Abstract
The histidine phosphotransfer (HPt) protein Ypd1 is an important participant in the Saccharomyces cerevisiae multistep two-component signal transduction pathway and, unlike the expanded histidine kinase gene family, is encoded by a single gene in nearly all model and pathogenic fungi. Ypd1 is essential for viability in both S. cerevisiae and in Cryptococcus neoformans. These and other aspects of Ypd1 biology, combined with the availability of structural and mutational data in S. cerevisiae, suggest that the essential interactions between Ypd1 and response regulator domains would be a good target for antifungal drug development. The goal of this minireview is to summarize the wealth of data on S. cerevisiae Ypd1 and to consider the potential benefits of conducting related studies in pathogenic fungi.
INTRODUCTION
Two-component signal transduction (TCST) pathways regulate many aspects of bacterial life, including stress responses (1, 2), the switch from free-living to biofilm type growth (3–5), cell division (6), and the transition to stationary phase and to sporulation (7). These pathways are most abundant in bacteria with some species sporting over 300 two-component proteins (8). The prototypical bacterial two-component pathway consists of two proteins, a transmembrane sensor histidine kinase (HK) and a soluble response regulator (RR). Most sensor HKs exist in the cell membrane as dimers in which one monomer is able to phosphorylate the other in an initial stimulus-regulated autophosphorylation step (9–13). A phosphotransfer step occurs between the phosphorylated histidine in the sensor histidine kinase and a conserved aspartate within the receiver domain of the RR protein. Phosphorylation of the RR leads to a change in its activity, and an associated or downstream effector domain dictates the nature of the output response. More complex TCST pathways are known in bacteria that include, for example, hybrid proteins with both kinase and receiver domains, more than two proteins in the pathway, and multiple His-Asp phosphotransfer events (14, 15). Phosphotransfer to or from a receiver domain Asp typically involves a histidine-containing phosphotransfer (HPt) domain (15, 16). Although the number of phosphotransfer events in a pathway can vary, the phosphotransfer events in any given pathway culminate in aspartyl phosphorylation and consequent change in response regulator activity.
TCST pathways have been characterized in some detail in fungi, plants, and slime mold. The eukaryotic pathways resemble the more intricate versions of bacterial pathways. Almost all of the eukaryotic two-component pathways involve a hybrid histidine kinase with both kinase and receiver domains (17, 18). In Saccharomyces cerevisiae, the pathway involves autophosphorylation of a membrane-associated histidine kinase followed by an intramolecular phosphotransfer event between the conserved histidine in the HK domain and a conserved aspartate in an attached receiver domain (19). A second step involves transfer of the phosphoryl group on the receiver domain of the hybrid kinase to a conserved histidine on the HPt protein. The final step(s) involves phosphotransfer from the HPt protein to one or more downstream response regulator proteins. Most TCST pathways in eukaryotes have at least two response regulators, one nuclear and one cytoplasmic.
Characterization of various fungal two-component signal transduction pathways has revealed roles for these pathways in osmotic and oxidative stress responses, fungicide sensitivity, phase transition, dimorphism, secondary metabolite production, sporulation, cell wall integrity, hyphal morphogenesis, and sexual and asexual development (20). In addition, two-component pathways are important determinants of pathogenicity in animal pathogens, such as Candida albicans (21, 22), Cryptococcus neoformans (23), Penicillium marneffei (24, 25), and the endemic mycoses, Blastomyces dermatitidis and Histoplasma capsulatum (26), and plant pathogens including Fusarium oxysporum (tomato) (27, 28), Monilinia fructicola (brown rot of stone fruit) (29), Botrytis cinerea (bean, tomato, and apple) (30–32), Alternaria brassiciola (black spot disease on brassicas) (33, 34), Cochliobolus heterostrophus (maize), and Gibberella zeae (cereal) (35). The involvement of two-component pathways in bacterial and fungal pathogenesis has generated significant interest in using these pathways as targets for antimicrobial drug development. Efforts have thus far centered on the histidine kinase protein; however, the HPt and RR domains are also suitable targets, since they are absent from animal genomes.
