Abstract
Host immune responses to Mycobacterium tuberculosis are generally able to contain infection and maintain a delicate balance between protection and immunopathology. A shift in this balance appears to underlie active disease observed in about 10% of infected individuals. Effects of local inflammation, combined with anti-M. tuberculosis systemic immune responses, are directly detectable in peripheral circulation, without ex vivo stimulation of blood cells or biopsy of the affected organs. We studied plasma immunomodulator and antibody biomarkers in patients with active pulmonary tuberculosis (TB) by a combination of multiplex microbead immunoassays and computational tools for data analysis. Plasma profiles of 10 immunomodulators and antibodies against eight M. tuberculosis antigens (previously reported by us) were examined in active pulmonary TB patients in a country where TB is endemic, Pakistan. Multiplex analyses were performed on samples from apparently healthy individuals without active TB from the same community as the TB patients to establish the assay baselines for all analytes. Over 3,000 data points were collected from patients (n = 135) and controls (n = 37). The data were analyzed by multivariate and computer-assisted cluster analyses to reveal patterns of plasma immunomodulators and antibodies. This study shows plasma profiles that in most patients represented either strong antibody or strong immunomodulator biomarkers. Profiling of a combination of both immunomodulators and antibodies described here may be valuable for the analysis of host immune responses in active TB in countries where the disease is endemic.
INTRODUCTION
Tuberculosis (TB) is increasingly viewed as an imbalance of host immune responses that transition from protection against Mycobacterium tuberculosis infection to disease resulting from immunopathology in about 10% of untreated individuals (1, 2). M. tuberculosis primarily infects the lung, where it is taken up by alveolar macrophages and dendritic cells (DCs), triggering an inflammatory response (3). This is followed by the recruitment of monocytes and polymorphonuclear neutrophils to the site of infection; these cells express diverse antimicrobial effector molecules to activate macrophages and escalate the inflammatory process (4). Antigen-presenting DCs activate T lymphocytes in the lymph node, which then migrate to the site of infection and proliferate, leading to the formation of granulomas, a hallmark of M. tuberculosis infection (3). About 10% of infected individuals exhibit active TB, whereas the remaining M. tuberculosis-infected people harbor M. tuberculosis in a dormant (latent) state without clinical symptoms. In a small proportion of individuals with latent infection, the bacterium may reactivate months or years later and produce disease (5). In active pulmonary TB, areas of high lymphoid cell activity, arranged in tertiary lymphoid structures, develop around lung granulomas and have been suggested to mimic lymphoid organs in their function (1). To define pathological mechanisms of TB, the roles of cytokines and chemokines have been extensively studied (1, 6). Cytokines are immunomodulating agents secreted by specific cells of the immune system that mediate interactions between cells and are thus required for an integrated response to a variety of stimuli in immune and inflammatory processes (7). Cytokines are grouped into different classes, such as interleukins, lymphokines, and cell signaling molecules. They play a role in many important biological activities, including cell proliferation, activation, death, and differentiation. Cytokines can be pro- or anti-inflammatory and are involved in both paracrine and autocrine pathways (8).
The outcome of M. tuberculosis infection is strongly influenced by cytokines and lipid mediators produced by cells of the innate immune system and the effect of these cytokines on the host cell and M. tuberculosis (6). Proinflammatory cytokines help in the control of M. tuberculosis infection, but they also play a crucial role during the chronic infection stage, dictating the pathogenesis of the disease (9). Tumor necrosis factor alpha (TNF-α), interleukin-12 (IL-12), and gamma interferon (IFN-γ) are central cytokines in the regulatory and effector phases of the immune response to M. tuberculosis (10). Alveolar macrophages and dendritic cells release inflammatory cytokines such as TNF-α, IL-12, and IL-23 along with a variety of chemokines, including C-C motif ligand 2 (CCL2), CCL5, and C-X-C motif ligand 8 (CXCL8). The Th1 response, important for granuloma assembly, is triggered by the production of IL-12 and IL-23 by DCs (3). Activated T cells regulate this flow of inflammatory events by secreting IFN-γ and IL-2, which activate alveolar macrophages to produce a variety of substances involved in growth inhibition and killing of mycobacteria (11). Immune responses to M. tuberculosis infection are downregulated by the production of anti-inflammatory cytokines such as IL-4, IL-10, and transforming growth factor β (TGF-β) (12). In TB patients, patterns of cytokines and chemokines detected in the blood circulation can provide evidence of infection and/or disease without direct analysis of tissue from the affected organ(s) (e.g., lung biopsy) (13, 14).
We and others have recently reported that a majority of TB patients mount a strong humoral antibody response against several M. tuberculosis antigens that are efficiently detected in patient plasma by multiplex methods (15, 16). Well-defined profiles of such antibodies, based on the multiplex microbead immunoassay format, may have potential value for TB diagnosis (15).
A biomarker approach combining detection of immunomodulators and anti-M. tuberculosis antibodies offers a means to study host responses in plasma that reflect cellular and humoral immune mechanisms and can potentially predict disease progression. Patient plasma, compared to other biological specimens (e.g., sputum, lung lavage fluid, and pleural effusion fluid, etc.), enables more consistent sampling. In addition, analysis of plasma circumvents the need for tedious procedures of stimulation of whole blood in assays such as IFN-γ release assays (IGRAs) and the associated sample variation (17).
