::********* | ||||||||||||
| A3 | ||||||||||||
| a | ASEKDKDRAIQITTELANKL | − | − | − | − | + | + | + | − | − | − | − |
| A-ENSRDKAFAVSTELANKL | ||||||||||||
| * *:.:*:*: ::******* | ||||||||||||
| b | AEASRDKAFAVSKDLADKL | + | + | − | − | − | − | − | − | − | − | − |
| AEASRDKAFAVSKDLADKL | ||||||||||||
| ******************* | ||||||||||||
| c | ASEKDKNRAIQITTELANKL | − | − | − | − | − | − | − | + | − | − | − |
| A-ENSRDKAFAVSKDLADKL | ||||||||||||
| * *:.:::*: ::.:**:** | ||||||||||||
| B1 | QKVAEANRRGLRRDLEASREAKKKVEAELAD | − | − | − | − | − | − | − | − | + | + | − |
| QKISEANRRGLHRDLEASREAKKKVEAELAD | ||||||||||||
| **::*******:******************* | ||||||||||||
| B2 | AELQKQKDASDKALAE | + | + | + | + | + | + | + | + | − | − | − |
| AELEKQKAASDAKVAE | ||||||||||||
| ***:*** ***:** | ||||||||||||
Fig 3.

Homology between the B repeat of SzMNC78 and C repeats of the plasminogen binding proteins (MLC36 and MLG72) of human group C and G streptococci. Asterisks, conserved residues; colons, residues with strongly similar properties; periods, residues with weakly similar properties.
Expression of rSzM of NC78.
The purity of the recombinant SzM was confirmed by SDS-PAGE and staining with Coomassie brilliant blue. Immunoblotting also was performed with the equine convalescent-phase serum pool, to confirm that rSzM showed the same reactivity as native SzM (Fig. 1a and b). Antisera specific for rSzM that were raised in yearling goats had antibody titers greater than 1:106,400.
Expression of SzM by other equine isolates of S. zooepidemicus.
SzM expression was detected, by reactivity with rSzM-specific goat antisera, in mutanolysin extracts of S. zooepidemicus isolates (NC32 and NC88) obtained from different stables during the New Caledonian epizootic (Fig. 4). No signal was detected in extracts of other S. zooepidemicus strains except NH182 and 7e, indicating an absence of the protein or low levels of cross-reactivity. Reactivity of the SzM-specific antiserum with a hot acid extract of NC78 confirmed the resistance of SzM to acid and heat (Fig. 1c).
Fig 4.
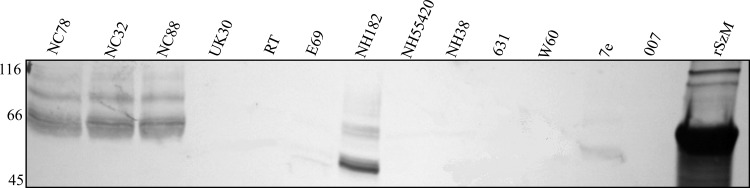
Expression of SzM by different isolates of S. zooepidemicus. Mutanolysin extracts of 18-h cultures of each isolate were separated by SDS-PAGE and blotted with goat antiserum specific for rSzMNC78. Numbers, molecular mass markers (in kDa).
SzM-specific antibody levels in convalescent-phase sera.