INNOVATIONS IN TWO-COMPONENT SIGNAL TRANSDUCTION PATHWAYS TO ACCOMMODATE EUKARYOTIC CELLS
Although the basic mechanism of the His-Asp phosphotransfer pathway is conserved, the compartmentalization and larger size of the eukaryotic cell have likely selected for a multistep phosphorelay rather than the simpler two-component pathway that is most common in bacteria. Eukaryotic TCST pathways with membrane-associated sensor histidine kinases require the presence of the small free-standing HPt protein, which is capable of shuttling between the cytoplasm and nucleus (36), permitting phosphotransfer to response regulators in different compartments. Of 12,638 nonredundant GenBank sequences (June 2013) containing the HPt domain, 2,177 have the stand-alone HPt domain architecture of the S. cerevisiae protein that has been so thoroughly characterized. Although there are examples of free-standing HPt domains in bacteria (e.g., ShpA and ChpT in Caulobacter crescentus and Spo0B in Bacillus subtilis), most are associated with additional signaling domains (e.g., HK and receiver). Only 14% of bacterial HPt domains (1,729 of 12,022 HPt domain-containing sequences) and 5.5% of archaeal HPt domains (9/162) are free-standing compared to 100% of fungal (177/177), 97% of plant (233/239), and 100% of amoebozoa (5/5) (CDART [37]).
Many fungal HKs are not membrane associated and could presumably become nuclearly localized in response to certain stimuli. However, the high conservation in fungal HPt size and structure suggest that this may not be necessary, as the HPt protein can translocate more easily. It will nonetheless be of interest to experimentally determine the localization of the large number of fungal HK proteins predicted to lack transmembrane domains.
Another interesting innovation in the eukaryotic pathways is the interface between TCST proteins and other types of signal transduction pathways. The best-studied example of this is the SLN1 TCST pathway in S. cerevisiae and the HOG1 mitogen-activated protein (MAP) kinase cascade. These pathways are joined by a physical interaction between the Ssk1 RR and the mitogen-activated protein kinase kinase kinase (MAPKKK) proteins, Ssk2 and Ssk22 (38, 39). The interaction is regulated by the phosphorylation state of Ssk1 (38). The interacting domain in Ssk2 was originally defined as the region between amino acids (aa) 294 and 413 (39) and later refined to the 19 amino acid stretch between aa 361 and 380. An SSK2 mutant lacking amino acids 361 to 380 fails to respond to hyperosmotic stress (38).
Eukaryotic two-component pathways also feature distinct compartmentalization of the RR proteins. In S. cerevisiae, the Skn7 response regulator is constitutively nuclear, while the Ssk1 response regulator is cytoplasmic (36, 40, 41). This organization requires the Ypd1 protein to shuttle in and out of the cytoplasm. In Arabidopsis thaliana, the five AHP-encoded HPts were originally thought to be cytoplasmic with nuclear relocalization in response to pathway activation by cytokinin (42, 43). However, recent studies show that the Arabidopsis HPt proteins are localized to both the nucleus and cytoplasm and that this distribution is not responsive to cytokinin (44) consistent with the unregulated nucleocytoplasmic shuttling of S. cerevisiae Ypd1 (36).
EXPANSION OF THE HK AND RR GENE FAMILIES IN EUKARYOTES
In many bacterial genomes, genes of related function like the HK and RR of two-component pathways are organized in operons. This ensures a one-to-one relationship of the components of the pathway. The arrangement of functionally related genes in eukaryotic genomes is less constrained, and this may have contributed to the origin of highly expanded HK and RR gene families in different taxa.