In this report, a patient population used for studies on immunomodulator and antibody (against M. tuberculosis antigens) profiles comprised of individuals diagnosed with active TB according to diagnostic criteria used in Pakistan and other countries where TB is endemic (WHO Global Tuberculosis Report 2012). The main diagnostic modality used in Pakistan is sputum smear acid-fast bacillus (AFB) microscopy (two to three sequential daily specimens). Accordingly, only patients positive by AFB microscopy were included in this study. Other TB tests have the following limitations in settings of endemicity: (i) although sputum culture is considered the gold standard for TB diagnosis, it takes several weeks to produce results and is therefore not commonly practiced in Pakistan, where the TB patient load in chest disease hospitals is very high, and (ii) tuberculin skin test (TST), chest X ray, and IGRAs may provide useful information in countries where TB is not endemic and where detection of latent TB is a main priority. In countries where TB is endemic, where latent M. tuberculosis infection, Mycobacterium bovis BCG vaccination, and exposure to environmental (nontuberculous) mycobacteria are widespread in the general population, these diagnostic modalities can produce confounding results (WHO Global Tuberculosis Report 2012). In addition, it is important to note that IGRA is a cost-prohibitive test in economically suppressed countries where TB is typically endemic. Because in Pakistan, active disease is the primary concern for the health care system overburdened with TB patients (fifth highest worldwide [WHO Global Tuberculosis Report 2012]), these tests are of limited value, and particularly because of poor resources in the country, they are not relied upon for TB diagnosis. Although under ideal conditions, TST, chest X ray, and IGRAs should be included in a study, their results were not available at the hospital where TB patient samples were collected. Therefore, AFB microscopy provided the most reliable method of confirmation of active TB for inclusion of patients in the study.
For immunomodulator and antibody profiling, as mentioned above, detection of multiple analytes is required, and therefore, multiplex methodologies are more suitable, as they allow simultaneous measurement of many analytes in a single sample under the same conditions (1). Multiplex data generated in this study were analyzed by computational algorithms, which revealed useful profiles of 10 immunomodulators (IL-6, IL-16, IL-18, IFN-γ, granulocyte colony-stimulating factor [G-CSF], GROα [CXCL1], IP-10 [CXCL10], MIG [CXCL9], VEGF, and PDGF-BB) and 8 M. tuberculosis antigens (Rv3881c, Rv2031c, Rv0054, Rv0934, Rv3804c, Rv1886c, Rv0129c, and Rv1860). In addition, the above-mentioned profiles suggested a dichotomy in the immune responses in patients with active pulmonary disease, indicating an inverse correlation between antibody and immunomodulator responses in TB patients.
MATERIALS AND METHODS
Blood sample collection, processing, and storage.
Samples were collected and handled as previously described (15). Briefly, 5-ml blood samples were collected from each individual, through venipuncture, into a Vaccutainer (EDTA, catalog number 367899; BD, NJ). Plasma was separated (1,000 × g for 10 min at room temperature) within 2 h of collection and immediately frozen in aliquots at −80°C until use. In all experiments, fresh, single aliquots were thawed on ice in order to make sure that there were no effects related to freeze-and-thaw cycles.
Multiplex immunomodulator kits.
Multiplex kits for measuring cytokines, chemokines, and growth factors, for use on the Luminex platform (Luminex Corp., Austin, TX), were obtained from Bio-Rad, Hercules, CA. Assays were performed according to the manufacturer's instructions. Two separate panels were used to measure a total of 48 analytes containing the following immunomodulators: (i) one kit contained a panel of 27 analytes, IL-1b, IL-1ra, IL-2, IL-4, IL-5, IL-6, IL-7, IL-8, IL-9, IL-10, IL-12(p70), IL-13, IL-15, IL-17, eotaxin (CCL11), basic fibroblast growth factor (FGF basic), G-CSF, granulocyte-macrophage colony-stimulating factor (GM-CSF), IFN-γ, IP-10 (CXCL10), monocyte chemoattractant protein 1 (MCP-1) (CCL2), MIP-1α (CCL3), MIP-1β (CCL4), PDGF-BB, RANTES (CCL5), TNF-α, and VEGF, and (ii) the second kit contained a panel of 21 analytes, IL-1α, IL-2Rα, IL-3, IL-12(p40), IL-16, IL-18, CTACK (CCL27), GROα (CXCL1), hepatocyte growth factor (HGF), IFN-α2, LIF, MCP-3 (CCL7), macrophage colony-stimulating factor (M-CSF), migration inhibition factor (MIF), MIG (CXCL9), bNGF, SCF, SCGFβ, SDF-1a, TNF-β, and TRAIL.
Sample groups.
Blood samples from TB patients and healthy individuals were obtained under protocols approved through the Institutional Review Board (IRB) at the University of California, Davis (UCD), and UAAR. HIV status was unknown by clinical testing, but AIDS was an exclusion criterion based on interview; patients and controls who stated that they did not have AIDS were included in the study. Samples were obtained from the groups described below.
Healthy control samples.
The healthy control group (n = 37) was comprised of individuals of mixed sex (21 to 30 years old) from the same geographical area (where TB is endemic) as the TB patients (Rawalpindi/Islamabad, Pakistan). BCG coverage in Pakistan since 1978 has been over 80% (18). Healthy donors were all BCG vaccinated and had no apparent clinical symptoms of respiratory/pulmonary disease, cough, or history of active TB. Donors were not tested for active disease by chest X ray, TST, and/or IGRAs because these tests are expected to produce confounding results in Pakistan, as mentioned above. The most appropriate test for TB in Pakistan is AFB microscopy, but due to a lack of any cough, the healthy donors were unable to produce sputum. Multiplex analyses of immunomodulators and antibodies in samples from healthy individuals were used for establishing baselines of analytes.
TB patient samples.