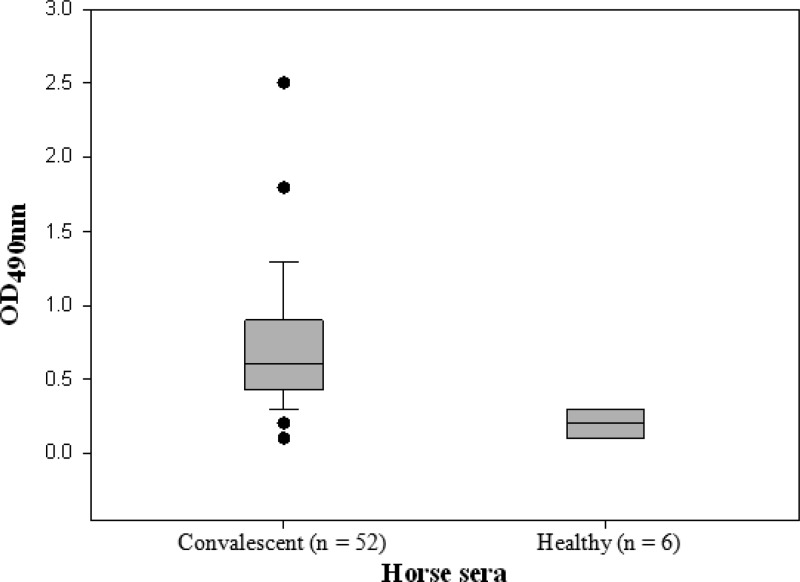
SzM-specific antibody levels in convalescent-phase sera from horses with S. zooepidemicus respiratory disease are shown in Fig. 5. Sera from many S. zooepidemicus-infected horses, including cases from the New Caledonia epizootic, had elevated levels of antibodies to recombinant SzMNC78 (rSzMNC78), indicating expression of this protein during lung infection. Sera from healthy horses showed very low reactivity in the ELISA, indicating low levels or an absence of nonspecific binding of equine IgG by SzM. This was confirmed by substituting HRP-conjugated mouse anti-horse IgG for protein G, which binds to the same domain on SeM as IgG (21).
Fig 5.
SzMNC78-specific antibody levels (ELISA) in equine convalescent-phase sera from cases of respiratory disease associated with S. zooepidemicus. Each value is the mean of 3 replicates. The difference between OD values for the 2 groups was significant (P ≤ 0.01, 2-tailed t test). Filled circles represent outlier values. Error bars represent 90th and 10th percentiles.
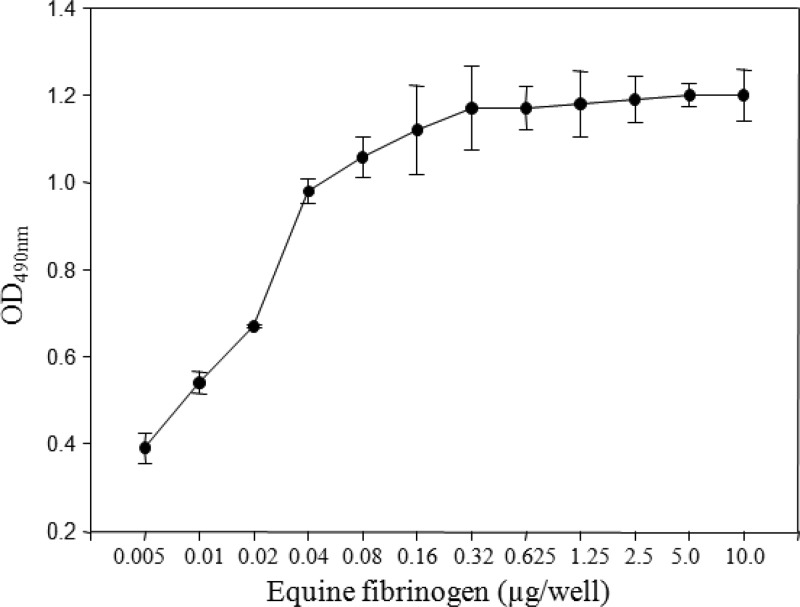
Fibrinogen binding.
SzM showed strong binding to equine fibrinogen immobilized on wells of ELISA plates (Fig. 6). Dose-dependent binding of equine fibrinogen to immobilized SzM revealed 320 ng of rSzM as the saturation concentration for 3 μg of fibrinogen.
Fig 6.
Dose-dependent binding of equine fibrinogen to rSzMNC78 of S. zooepidemicus NC78.