Although the S. cerevisiae genome has a single hybrid HK gene, other fungi have expanded HK gene families with a trend toward higher numbers of HK genes in filamentous fungi. Species of the Saccharomycetes class of the Ascomycota have between 1 and 5 HK genes per genome, while the filamentous ascomycetes species have 8 to 16 per genome (18). The plant fungal pathogen Stagonospora nodorum has 19 HK genes (18). Interestingly, the expanded HK gene families in fungi are not predominantly membrane associated. For example, of 254 Ascomycota HK proteins listed in the SMART database (February 2012), only 25 are predicted to have transmembrane (TM) domains, 14 have a single TM domain, and 11 have two, like the S. cerevisiae Sln1 HK. The expansion of the HK gene family may relate to the need for novel sensory activities related to pathogenesis. However, both the nonpathogenic Aspergillus oryzae used in the production of soy sauce, miso, and sake and the pathogenic Aspergillus flavus associated with aspergillosis of the lungs have 14 HK genes (SMART genome database in February 2012).
In contrast, higher plant genomes have a modest HK gene family size with 8 each in Arabidopsis thaliana and Oryzae sativum and 5 in Chlamydomonas reinhardtii, but a highly expanded RR gene family. While most fungal genomes include between 1 and 5 highly conserved RR genes, the Arabidopsis genome has 23 RR-encoding genes and O. sativum has 28, although some of these genes are encoding pseudo-RRs (18, 45), which contain receiver domains but lack key residues required for activity.
Perhaps consistent with the profusion of RRs, higher plants also contain more than one HPt-encoding gene. Arabidopsis contains 5 HPt genes, and O. sativum has 2. Plant genomes are also known to include divergent HPt genes, which are presumably incapable of phosphotransfer (18). The imbalance in the size of the HK and HPt versus the RR gene families suggests that plant RRs may mediate non-HK as well as HK-HPt signals.
TAXONOMIC DISTRIBUTION OF HPt GENES
YPD1 orthologs are found in numerous fungal genomes from Ascomycota and Basidiomycota and in the more basal Chytridiomycota (Gonapodya prolifera but not Batrachochytrium dendrobatidis). In addition, a set of paralogous genes were identified in the Rhizopus delemar genome from Zygomycota (see Fig. 2 and Table 2). No orthologs were found in available Microsporidia or Neocallimastigomycota genomes. HPt-encoding genes have also been identified in nonfungal genomes, including plants, Dictyostelium, green algae, and diatoms (18, 46). Recent dramatic growth in genomic databases has resulted in the occasional potential misannotation of genes in sporadic animal taxa as belonging to the family of two-component regulators. These misannotations are most common in early assemblies in which short contigs may be maintained until they can be definitively ascribed to contaminating bacterial sequences but could also be attributable to rare horizontal gene transfer events between eukaryotic lineages (47).
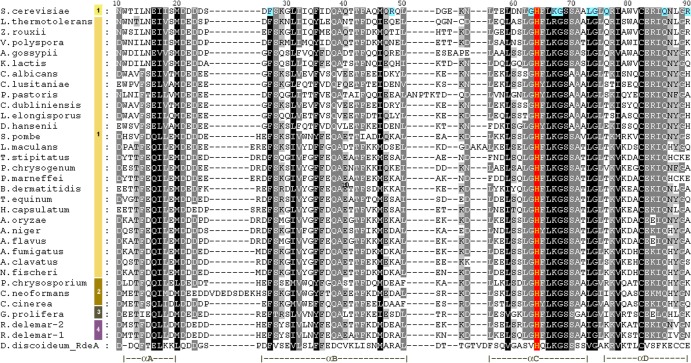
Fig 2.