Plasma samples from AFB microscopy-positive (AFB+) TB patients (n = 135) of mixed sex (21 to 35 years old) were collected at the Federal Government TB Hospital, Rawalpindi, Pakistan. All TB patients were primary TB cases (no history of TB). In Pakistan, like most other countries where TB is endemic, TB clinics rely on AFB microscopy (WHO Global Tuberculosis Report 2012). Accordingly, the main diagnostic test used to detect active TB at the Federal Government TB Hospital, Rawalpindi, is AFB microscopy. Although chest X ray may be examined by the attending physician, it is not considered specific for TB. Therefore, to make sure that only patients with active TB were included in this study, samples from AFB+ TB patients were obtained. Three consecutive daily sputum specimens were used to determine positivity by AFB microscopy. Chest X ray, TST, or IGRA was not performed because these procedures are not required for AFB+ patients at this hospital. All TB patients were enrolled in the study at the time of initial diagnosis and were treatment naive. Patients were divided into two main groups based on plasma sample reactivity to eight previously described M. tuberculosis antigens (Rv3881c, Rv2031c [HspX], Rv0054, Rv0934 [P38 or PstS1], Rv3804c [Ag85a], Rv1886c [Ag85b], Rv0129c [Ag85c], and Rv1860 [MPT32]) (15). It was observed that in general, certain patient samples displayed reactivity to multiple antigens, while others displayed reactivity to a limited number or no reactivity at all (Fig. 1). Patients were divided into two groups. One group consisted of patients with antibodies against several of the above-mentioned M. tuberculosis antigens and was designated the multiple-antibody group (>3 antigens; n = 52); this group was randomly selected from a group of 160 such multiple-antibody patients. The other group contained patients with antibodies against few antigens (≤3 antigens); this group was designated the limited-antibody group (n = 83). It was speculated that patients with limited antibody responses might have overall weak immune responses. It was further speculated that such weak immune responders may in addition display weak immunomodulator alterations in plasma. Therefore, the group with limited antibodies was analyzed in its entirety in order to examine immunomodulator profiles in as many samples as possible where the patients contained limited antibodies against the eight antigens. Healthy control plasma samples were obtained from the same geographical area (where TB is endemic) as the TB patients (n = 37).
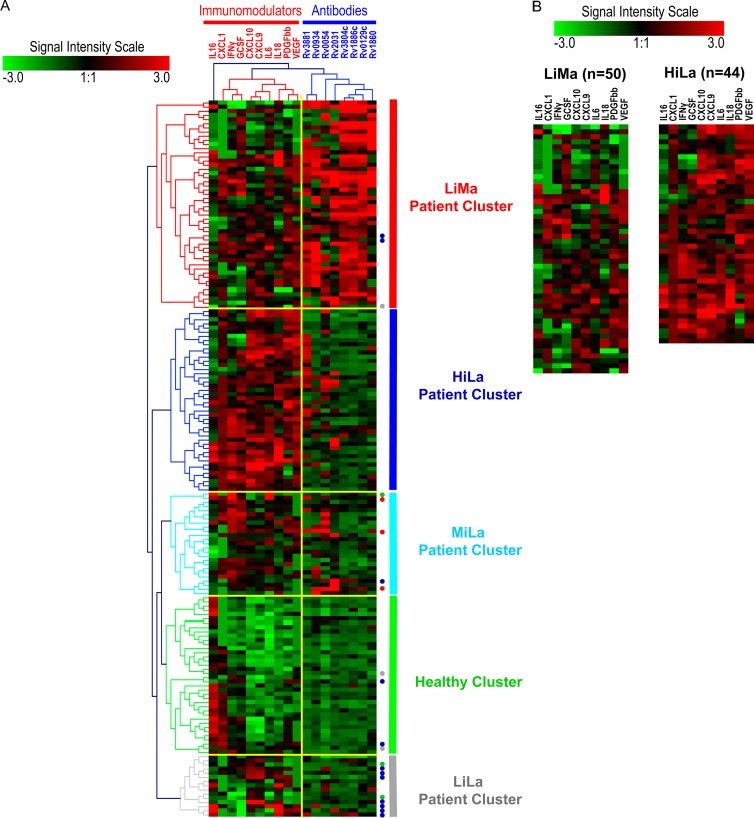
Fig 1.
(A) All samples were classified using computer algorithms based on two sets of variable biomarkers (antibodies and immunomodulators) into different computationally classified clusters. Analytes and their clusters are indicated by dendrograms at the top, and samples and their clusters are indicated by color-coded dendrograms on the left side of the heat map. Analyte and patient clusters are entitled as shown at the top and on the right side of the heat map, respectively. The extent of signal for each analyte, in each sample, is depicted by the intensity scale. The signal level in arbitrary units ranged from “−3 to 3,” with green as the minimum and red as the maximum signal intensities. Three sample clusters were most prominent: healthy individuals (healthy cluster) (green), the patient cluster with low levels of immunomodulators and multiple antibodies (≥3 antigens) (LiMa cluster) (red), and the patient cluster with high levels of immunomodulators and limited antibodies (<3 antigens) (HiLa cluster) (blue). Two minor patient clusters contained limited antibodies and medium levels (MiLa) (light blue) or low levels (LiLa) (gray) of immunomodulators. Misclassified samples are represented by colored dots on the right side. The color of the dot denotes the group that each misclassified sample belongs to. (B) Direct comparison of two major patient clusters (LiMa and HiLa) shows differences in the numbers and levels of immunomodulators in each sample. Notice that the orders of immunomodulators in panels A and B are different. This is because the results represent two independently performed cluster analyses.
Multiplex analysis and data collection.
Multiplex antibody microbead immunoassays against the eight antigens (see above) were performed, and antibody data were collected as median fluorescence intensity (MFI) units, as previously described (15). For immunomodulators, multiplex microbead immunoassays were performed according to the manufacturer's instructions. Data for immunomodulators were collected as plasma concentrations (pg/ml), calculated from MFI data by using the software package BioPlex Manager 5.0 (Bio-Rad) according to the manufacturer's instructions.
To screen immunomodulators of potential value, a pilot experiment with 48 analytes was first performed on a limited number of TB patients (n = 12 for each of the two patient groups) and healthy individuals (n = 15); samples from healthy individuals were used to establish analyte assay baselines. Based on the results, 10 immunomodulators were identified for further evaluation (IL-6, IL-16, IL-18, IFN-γ, GROα [CXCL1], G-CSF, IP10 [CXCL10], MIG [CXCL9], VEGF, and PDGF-BB). The selected immunomodulators were grouped as a single multiplex panel. Custom-assembled kits, prepared as a single batch by Bio-Rad, were obtained for multiplex plasma immunomodulator analysis of all samples from patients and healthy individuals.