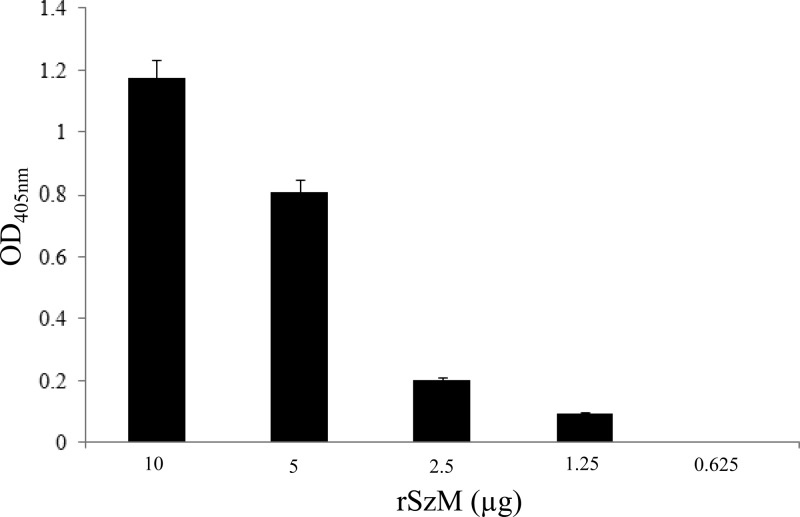
Plasminogen activation.
Equine plasminogen showed strong affinity for rSzMNC78 immobilized on wells of ELISA plates (Fig. 7). The levels of plasmin released decreased with decreasing concentrations of rSzMNC78, suggesting that SzM contributes to the protease activity of S. zooepidemicus in tissue.
Fig 7.
Dose-dependent plasmin activity following the addition of equine plasminogen and streptokinase to rSzMNC78. Plasmin activity was measured using the hydrolysis of d-VLK-pNA.
Efficiency of SzM as a protective antigen in mice.
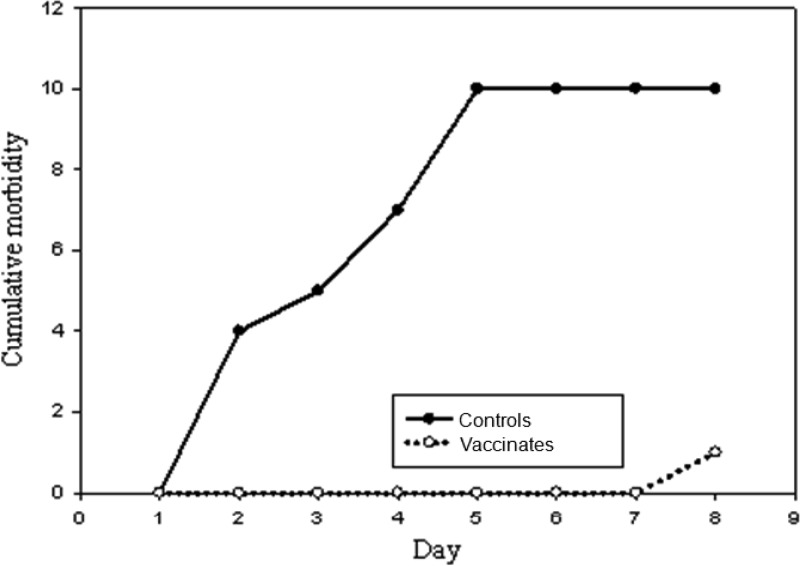
Preliminary dose titration revealed that 104 CFU of NC78 administered intraperitoneally caused illness (ruffled coat, crouching, and depression) followed by death within a few hours in 100% of normal mice. Therefore, euthanasia was performed promptly once signs of illness were observed in the vaccination-challenge study. Cultures of heart blood (10 μl) from sick mice consistently yielded heavy growth of S. zooepidemicus. No immunized mice became sick, and cultures of heart blood from those mice were negative for S. zooepidemicus following euthanasia after 8 days. A highly significant difference (P ≤ 0.01) in susceptibility (illness/bacteremia) was observed in immunized mice compared with control mice (Fig. 8).
Fig 8.
Cumulative morbidity curves for groups of 10 normal mice and 10 mice vaccinated with recombinant SzM protein of S. zooepidemicus NC78 and subsequently challenged intraperitoneally with 3 ×103 CFU (males) or 3 ×104 CFU (females) of S. zooepidemicus NC78.
Opsonophagocytic activity of mouse antiserum to rSzM in horse blood.