Multiple-sequence alignment of the HPt domain of diverse fungal Ypd1 orthologs. Orthologs were retrieved by BLASTp analysis using the National Center for Biotechnology Information (NCBI) and Joint Genome Institute (JGI) web servers and confirmed when possible using orthology calls from the Ensembl Fungi database and phylogenetic analysis (not shown). The full species names, NCBI gi numbers, other accession numbers, and systematic gene names are provided in Table 2. Complete peptide sequences were aligned using MUSCLE (93). Aligned sequences were imported into GeneDoc (94). Four levels of conservation with default conservation groups enabled were used and are indicated as follows: 100% conservation, black background; 80% conservation, gray background with white lettering; 60% conservation, gray background with black lettering; less than 60% conservation, white background). The image was then imported into Adobe Photoshop for further annotation. Taxonomic representation is indicated using color and numerical codes shown to the left of the sequence alignment: S. cerevisiae (light yellow 1); other Ascomycetes (dark yellow 1); Basidiomycetes (mustard 2), Chytrids (brown 3); Zygomycetes (magenta 4). The numbering across the top corresponds to the residues in the S. cerevisiae Ypd1 protein. The conserved phosphoaccepting histidine (H64 in S. cerevisiae) is shown using a red background. Additional residues in the S. cerevisiae protein that have been genetically or biochemically characterized and are discussed in the text are shown on a blue background. Secondary structure elements derived from the S. cerevisiae Ypd1 protein structures (Protein Data Bank [PDB] identifications [IDs] 1QSP and 1CO2) (95, 96) are shown across the bottom of the alignment. Gaps introduced to maximize alignment are indicated by dashes.
Table 2.
Sequence identifiers and features
| Species | gi | Other identifier (GenBank, EMBL, or Refseq) | Gene | Length (aa) of protein | Hpt domain (aa) |
|---|---|---|---|---|---|
| Fungal species | |||||
| Ashbya gossypii ATCC 10895 | 44980684 | AAS50589.1 | ABL182Cp | 138 | 25–90 |
| Aspergillus clavatus NRRL 1 | 121707662 | XP_001271903.1 | ACLA_049490 | 168 | 85–109 |
| Aspergillus flavus NRRL3357 | 220699018 | EED55357.1 | AFLA_026290 | 166 | 87–111 |
| Aspergillus fumigatus A1163 | 159125284 | EDP50401.1 | AFUB_067390 | 171 | 61–143 |
| Aspergillus niger CBS 513.88 | 317038304 | XP_001402021.2 | ANI_1_1060184 | 162 | 59–109 |
| Aspergillus oryzae RIB40 | 317144662 | XP_001820278.2 | AOR_1_2120154 | 166 | 87–111 |
| Blastomyces dermatitidis | 261205810 | XP_002627642.1 | BDBG_02313 | 165 | 59–145 |
| Coprinopsis cinerea Okayama 7#130 | 299743631 | XP_002910687.1 | CC1G_15018 | 229 | 99–178 |
| Candida albicans | 9886962 | AF213247_1 | CaYPD1 | 184 | 35–108 |
| Candida dubliniensis CD36 | 223640675 | CAX44979.1 | CD36_06780 | 183 | 35–108 |
| Clavispora lusitaniae | 170877388 | ACB38709.1 | C1YPD1 | 148 | 38–109 |
| Cryptococcus neoformans | 58262068 | XP_568444.1 | CNM01530 | 209 | 110–160 |
| Debaryomyces hansenii CBS767 | 50419265 | XP_458156.1 | DEHA2C10890p | 146 | 35–97 |
| Gonapodya prolifera | JGI-128095 | 175 | 53–133 | ||
| Histoplasma capsulatum | 240281859 | 240281859 | HCDG_00941 | 161 | 33–115 |