Statistical methods.
For the commercial multiplex panels of immunomodulators, masterPlex QT software for quantitation was used with 5-point curve fitting to generate standard curves and analyze data according to the manufacturer's instructions (MiraiBio, Alameda, CA). This software uses Levenberg-Marquardt nonlinear least-squares minimization algorithms for curve fitting and determines the high and low limits of detection (MiraiBio user's manual). Concentrations (pg/ml) of different analytes in the samples were determined by using the standard curves. Data points for analytes that were above or below the detection range were discarded. The analyte concentrations were transformed to a log2 scale for further analysis to accommodate the dynamic range in the concentration values. The MFI level of the antibodies was used without any additional modifications.
Multivariate analysis was performed to reveal biomarker profiles (immunomodulators and antibodies) in patient plasma samples. Multivariate analysis comprises a set of algorithms dedicated to the analysis of data sets with more than one variable. These algorithms require the computational power of modern computers to process and analyze large volumes of data such as proteomics/genomics. Traditional univariate analysis is concerned with just one dependent variable of interest. Although any analysis of data involving more than one variable could be seen as “multivariate,” this term is typically reserved for multiple dependent variables. The current experimental design is inherently multivariate; typically, subjects are assigned to groups, and the experimental measurements relate corresponding changes to a combination of outcomes (e.g., healthy individuals or TB patients with multiple antibodies). The data are then analyzed to expose the inherent structure and meaning revealed within these sets of immunomodulator and antibody variables through application and interpretation of various statistical methods described below. Calculations were performed for plasma concentrations (pg/ml) of immunomodulators and the raw MFI values for plasma antibodies. The data for immunomodulators and antibodies were combined and analyzed simultaneously. First, 10 immunomodulators were selected from a total of 48 in the pilot study based on fold changes calculated by multivariate analysis in patients compared to analyte assay baselines (see below). Second, cluster analysis of the data was performed to profile immunomodulators and antibodies.
Robust linear regression was used on the observed versus theoretical quantiles to determine which linear transformation of the t statistics would confer a normal distribution and then scaled accordingly by using a quantile normalization procedure. Data were visualized with box-and-whisker plots and scatter plots prior to analysis. Cytokine and chemokine concentrations were adjusted to the same interquartile range. A linear model fit was determined for each analyte by using the LIMMA package in R. The data corresponding to immunomodulators (and antibodies) with a positive signal were combined in a two-step process to obtain an initial analytical data set. Relative fold changes (increase or decrease) of the levels of both immunomodulators and antibodies between the healthy controls and patient groups in the pilot as well as the expanded experiments were calculated according to a previously described procedure (19). Briefly, fold changes were calculated across conditions by fitting a linear model to the data, and the statistical significance was estimated by a two-step process. First, an analytical data set that encompassed all the plasma antibodies for which a significantly positive signal was detected in at least one comparison between baselines and patient groups was obtained. This was followed by differential expression across the multiple comparisons detected by an F test along with separate F tests for each immunomodulator and antibody. For the selection of useful antibodies, P values were adjusted by using the Benjamini-Hochberg procedure. Immunomodulators were selected based on the fold change as well as adjusted P values for multiple comparisons. Immunomodulators with a fold change of at least 2 and a P value of <0.01 were considered significant (19).
To define and visually depict natural groupings of immunomodulators and antibodies, with reference to different healthy individual and TB patient samples, hierarchical cluster analysis was performed by using a Euclidean distance metric, without standardization. Clustered data were further scaled to a relative intensity scale ranging from −3 to 3 for presentation as a heat map.
For visual depiction, results of the cluster analysis were presented as dendrograms and heat maps that were generated by a combination of codes using the Matlab (20) and SigmaPlot (21) programs. All the statistical analyses were performed by using standard procedures in “R” (R Development Core Team [http://www.R-project.org/]). The complete set of data used for the analysis is provided in Table S1 in the supplemental material.
RESULTS
Selection of the multiplex plasma immunomodulator panel.
In order to identify immunomodulators with significantly altered concentrations in patient plasma, 48 analytes were evaluated by multiplex analysis. Plasma samples randomly picked from TB patients (multiple-antibody [n = 12] and limited-antibody [n = 12] samples) and indigenous healthy individuals (n = 15) were tested in duplicate. Eighteen immunomodulators were significantly different between healthy individuals and TB patients (Table 1). To focus on the most valuable candidates, 10 immunomodulators were selected. This selection was based on the following criteria: (i) at least a 2-fold difference (P value of <0.001) in the levels of the respective immunomodulators in patients with multiple antibodies (>3 antigens) in comparison to baseline (healthy individuals) and/or (ii) at least a 2-fold difference (P value of <0.001) in the level of immunomodulators between the patient group with multiple antibodies and that with limited antibodies (≤3 antigens). In the pilot experiment, a difference in the level of immunomodulators between healthy individuals and limited-antibody patients (≤3 antigens) and the baseline was not considered for selection. The exceptions were three immunomodulators: IL-2, IL-18, and IFN-γ. Based on statistical analysis alone, IL-2 appeared to be a valuable immunomodulator (Table 1), but absolute plasma concentrations in patient samples were close to the limit of detection (10 pg/ml). Therefore, IL-2 was not considered for further analysis. In contrast, IL-18 did not appear to be a useful biomarker based on the value obtained in the composite statistical analysis, but its level was high in plasma samples of many individual patients who contained limited antibody levels and was therefore included for further analysis. The difference in the levels of IFN-γ in healthy individuals and patients was slightly below 2-fold (Table 1). However, because this immunomodulator has importance in TB patients, as tested by IGRAs, it was included for further analysis. Of the 10 selected analytes, 6 were cytokines (IL-6, IL-16, IL-18, IFN-γ, CXCL1, and G-CSF), two were chemokines (CXCL10 and CXCL9), and two were growth factors (VEGF and PDGF-BB). All samples were analyzed with these 10 immunomodulators regardless of their inclusion in the pilot experiment.