Sera from mice immunized with purified rSzM with high levels (>1:106,400) of antibodies to rSzMNC78 in ELISA were opsonic for both NC78 and W60 (Table 3). The numbers of bacteria decreased 9- to 21-fold following opsonization with immune serum but increased following opsonization with normal mouse serum.
Table 3.
Opsonophagocytosis of S. zooepidemicus NC78 and W60 pretreated with mouse antiserum specific for rSzMNC78 or with normal mouse serum and incubated for 90 min in horse blood
| Isolate | Serum | CFU values for triplicate samples at: |
Change in CFU | |
|---|---|---|---|---|
| T0 | T90 | |||
| W60 | Normal | 248, 288, 300 | 492, 560, 624 | 2-fold increase |
| Anti-SzM | 272, 292, 328 | 13, 17, 12 | 21.2-fold decrease | |
| NC78 | Normal | 220, 216, 228 | 352, 272, 344 | 1.5-fold increase |
| Anti-SzM | 252, 224, 260 | 30, 25, 30 | 8.8-fold decrease | |
DISCUSSION
Early Ouchterlony studies of S. equi and S. zooepidemicus demonstrating a hot acid-resistant M-like antigen, together with evidence that extracts of the closely related S. zooepidemicus did not react with antiserum to SeM of S. equi, suggested that this antiphagocytic protein was uniquely expressed by S. equi (19, 22). However, later genomic studies revealed a homolog of SeM (SzM) in H70 (18). Our study documents for the first time the expression of SzM by a strain of S. zooepidemicus (NC78) from a clonal outbreak of equine respiratory disease and describes its molecular features and functional characteristics. SzMNC78 resembles SeM in its mainly secondary alpha-helical structure, nearly identical signal sequence, and fibrinogen binding, opsonogenic, and mouse-protective properties.
Comparisons of the amino acid sequences of SzMNC78 and SeM and SzM proteins from different strains of S. zooepidemicus and S. equi revealed 2 clades, with clade 1 being composed of S. zooepidemicus strains RT, NH55426, NH38, H70, ATCC 35246, and UK30 and S. equi strains CF32 and 4047 and clade 2 being composed of S. zooepidemicus strains NC78, NH182, and W60. SzM sequences in clades 1 and 2 shared 43 to 55% and 20 to 24% homology, respectively, of their C-terminal halves with SeM, suggesting that clade 1 strains may be more closely related to the putative ancestor of the almost clonal S. equi. Comparisons of repeat sequences (Table 2) in SeM and SzM proteins from the 9 strains of S. zooepidemicus revealed some interesting similarities and differences. Nine or 10 proline-rich repeats in the C termini of SzMNC78 and the W60 and NH182 SzM proteins were absent from SeM and from the SzM proteins from all S. zooepidemicus strains of clade 1. SzMNC78 also contained a set of tandem repeats (repeat A, amino acids 308 to 332 and 336 to 360; repeat B, amino acids 374 to 404 and 416 to 446). Clade 1 S. zooepidemicus strains all had B repeats similar to the B repeats of S. equi. The three different A3 repeats (A3a, A3b, and A3c) in clade 1 strains probably represent degenerate versions of each other. Taken together, the repeat sequence data illustrate well the effects of recombination in the S. zooepidemicus genome, as noted previously for the SzP protein (8). An interesting feature of the B repeat of SzMNC78 is its homology to the C2, C3, and C4 repeats of the plasminogen binding proteins of human group C and G streptococci (23). These plasminogen binding proteins (MLC36 and MLG72) are M-like in their resistance to hot acid but share no homology with SzMNC78 other than that of the C repeats. Although the Lancefield group C and G streptococci usually isolated from clinical specimens from humans represent a distinct genetic set, compared with strains isolated from animals, their common ancestry and ability to receive DNA horizontally suggest great potential for sequence rearrangement and acquisitions that explain the emergence of more-virulent clones.