| Kluyveromyces lactis NRRL Y-1140 | 50307853 | XP_453920.1 | KLLA0D19338g | 135 | 32–121 |
| Lachancea thermotolerans | 255711708 | XP_002552137.1 | KLTH0B08030p | 138 | 33–124 |
| Leptosphaeria maculans | 312213483 | CBX93565.1 | LEMA_P044660.1 | 141 | 32–95 |
| Lodderomyces elongisporus NRRL YB-4239 | 149247273 | XP_001528049.1 | LELG_00569 | 243 | 37–112 |
| Neosartorya fischeri NRRL 181 | 119500460 | XP_001266987.1 | NFIA_105780 | 171 | 61–143 |
| Penicillium chrysogenum Wisconsin 54-1255 | 211592212 | CAP98539.1 | Pc22g12510 | 201 | 51–133 |
| Penicillium marneffei ATCC 18224 | 212542051 | XP_002151180.1 | PMAA_040370 | 177 | 54–118 |
| Phanerochaete chrysosporium | 132047 | e_gww2.9.411.1 | 145 | 12–92 | |
| Pichia pastoris CBS 7435 | 328351330 | CCA37729.1 | PP7435_Chr2-0030 | 158 | 42–102 |
| Rhizopus oryzae (delemar)-1 RA 99-880 | 384499124 | EIE89615.1 | RO3G_14326 | 168 | 68–147 |
| Rhizopus oryzae (delemar)-2 RA 99-880 | 384485568 | EIE77748 | RO3G_02452 | 165 | 40–120 |
| Saccharomyces cerevisiae | 6319966 | NP_010046.1 | ScYPD1 | 167 | 31–108 |
| Schizosaccharomyces pombe | 3925752 | CAA22174.1 | MPR1 | 295 | 187–267 |
| Talaromyces stipitatus ATCC 10500 | 218725057 | EED24474.1 | TSTA_078330 | 179 | 56–136 |
| Trichophyton equinum CBS 127.97 | 326480990 | EGE05000.1 | TEQG_03843 | 152 | 51–133 |
| Vanderwaltozyma polyspora DSM 70294 | 156845926 | XP_001645852.1 | Kpol_1054p41 | 137 | 30–95 |
| Zygosaccharomyces rouxii | 238940750 | CAR28924.1 | ZYRO0F15114p | 163 | 32–108 |
| Nonfungal species | |||||
| Dictyostelium discoideum | 3513528 | AAC61850.1 | RDEA | 254 | 32–112 |
BIOLOGY OF THE FUNGAL HPt PROTEIN Ypd1
S. cerevisiae YPD1 is an essential gene required for transmission of the Sln1 HK signal to the Ssk1 RR. Signaling to Ssk1 is necessary for viability because Ssk1∼P is needed to suppress lethal activation of the Hog1 MAPK pathway under normal osmotic conditions (48). Ypd1 also transmits the Sln1 HK signal to the Skn7 RR; however, viability of the nonphosphoaccepting skn7D427N mutant and the skn7Δ mutants (40, 49) indicate that loss of this activity is not lethal.
Several fungal YPD1 orthologs have been shown to complement the lethal phenotype of the S. cerevisiae ypd1 mutant, including the Schizosaccharomyces pombe MPR1 gene (also known as SPY1), and the C. albicans YPD1 gene, while H/Q nonphosphorylatable mutants of YPD1 orthologs fail to complement (50, 51). This type of in vivo data together with the results of in vitro phosphorelay reconstitution experiments (52, 53) indicates that YPD1 orthologs share the TCST phosphotransfer function at the biochemical level. Given the many documented functions of TCST pathways, the YPD1 orthologs are likely to differ biologically. For example, although the S. cerevisiae and C. neoformans YPD1 genes are essential (19, 54), both the S. pombe and Dictyostelium genes are dispensable for viability (50, 53, 55, 56). The nonessential S. pombe Mpr1 HPt transmits oxidative stress signals to a MAPK cascade (55, 57, 58) and plays a role in regulation of the G2/M cell cycle progression (50) in contrast to the osmotic stress-sensing function of S. cerevisiae Ypd1. Table 1 summarizes the phenotypes of fungal hpt mutants. In general, two-component pathways involved in osmoregulation via the Hog1 MAPK signaling have an essential function.
Table 1.