Table 1.
Fold changes in immunomodulators in limited- and multiple-antibody patientsa
| Immunomodulator | Fold change |
||
|---|---|---|---|
| Multiple-antibody patients vs baseline | Limited-antibody patients vs baseline | Multiple- vs limited-antibody patients | |
| MIG (CXCL9) | 3.7 | 6.0 | (2.4) |
| VEGF | 2.5 | 3.9 | (1.4) |
| IP-10 (CXCL10) | 4.0 | 9.4 | (5.4) |
| IL-2 | 3.1 | 3.9 | (0.8) |
| PDGF-BB | 2.2 | 2.4 | (0.1) |
| IL-6 | 1.8 | 4.6 | (2.8) |
| HGF | 1.9 | 2.8 | (0.9) |
| IL-16 | (1.7) | (3.8) | 2.1 |
| IL-12(p70) | 1.4 | 2.2 | (0.7) |
| FGF basic | 1.5 | 2.6 | (1.1) |
| MIP-1α (CCL3) | 1.3 | 2.2 | (0.9) |
| G-CSF | 0.0 | 2.4 | (2.4) |
| IL-17 | 1.3 | 2.4 | (1.1) |
| IL-15 | 1.6 | 2.9 | (1.4) |
| IL-9 | 1.3 | 2.3 | (1.0) |
| GROα (CXCL1) | 3.0 | 8.9 | (6.0) |
| IL-13 | 1.5 | 2.2 | (0.7) |
| IFN-γ | 1.1 | 2.1 | (1.0) |
In the pilot experiment, 50 immunomodulators were tested in a multiplex format. Patient groups with multiple antibodies (>3 antigens) and limited antibodies (≤3 antigens) are shown. Immunomodulator fold changes with respect to analyte assay baselines (see Materials and Methods) were determined by multivariate analysis. The difference between the two patient groups is also shown. Values in parentheses indicate negative fold changes.
Patient and analyte cluster analysis.
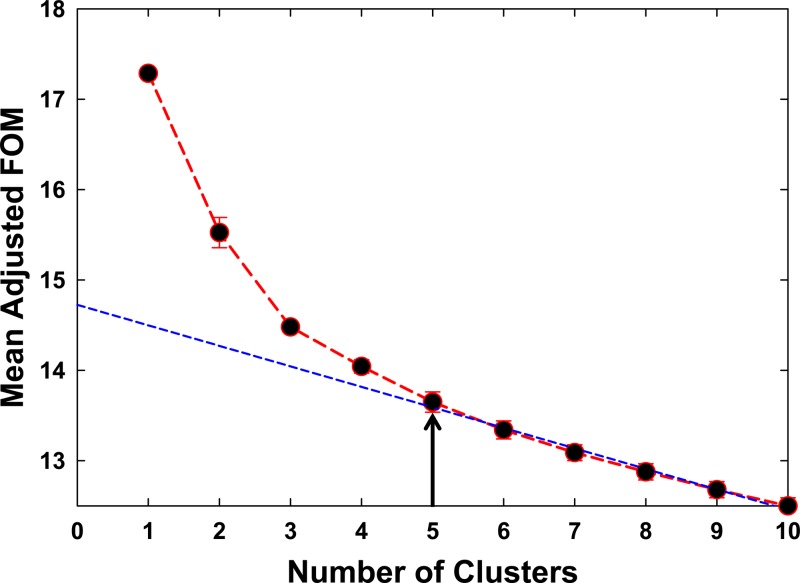
Two key variables, antibodies and immunomodulators, were the focus of the computational classification of sample clusters. Data from patient groups containing either multiple antibodies (>3 antigens) or limited antibodies (≤3 antigens) (n = 52 and n = 83, respectively) were analyzed. Clusters and relative signals for the detection of each antibody and immunomodulator concentration in individual samples are shown in a heat map (Fig. 1A). Cluster analysis led to the computer-generated natural groupings of data obtained from all analytes (antibodies and immunomodulators) and samples (healthy individuals and TB patients) combined together. These natural groupings automatically classified all data into two clusters of analytes and five clusters of samples (Fig. 1A). A frequency-of-merit (FOM) analysis was performed to confirm the number of groups of samples that could be clearly classified (Fig. 2). The FOM analysis confirmed the above-mentioned observation (Fig. 1A) that computer algorithms could classify only 5 meaningful sample clusters for the data sets in this study. One sample cluster was classified to contain few or no analytes above background, and this cluster was interpreted to represent samples from healthy individuals and was termed the “healthy cluster” (n = 38). Although this cluster contained no (or negligible) M. tuberculosis-specific antibodies, two cytokines, IL-16 and CXCL1, were detectable in many samples. Four clusters of data were interpreted to represent patient groups. One contained antibodies against several antigens but low levels of immunomodulators. This cluster represented the patient group with multiple antibodies (>3 antigens). Patient samples in this cluster contained low levels of immunomodulators, and this cluster was termed the “low immunomodulators, multiantibodies,” or “LiMa,” cluster (n = 50). The second major cluster contained limited antibodies (≤3) but high levels of cytokines and was termed the “high immunomodulators, limited antibodies,” or “HiLa,” cluster (n = 44). The remaining two small patient clusters represented patient samples with limited antibodies. One cluster contained medium levels and the other contained low levels of immunomodulators and were termed “MiLa” (n = 25) and “LiLa” (n = 15), respectively.
Fig 2.