S. zooepidemicus has been implicated in a wide range of opportunistic infections of the respiratory and reproductive tracts in many vertebrate hosts. There also is evidence of the emergence of specific clones associated with outbreaks of severe respiratory disease in shelter dogs, horses, pigs, and monkeys (7, 24, 25). In the early 1900s, pneumonia associated with S. zooepidemicus caused great losses in civil and military horse populations, with mortality rates as high as 15% and prolonged periods of convalescence (26, 27). Theoretically, the association of disease outbreaks with large numbers of horses and dogs in close confinement suggests a scenario wherein a clone of S. zooepidemicus with enhanced virulence is selected and rapidly propagated and transmitted within the group. Effective control of in-house outbreaks in earlier times often was achieved by segregating (picketing out) affected horses (27), which indicated that the epizootiology was that of a transmissible and not opportunist infection model. Enhanced virulence, for example, might be explained by the emergence and selection of a clone in which recombination and addition of sequences to szm resulted in greater resistance to phagocytosis. The addition of a plasminogen binding sequence would be expected to increase the pathogenic potential by enhancing the capture of a potential protease for activation by streptokinase (3). The generation of plasmin is known to enhance the virulence of S. pyogenes in mice (28). Also, the survival of Streptococcus canis in phagocytic analyses has been shown to be enhanced by plasminogen recruitment to the bacterial surface by M protein and enolase (29).
Mouse antiserum to rSzMNC78 reduced the proliferation of both NC78 and W60 in horse blood (Table 3), suggesting that opsonins may be specific for an epitope encoded by the secondary structure, since the NC78 and W60 amino acid sequences share only 43.52% similarity and have few predicted linear B cell epitopes in common. Consistent with this interpretation was the failure of goat antiserum specific for SzMNC78 to react with SzM of strain W60 on an immunoblot (Fig. 4). The greater decrease (21.1-fold versus 8.8-fold) in CFU after 90 min that was observed for the unencapsulated W60 opsonized with SzMNC78-specific antibody is explained by the greater resistance to phagocytosis of the encapsulated NC78.
Antibodies induced by immunization of mice with rSzMNC78 also were strongly protective against homologous challenge. Cultures of heart blood from all mice showing signs of infection (coat ruffling and depression) showed heavy growths of S. zooepidemicus. Thus, the readout parameter was the approximate incubation period. For mice in the control group, this period ranged from 2 to 5 days, which may reflect the outbred genetic status of the Hsd:ICR(CD-1) strain.
The equine sera tested for reactivity with rSzMNC78 were from cases of rhinitis/bronchiolitis and pneumonia on Kentucky farms in which S. zooepidemicus was cultured in large numbers from clinical samples. Information on the presence and extent of pneumonia was not available in all cases. It is likely, however, that clinically evident respiratory disease requiring sampling for laboratory testing involved the lung in many instances (30, 31). Since it is probable that SzM sequences varied from S. zooepidemicus strain to strain, the moderate to low reactivity of many sera in ELISAs may be explained by a lack of homology of SzM of the infecting strain to SzMNC78, the screening antigen used in the ELISA. It is also likely that some sera were collected at an early stage of the acquired immune response. It is noteworthy that the very low ELISA OD values obtained with sera from healthy horses with no history of respiratory disease indicate that SzMNC78, unlike SeM, does not have an IgG-binding domain (32). ELISA OD values ranging between 1.0 and 1.8 for 11 sera (20%) confirm robust SzM-specific antibody responses to infection of the respiratory tract by some S. zooepidemicus strains. Nonetheless, more-detailed study of convalescent-phase responses using SzM of an infecting S. zooepidemicus strain will be required for more-complete evaluation of this antigen as a serological tool and as a correlate of protection engendered by a clonal epizootic. Future studies also should address the level of expression of SzM by different strains of S. zooepidemicus and whether different SzM proteins vary in the degree of antiphagocytic/plasminogen binding function and thus affect the virulence of clones associated with severe respiratory disease.
ACKNOWLEDGMENTS
Income from the Endowment for the Keeneland Chair of Infectious Diseases (J.F.T.) and a grant from the Equine Drug Research Council supported this research.