Phenotypes of fungal hpt mutants
| Organism | HPt gene | Deletion phenotype | In vivo phenotype of missense mutant | In vivo phenotype of ypd1 (hpt) hog1 pathway double mutant | Complementation of S. cerevisiae ypd1 deletion strain in vivo or in vitro |
|---|---|---|---|---|---|
| S. cerevisiae | YPD1 | Inviable (19) | Pradimicin resistance (69) | Not applicable | |
| Conditional lethality (Fassler, unpublished) | |||||
| SLN1-SKN7 pathway activation (Fassler, unpublished) | |||||
| Inviable (Fassler, unpublished) | |||||
| N. crassa | hpt-1 | Inviable (59) | Suppression of the osmotic stress sensitivity of os mutants (59) | No information | |
| Iprodione resistance equivalent to os mutant (97) | |||||
| S. pombe | mpr1/spy1 | Viable (50, 55) | Defective oxidative stress signaling (55) | Wild type and mpr1ΔN167 complement in vivo and in vitro (53) | |
| Precocious entry into M phase (50) | |||||
| C. albicans | CaYPD1 | Viable (98) | In vivo complementation by wild type but not H69Q (51) | ||
| A. nidulans | ypdA | Inviable (60, 99) | No mutant analysis reported | Reconstitution of phosphorelay from YpdA to RR (52) |
Consistent with the observation of Hog1-independent as well as the essential Hog1-dependent role for Ypd1 signaling known in S. cerevisiae, ypd1 hypomorphs in other fungi exhibit complex phenotypes. For example, Cryptococcus ypd1Δ hog1Δ strains, viable because the hog1 mutation prevents the lethal effects of inappropriate Hog1 pathway activation, exhibit elevated levels of diamide resistance relative to hog1, ssk1, or skn7 mutants (54). This suggests that Hog1-independent Ypd1-mediated signaling in Cryptococcus may involve a third RR or some other type of signaling. Analysis of viable Neurospora crassa hpt os-2 double mutants likewise revealed reduced osmotic stress sensitivity and increased oxidative stress sensitivity compared to the os-2 MAPK mutant, suggesting complex regulation of stress responses that involve both TCST-dependent and TCST-independent regulation (59). Finally, reduction in Aspergillus nidulans ypdA function, evaluated in ypdAΔ/ypdA+ heterokaryons, caused reduced viability and increased sensitivity to osmotic stress (60), although the dependence of these phenotypes on a downstream MAPK has not been directly tested.
Fungal TCST proteins are important for pathogenesis in both plant and animal fungal pathogens. Many HK genes have been implicated (21, 28, 29, 32, 35, 61); however, the role that His-Asp phosphotransfer plays in pathogenesis has not been clearly established. In the animal pathogen C. neoformans, loss of YPD1 function leads to changes in melanin production, a major virulence factor (54). Additional experiments directly testing the role of Ypd1 or the phosphorylated histidine and phosphoaccepting aspartate in HKs or RRs in plant or animal virulence are needed to further establish the requirement for TCST pathway activity in fungal pathogenesis.
STRUCTURAL FEATURES OF Ypd1 PROTEINS
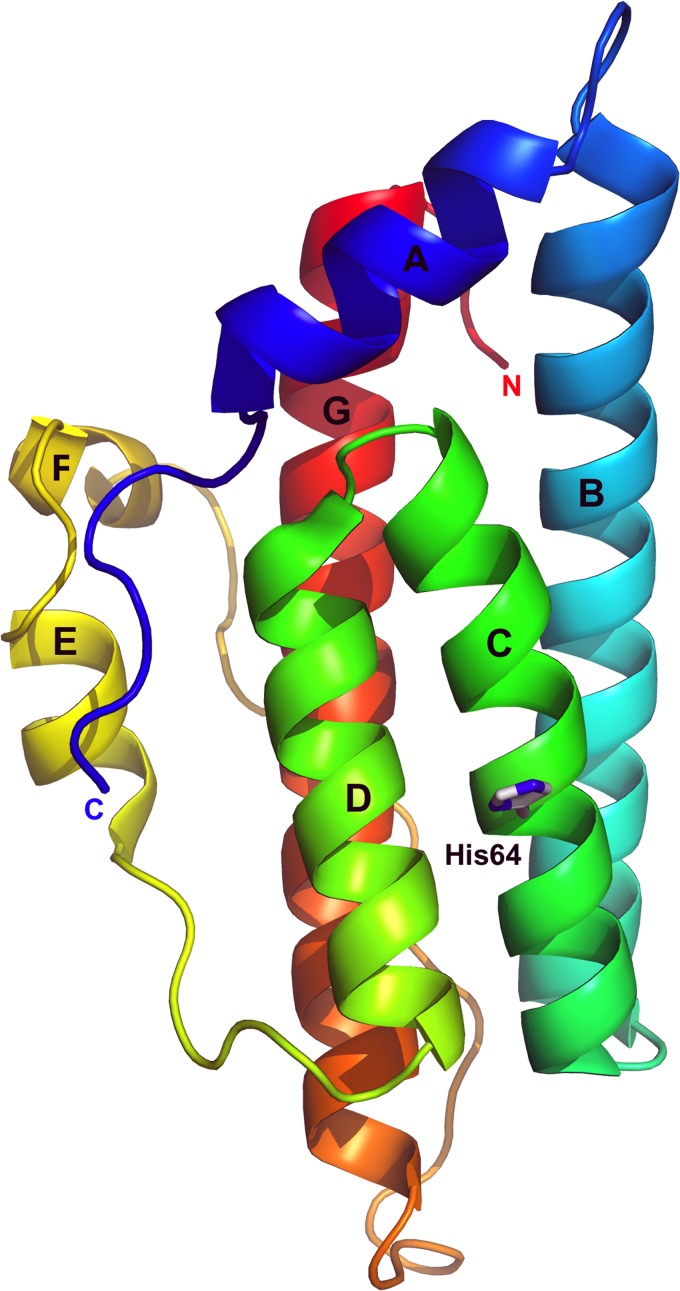