Frequency-of-merit (FOM) analysis for the number of valuable clusters, classified by multivariate analysis. FOM analysis shows that there are a maximum of five valuable clusters (arrow). The three prominent clusters in this figure (1, 2, and 3) are represented by the LiMa, HiLa, and healthy clusters in Fig. 1A, respectively. The two minor clusters (4 and 5) are represented by MiLa and LiLa in Fig. 1A, respectively.
The accuracy of automatic, natural sample clustering, based on multianalyte (immunomodulators and antibodies) profiles, by computer algorithms was assessed by comparing computationally classified clusters to the raw data for each individual sample. Among the three major clusters, samples in the HiLa cluster were most accurately classified, without inclusion of any inappropriately classified samples. In the cluster representing healthy samples, i.e., the healthy cluster, four patient samples with limited antibodies were inappropriately classified. Based on the immunomodulator profile, their correct classification should have been into the HiLa (n = 2) or LiLa (n = 2) cluster. In the cluster representing patients with low levels of immunomodulators and multiple antibodies, LiMa, three samples containing limited antibodies were classified. These should have been classified into the HiLa (n = 2) and LiLa (n = 1) clusters. A total of five samples were inappropriately classified into the MiLa cluster and should have been classified as healthy (n = 1), LiMa (n = 3), or HiLa (n = 1). The largest number of inappropriate classifications was in the LiLa cluster, which included samples that should have been classified into either the healthy (n = 2) or HiLa (n = 7) cluster. Thus, the overwhelming majority of samples with strong analyte signals (immunomodulator levels or multiple antibodies) or weak analyte signals (healthy cluster) were appropriately classified by computer algorithms as natural groups.
Immunomodulator profiles. (i) Visual profiles.
In most patient samples, levels of the majority of plasma immunomodulators were clearly detectable, with the exception of IL-16 (Fig. 1A). The HiLa cluster, with high levels of immunomodulators and limited antibodies, was dominated by IL-18, IFN-γ, CXCL10, CXCL9, IL-6, G-CSF, VEGF, PDGF-BB, and CXCL1. In contrast, the profile of the LiMa cluster did not appear to be dominated by a particular set of immunomodulators. In addition, in the LiMa cluster, fewer immunomodulators were detectable in each sample and at lower levels. Among the other two patient clusters with samples containing limited antibodies, the MiLa cluster contained IFN-γ and G-CSF as dominating immunomodulators, whereas CXCL10 and CXCL9 were dominant in the LiLa cluster.
(ii) Fold changes in HiLa and LiMa clusters.
In the HiLa cluster, plasma levels of all immunomodulators were at least about 2-fold higher than the baseline (Table 2). In particular, the levels of three immunomodulators, CXCL10, CXCL9, and VEGF, were more than 6-fold higher. Samples in the LiMa cluster also contained immunomodulators with higher plasma levels. However, there was generally less of a difference in levels between TB patients and the baseline. In this cluster, VEGF was the only analyte with a level that was more than 6-fold higher. Thus, a clear difference was observed in fold changes among the two main patient clusters. This is emphasized by the fact that the levels of several immunomodulators (CXCL10, CXCL9, IL-18, CXCL1, and VEGF) were about 2- to 8-fold higher in the HiLa cluster than in the LiMa cluster (Table 2). Interestingly, the level of IL-16 was about 3-fold lower in TB patient samples.
Table 2.
Fold changes in subclusters of patients for 10 selected immunomodulatorsa
| Immunomodulator | Fold change |
||
|---|---|---|---|
| HiLa vs baseline (A) | LiMa vs baseline (B) | Difference (A − B) | |
| IP-10 (CXCL10) | 9.0 | 3.8 | 5.2 |
| MIG (CXCL9) | 6.6 | 3.8 | 2.8 |
| IL-18 | 3.8 | 2.2 | 1.6 |
| IL-6 | 4.1 | 3.6 | 0.5 |
| IL-16 | (2.7) | (2.9) | (0.2) |
| PDGF-BB | 2.8 | 2.3 | 0.5 |
| IFN-γ | 2.3 | 1.7 | 0.6 |
| GROα (CXCL1) | 3.2 | 0.0 | 3.2 |
| G-CSF | 1.8 | 1.6 | 0.2 |
| VEGF | 14.2 | 6.5 | 8.1 |
Immunomodulators (10 analytes selected from 50) in the two main patient subclusters with respect to analyte assay baselines (see Materials and Methods) were determined by using multivariate analysis. Immunomodulators in the HiLa cluster altered by about 2-fold (P value of <0.01) compared to the LiMa cluster are shaded. Negative fold changes are given in parentheses.
DISCUSSION
This study describes immune responses in plasma from active pulmonary TB patients in a country where TB is endemic. The samples were collected in Pakistan from TB patients with active disease. Patients were included based on the standard diagnostic procedure (AFB microscopy) used in Pakistan, according to WHO guidelines. A potential limitation of the study is that culture, TST, X ray, and IGRA results were not included due to their unavailability at the hospital where samples were collected. Nevertheless, this study is consistent with practice in resource-poor countries, where the focus is typically on efficient diagnosis of active disease using cost-effective diagnostic procedures. Healthy controls were individuals from the same community as the TB patients. Therefore, they were exposed to the same environmental insults, including microbiological and parasitic exposures (e.g., to nontuberculous mycobacteria and intestinal pathogens, etc.), as the patients, with one key difference: they did not have active TB. Accordingly, they were relevant as a healthy control group for valid use in the determination of assay baselines.