Footnotes
Published ahead of print 5 June 2013
REFERENCES
- 1. Anzai T, Walker JA, Blair MB, Chambers TM, Timoney JF. 2000. Comparison of the phenotypes of Streptococcus zooepidemicus isolated from tonsils of healthy horses and specimens obtained from foals and donkeys with pneumonia. Am. J. Vet. Res. 61:162–166 [DOI] [PubMed] [Google Scholar]
- 2. Skjold SA, Quie PG, Fries LA, Barnham M, Cleary PP. 1987. DNA fingerprinting of Streptococcus zooepidemicus (Lancefield group C) as an aid to epidemiological study. J. Infect. Dis. 155:1145–1150 [DOI] [PubMed] [Google Scholar]
- 3. Beres SB, Sesso R, Pinto SWL, Hoe NP, Porcella SF, DeLeo FR, Musser JM. 2008. Genome sequence of a Lancefield group C Streptococcus zooepidemicus strain causing epidemic nephritis: new information about an old disease. PLoS One 3:e3026. 10.1371/journal.pone.0003026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Timoney JF. 2004. The pathogenic equine streptococci. Vet. Res. 35:397–409 [DOI] [PubMed] [Google Scholar]
- 5. Seastone CV. 1939. Hemolytic streptococcus lymphadenitis in guinea pigs. J. Exp. Med. 70:347–359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Ural O, Tuncer I, Dikici N, Aridogan B. 2003. Streptococcus zooepidemicus meningitis and bacteraemia. Scand. J. Infect. Dis. 35:206–207 [DOI] [PubMed] [Google Scholar]
- 7. Pesavento PA, Hurley KF, Bannasch MJ, Artiushin S, Timoney JF. 2008. A clonal outbreak of fatal hemorrhagic pneumonia in intensively housed (shelter) dogs caused by Streptococcus equi subsp. zooepidemicus. Vet. Pathol. 45:51–53 [DOI] [PubMed] [Google Scholar]
- 8. Walker JA, Timoney JF. 1998. Molecular basis of variation in protective SzP proteins of Streptococcus zooepidemicus. Am. J. Vet. Res. 59:1129–1133 [PubMed] [Google Scholar]
- 9. Timoney JF, Walker J, Zhou M, Ding J. 1995. Cloning and sequence analysis of a protective M-like protein gene from Streptococcus equi subsp. zooepidemicus. Infect. Immun. 63:1440–1445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Causey RC, Artiushin SC, Crowley IF, Weber JA, Homola AD, Kelley A, Stephenson LA, Opitz HM, Gilmain S, Timoney JF. 2010. Immunization of the equine uterus against Streptococcus equi subspecies zooepidemicus using an intranasal attenuated Salmonella vector. Vet. J. 184:156–161 [DOI] [PubMed] [Google Scholar]
- 11. Fan H-J, Tang F-Y, Mao Y, Lu C-P. 2009. Virulence and antigenicity of the szp-gene deleted Streptococcus equi ssp. zooepidemicus mutant in mice. Vaccine 27:56–61 [DOI] [PubMed] [Google Scholar]
- 12. Boschwitz JS, Timoney JF. 1994. Characterization of the antiphagocytic properties of fibrinogen for Streptococcus equi subsp. equi. Microb. Pathog. 17:121–129 [DOI] [PubMed] [Google Scholar]
- 13. Sheoran AS, Sponseller B, Holmes TMA, Timoney JF. 1997. Serum and mucosal antibody isotype responses to M-like protein (SeM) of Streptococcus equi in convalescent and vaccinated horses. Vet. Immunol. Immunopathol. 59:239–251 [DOI] [PubMed] [Google Scholar]
- 14. Hoffman AM, Staempfli HR, Prescott JF. 1991. Field evaluation of a commercial M protein vaccine against Streptococcus equi infection in foals. Am. J. Vet. Res. 52:589–595 [PubMed] [Google Scholar]
- 15. Bazely PL. 1942. Studies with equine streptococci. 4. Cross-immunity to S. equi. Aust. Vet. J. 18:189–194 [Google Scholar]
- 16. Galán JE, Timoney JF. 1988. Immunologic and genetic comparison of Streptococcus equi isolates from the United States and Europe. J. Clin. Microbiol. 26:1142–1146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Timoney JF, DeNegri R, Sheoran A, Foster N. 2010. Affects of N-terminal variation in the SeM protein of Streptococcus equi on antibody and fibrinogen binding. Vaccine 28:1522–1527 [DOI] [PubMed] [Google Scholar]