S. cerevisiae Ypd1 is an all-helical protein with six α-helices and a seventh more compact single-turn 310-helix (designated A to G) (Fig. 1) (62). The HPt domain contains a four-helix bundle (αB-αC-αD-αG) as a minimal core structure. The αC-αD helical hairpin motif of Ypd1 with its centrally located and solvent-exposed histidine is an important conserved structural feature of HPt domains.
Fig 1.
Ribbon representation of Ypd1. The core of the Ypd1 molecule is a compact four-helical bundle composed of the α-helix B (αB), αC, αD, and αG helices. Residues from Ypd1 that make contact with Sln1 come from helices αA, αB, αC, and αD (66, 91). The H64 side chain of Ypd1 that is involved in phosphotransfer is shown as a stick model.
Proteins encoded by the fungal YPD1 gene family range in size from 137 to 295 aa (Table 2). The S. cerevisiae protein is 167 aa. Ypd1 orthologs in nonfungal eukaryotes are similarly compact. Dictyostelium RdeA is 254 aa, and the Arabidopsis Ahp2 protein is 156 aa. Most ascomycetes HPt proteins have short N-terminal extensions of less than 100 aa prior to the start of the HPt domain. S. pombe Mpr1 is an exception with an N-terminal extension of 186 aa. Among the basidiomycete HPts, Phanerochaete chrysosporium and Postia placenta have relatively short N-terminal extensions, while the C. neoformans and Puccinia graminis HPt proteins have longer extensions of 109 and 182 aa, respectively. The role of this N-terminal domain has not been thoroughly investigated, although yeast two-hybrid interaction data suggest that the N-terminal region of the S. pombe Mpr1 protein is involved in the interaction with RR receiver domains (53).
The sequences of the most conserved portion of the HPt domains corresponding to the fungal Ypd1 orthologs listed in Table 2 were aligned (Fig. 2). Gaps in the sequence alignment fall at the edges or between known secondary structure elements, thus validating the alignment. The C. neoformans sequence introduces a small insertion between helices A and B, while Pichia pastoris and others introduce small insertions between helices B and C. The spacing between helices C and D is completely conserved, suggesting that the spatial relationship between these two helices or between these helices and other parts of the protein or its interactors may be functionally important.
STRUCTURE-FUNCTION ANALYSIS OF Ypd1
Based on structure models of the S. cerevisiae Ypd1 protein, a variety of Ypd1 residues in the vicinity of the phosphorylatable H64 are predicted to have important roles in phosphoryl transfer to and from Ypd1. Many of these residues have been functionally characterized (63). For example, an alanine substitution of the highly conserved K67 residue, located one turn of the helix away from H64 in helix C, revealed that K67 is important for efficient histidyl phosphorylation and for His∼P stability (64, 65). K67 is conserved in all aligned fungal Ypd1 orthologs as well as in the HPt from the bacterium Anaerofustis stercorihominis, Arabidopsis Ahp2, and Dictyostelium RdeA (Fig. 2) and is expected to function similarly in all HPt proteins.