Pulmonary TB is an inflammatory disease with an involvement of many immunomodulators (1, 2, 6). To date, several studies have demonstrated that cytokines (e.g., IFN-γ) measured in cell-based assays are useful to study immune responses in TB infection (6). However, these assays require overnight stimulation of a patient blood sample with M. tuberculosis-specific antigens. Importantly, handling and treatment conditions of blood introduce additional variables that are difficult to control and thus may adversely affect the consistency of analysis (17). Therefore, direct measurement of immunomodulators in patient plasma offers a more practical means to study host immune responses in TB infection. Based on multiplex analysis of 10 immunomodulators, we report alterations in their levels in patient plasma samples. Profiles comprising several antibodies and immunomodulators in TB patients were studied. Previously, we described profiles of plasma anti-M. tuberculosis antibodies in TB patients with active pulmonary disease (15). In the current study, patients with limited or multiple antibodies against M. tuberculosis antigens (≤3 or >3 antigens, respectively) were included (n = 83 and n = 52, respectively). A large proportion of TB patients had elevated levels of several plasma immunomodulators (Fig. 1A). This observation demonstrates that direct studies of plasma immunomodulator profiles are useful to study immune responses in TB patients. Another important finding was that the multianalyte (immunomodulators and antibodies) profiles were consistent enough for the computer algorithms to generate clusters of both analytes and samples. Samples from patients with limited antibody responses contained higher levels of immunomodulators and in a larger proportion of patients (HiLa cluster) than samples from patients with multiple antibodies (LiMa cluster) (Table 2 and Fig. 1B). The profile of dominating immunomodulators consisted of IL-18, IFN-γ, CXCL10, CXCL9, G-CSF, IL-6, CXCL1, VEGF, and PDGF-BB. Importantly, there were no misclassified samples in the HiLa cluster. This result indicates that although samples in this cluster contained limited antibodies, the sample immunomodulator profiles displayed a strong and consistent pattern, which enabled computational classification to be accurate (Fig. 1A). In addition, this result shows that our earlier speculation that patients with limited antibodies might overall be weak immune responders such that levels of immunomodulators may not be elevated was wrong. Two smaller clusters of patients with limited antibody responses contained fewer immunomodulators. Nevertheless, they too displayed profiles (IFN-γ and G-CSF, and CXCL10 AND CXCL9, in the MiLa and LiLa clusters, respectively) consistent with inflammatory responses.
The immune response to infection is a complex phenomenon involving many classes of immune cells and immunomodulatory molecules secreted by them. This is further complicated by redundancies and pleiotropic effects of both the cells and their secreted immunomodulators. In TB patients, a delicate balance has been proposed to exist between immune responses in the lung that successfully contain the infection and those leading to immunopathology (1, 6). After infection with M. tuberculosis, downstream of innate immunity, IL-18 plays a key role in the polarization of CD4 cells into Th1 cells (1, 6). The Th1 cells in turn secrete IFN-γ and other inflammatory molecules at the site of inflammation. In response, lung epithelial cells secrete IP-10 and MIG, both of which are potent chemoattractants that play a key role in the recruitment of T cells and monocytes/macrophages (22). The other immunomodulators identified in this study are also inflammatory molecules. Together, they appear to fuel a strong Th1 response. The profile of inflammatory immunomodulators identified in this study and their levels in plasma samples of patients with weak antibody responses suggest that a differential cellular and antibody response occurs in patients with active TB. Patients who mount a strong Th1 response (cellular) may not have a concomitantly strong Th2-driven B-cell (humoral antibody) response, and vice versa.
It is generally believed that cellular responses are more important than humoral antibody responses in controlling infection with M. tuberculosis to levels below the threshold of disease (1, 2, 23). If so, our findings would support the hypothesis that biomarker profiling as described above can predict therapeutic success. However, in this exploratory study, the intent was not to definitively identify the relationship of the immune profiles to clinical outcomes. In future studies, it would therefore be useful to analyze plasma antibody and immunomodulator profiles over time in patients undergoing therapy. In addition, in evaluations of vaccine candidates, such immune biomarker profiles may help identify immune correlates of protection.
This study suggests that plasma immunomodulators, in combination with antibody responses, are useful in investigating immune mechanisms involved in active pulmonary TB. The data presented here do not suggest the utility of these biomarkers in studying latent TB; we plan to perform a separate study on latent TB. A better understanding of underlying immune mechanisms of this disease may influence the design and development of not only better diagnostics but also more efficacious vaccines. In this regard, it is conceivable that the expression of certain M. tuberculosis antigens during the course of infection and disease (or due to certain vaccines) may influence the immune responses resulting in the elevation of levels of certain immunomodulators locally as well as in the general blood circulation (24, 25). In addition, the monitoring of host immune mechanisms through analysis of both immunomodulatory proteins and antibody profiles may be valuable for assessing an individual patient's progress during therapy. This is important because TB therapy is a long process (about 6 months) involving multiple drugs and changes in drug regimens that currently are not tailored to individuals.
Supplementary Material
ACKNOWLEDGMENTS
We thank Daniel Vang (UCD) for expert technical assistance in the multiplex microbead immunoassay analysis of plasma immunomodulators. We are thankful to Irum Nawaz, Mirza Imran Shahzad, and Arif Nadeem (Pir Mehr Ali Shah Arid Agriculture University, Rawalpindi, Pakistan) for their dedicated effort in the collection of blood/plasma samples and clinical information.
V.V.K. was supported in part by a RIMI (Research Infrastructure for Minority Institutions) grant (P20 MD 002732). This work was supported by funding (to I.H.K., P.A.L., and A.K.), in whole or in part, from the National Academy of Sciences, the U.S. Agency for International Development, the U.S. Department of State, and the Higher Education Commission of Pakistan, and any opinions, findings, conclusions, or recommendations expressed are those of the authors and do not necessarily reflect the views of the project sponsors.
Footnotes
Published ahead of print 12 June 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/CVI.00213-13.