- 18. Holden MT, Heather Z, Paillot R, Steward KF, Webb K, Ainslie F, Jourdan T, Bason NC, Holroyd NE, Mungall K, Quail MA, Sanders M, Simmonds M, Willey D, Brooks K, Aanensen DM, Spratt BG, Jolley KA, Maiden MC, Kehoe M, Chanter N, Bentley SD, Robinson C, Maskell DJ, Parkhill J, Waller AS. 2009. Genomic evidence for the evolution of Streptococcus equi: host restriction, increased virulence, and genetic exchange with human pathogens. PLoS Pathog. 5:e1000346. 10.1371/journal.ppat.1000346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Timoney JF, Artiushin SC, Boschwitz JS. 1997. Comparison of the sequences and functions of Streptococcus equi M-like proteins SeM and SzPSe. Infect. Immun. 65:3600–3605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Lancefield RC, Perlmann GE. 1952. Preparations and properties of type-specific M antigen isolated from group A type I haemolytic streptococci. J. Exp. Med. 96:71–82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Lewis MJ, Meehan M, Owen P, Woof JM. 2008. A common theme in interaction of bacterial immunoglobulin-binding proteins with immunoglobulins illustrated in the equine system. J. Biol. Chem. 283:17615–17623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Moore BO, Bryans JT. 1970. Type specific antigenicity of group C streptococci from disease of the horse, p 231–238 In Bryans JT, Gerber H. (ed), Equine infectious diseases II. S. Karger, Basel, Switzerland [Google Scholar]
- 23. Ben Nasr A, Wistedt A, Ringdahl U, Sjobring U. 1994. Streptokinase activates plasminogen bound to human group C and G streptococci through M-like proteins. Eur. J. Biochem. 222:267–276 [DOI] [PubMed] [Google Scholar]
- 24. Soedarmanto I, Pasaribu FH, Wibawan IW, Lammler C. 1996. Identification and molecular characterization of serological group C streptococci isolated from diseased pigs and monkeys in Indonesia. J. Clin. Microbiol. 34:2201–2204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Bjorndottir S. 2012. Streptococcus zooepidemicus: more than just an opportunist. J. Equine Vet. Sci. 32(Suppl):S8 [Google Scholar]
- 26. Hignett SL, King WS. 1940. Streptococcal infection in the commercial horse. Vet. J. 96:81–86 [Google Scholar]
- 27. Stableforth AW, Gallaway IA. 1959. Streptococcal diseases. In Infectious diseases of animals, diseases due to bacteria, vol 2, 589–650 Academic Press, New York, NY [Google Scholar]
- 28. Sun H, Ringdahl U, Homeister JW, Fay WP, Engleberg NC, Yang AY, Rozek LS, Wang X, Sjobring U, Ginsburg D. 2004. Plasminogen is a critical host pathogenicity factor for group A streptococcal infection. Science 305:1283–1286 [DOI] [PubMed] [Google Scholar]
- 29. Fulde M, Rohde M, Polok A, Preissner KT, Chhatwal GS, Bergmann S. 2013. Cooperative plasminogen recruitment to the surface of Streptococcus canis via M protein and enolase enhances bacterial survival. mBio 4:e00629–12. 10.1128/mBio.00629-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Hoffmann AM, Viel L, Juniper E, Prescott JF. 1993. Clinical and endoscopic study to estimate the incidence of distal respiratory tract infection in thoroughbred foals on Ontario breeding farms. Am. J. Vet. Res. 54:1602–1607 [PubMed] [Google Scholar]
- 31. Hoffmann AM, Viel L, Prescott JF, Rosendal S, Thorsen J. 1993. Association of microbiologic flora with clinical, endoscopic, and pulmonary cytologic findings in foals with distal respiratory tract infection. Am. J. Vet. Res. 54:1615–1622 [PubMed] [Google Scholar]
- 32. Meehan M, Lewis MJ, Byrne C, O'Hare D, Woof JM, Owen P. 2009. Localization of the equine IgG-binding domain in the fibrinogen-binding protein (FgBP) of Streptococcus equi subsp. equi. Microbiology 155:2583–2592 [DOI] [PubMed] [Google Scholar]