REFERENCES
- 1.Kaufmann SH, Parida SK. 2008. Tuberculosis in Africa: learning from pathogenesis for biomarker identification. Cell Host Microbe 4:219–228 [DOI] [PubMed] [Google Scholar]
- 2.Dorhoi A, Reece ST, Kaufmann SH. 2011. For better or for worse: the immune response against Mycobacterium tuberculosis balances pathology and protection. Immunol. Rev. 240:235–251 [DOI] [PubMed] [Google Scholar]
- 3.Sasindran SJ, Torrelles JB. 2011. Mycobacterium tuberculosis infection and inflammation: what is beneficial for the host and for the bacterium? Front. Microbiol. 2:2. 10.3389/fmicb.2011.00002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Korbel DS, Schneider BE, Schaible UE. 2008. Innate immunity in tuberculosis: myths and truth. Microbes Infect. 10:995–1004 [DOI] [PubMed] [Google Scholar]
- 5.Kleinnijenhuis J, Oosting M, Joosten LA, Netea MG, Van Crevel R. 2011. Innate immune recognition of Mycobacterium tuberculosis. Clin. Dev. Immunol. 2011:405310. 10.1155/2011/405310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dheda K, Schwander SK, Zhu B, van Zyl-Smit RN, Zhang Y. 2010. The immunology of tuberculosis: from bench to bedside. Respirology 15:433–450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Feghali CA, Wright TM. 1997. Cytokines in acute and chronic inflammation. Front. Biosci. 2:d12–d26 [DOI] [PubMed] [Google Scholar]
- 8.Dinarello CA. 2000. Proinflammatory cytokines. Chest 118:503–508 [DOI] [PubMed] [Google Scholar]
- 9.Flynn JL, Chan J. 2001. Immunology of tuberculosis. Annu. Rev. Immunol. 19:93–129 [DOI] [PubMed] [Google Scholar]
- 10.Boom WH, Canaday DH, Fulton SA, Gehring AJ, Rojas RE, Torres M. 2003. Human immunity to M. tuberculosis: T cell subsets and antigen processing. Tuberculosis (Edinb.) 83:98–106 [DOI] [PubMed] [Google Scholar]
- 11.Cooper AM. 2009. Cell-mediated immune responses in tuberculosis. Annu. Rev. Immunol. 27:393–422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sharma S, Bose M. 2001. Role of cytokines in immune response to pulmonary tuberculosis. Asian Pac. J. Allergy Immunol. 19:213–219 [PubMed] [Google Scholar]
- 13.Yu Y, Zhang Y, Hu S, Jin D, Chen X, Jin Q, Liu H. 2012. Different patterns of cytokines and chemokines combined with IFN-gamma production reflect Mycobacterium tuberculosis infection and disease. PLoS One 7:e44944. 10.1371/journal.pone.0044944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mihret A, Bekele Y, Bobosha K, Kidd M, Aseffa A, Howe R, Walzl G. 2013. Plasma cytokines and chemokines differentiate between active disease and non-active tuberculosis infection. J. Infect. 66:357–365 [DOI] [PubMed] [Google Scholar]
- 15.Khan IH, Ravindran R, Krishnan VV, Awan IN, Rizvi SK, Saqib MA, Shahzad MI, Tahseen S, Ireton G, Goulding CW, Felgner P, DeRiemer K, Khanum A, Luciw PA. 2011. Plasma antibody profiles as diagnostic biomarkers for tuberculosis. Clin. Vaccine Immunol. 18:2148–2153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kunnath-Velayudhan S, Salamon H, Wang HY, Davidow AL, Molina DM, Huynh VT, Cirillo DM, Michel G, Talbot EA, Perkins MD, Felgner PL, Liang X, Gennaro ML. 2010. Dynamic antibody responses to the Mycobacterium tuberculosis proteome. Proc. Natl. Acad. Sci. U. S. A. 107:14703–14708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Detjen AK, Loebenberg L, Grewal HM, Stanley K, Gutschmidt A, Kruger C, Du Plessis N, Kidd M, Beyers N, Walzl G, Hesseling AC. 2009. Short-term reproducibility of a commercial interferon gamma release assay. Clin. Vaccine Immunol. 16:1170–1175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pakistan Institute of Legislative Development and Transparency 2010. Immunization in Pakistan—briefing paper. May 2010. Pakistan Institute of Legislative Development and Transparency, Islamabad, Pakistan [Google Scholar]
- 19.Khan IH, Krishnan VV, Ziman M, Janatpour K, Wun T, Luciw PA, Tuscano J. 2009. A comparison of multiplex suspension array large-panel kits for profiling cytokines and chemokines in rheumatoid arthritis patients. Cytometry B Clin. Cytom. 76:159–168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mathworks Inc 2005. Matlab, 12th ed Mathworks Inc, Natick, MA [Google Scholar]
- 21.Systat Software Inc 2005. SigmaPlot, 9th ed Systat Software Inc, San Jose, CA [Google Scholar]
- 22.Sauty A, Dziejman M, Taha RA, Iarossi AS, Neote K, Garcia-Zepeda EA, Hamid Q, Luster AD. 1999. The T cell-specific CXC chemokines IP-10, Mig, and I-TAC are expressed by activated human bronchial epithelial cells. J. Immunol. 162:3549–3558 [PubMed] [Google Scholar]
- 23.Lin PL, Flynn JL. 2010. Understanding latent tuberculosis: a moving target. J. Immunol. 185:15–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bansal K, Elluru SR, Narayana Y, Chaturvedi R, Patil SA, Kaveri SV, Bayry J, Balaji KN. 2010. PE_PGRS antigens of Mycobacterium tuberculosis induce maturation and activation of human dendritic cells. J. Immunol. 184:3495–3504 [DOI] [PubMed] [Google Scholar]
- 25.Bansal K, Sinha AY, Ghorpade DS, Togarsimalemath SK, Patil SA, Kaveri SV, Balaji KN, Bayry J. 2010. Src homology 3-interacting domain of Rv1917c of Mycobacterium tuberculosis induces selective maturation of human dendritic cells by regulating PI3K-MAPK-NF-kappaB signaling and drives Th2 immune responses. J. Biol. Chem. 285:36511–36522 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.