In contrast, the positively charged R90 residue in helix D, postulated to be involved in stabilizing the antiparallel arrangement of helices C and D via ionic interactions, was found to have a modest effect on the levels of Ypd1 phosphorylation and a twofold decrease in the stability of the phospho-imidazole linkage but no effect on phosphorelay efficiency (64, 65). Its interaction with Sln1 is normal, and the interactions with Ssk1 and Skn7 are only slightly compromised (66). Interestingly, this position is not conserved among fungi; only 7 of the 32 fungal species in the alignment have K or R at this position (Fig. 2), suggesting that the positive charge at this position may be one of several possible mechanisms for stabilizing helices C and D in fungal Ypd1 proteins.
Analysis of the G68Q substitution mutant confirmed that the small size of glycine at position 68, just 4 residues downstream of H64, is important for access by receiver domains to the H64 residue. The G68Q Ypd1 protein exhibits reduced levels of phosphorylation (63), severe inhibition in receiver domain interactions (66), and no detectable phosphotransfer (64, 65). G68 is conserved in all fungal Ypd1 proteins (Fig. 2). G63, a second conserved glycine adjacent to H64, is also conserved in fungi (Fig. 2). Although the G63 residue has not yet been characterized, the conservation of glycines flanking H64 may suggest that the functionality of a fungal HPt protein requires this negative space/pocket to facilitate its phosphorylation.
The Ypd1-receiver domain interaction surface was also interrogated by alanine-scanning mutagenesis (66, 67) (Table 3). Each mutation was tested for its ability to interact with the Sln1, Ssk1, or Skn7 receiver domain in two-hybrid experiments. A core set of 10 surface residues, including E16, M20, D21, F27, L31, D60, F65, G68, S69, and L63, mapping to helices A, B, and C, were found to be required for all receiver domain interactions. This residue cluster forms a classic hydrophobic binding site for RR docking. Flanking the core set of 10 surface residues are additional residues involved in interactions with specific receiver domains. These residues map to the distal part of helix C and the proximal part of helix D as well as to helices A and B. It remains to be determined which residues dictate the observed Ypd1 phosphotransfer bias for the Ssk1 versus Skn7 RR (68; A. H. West, unpublished data).
Table 3.
Structure-function characterization of S. cerevisiae Ypd1
| Helixa | Residueb | Fungal conservation (%)c | Receiver domain (R1, R2, and R3d) interactionse |
|---|---|---|---|
| αA | T12 | X | + + + |
| I13 | I, V, T ✓ | + − − | |
| E16 | E or Q | − − − | |
| S19 | X | + + + | |
| M20 | M or L ✓ | − − − | |
| D21 | ✓ | − − − | |
| D23 | D or E ✓ | + + − | |
| D24 | D or E ✓ | + − − | |
| αB | F27 | ✓ | − − − |
| L31 | L, I, M ✓ | − − − | |
| Q38 | ✓ | + − − | |
| Q45 | X | + + + | |
| R48 | X | + + + | |
| E53 | X | + + + | |
| αC | N55 | D, N (EK) ✓ | + + + |
| T57 | X | + + + | |
| D60 | S/T (75) | − − − | |
| N61 | S (56) | + + + | |
| H64f | ✓ | + + + | |
| F65 | F or Y ✓ | − − − | |
| K67 | K or R ✓ | + + − | |
| G68Q | ✓ | − − − | |
| S69 | ✓ | − − − | |
| S70 | S or A ✓ | + + − | |
| L73 | ✓ | − − − | |
| G74C | ✓ | Not tested | |
| αD | Q76 | S/T (50) | + − − |
| W80 | X | − + − | |
| E83 | ✓ | − + − | |
| Q86 | ✓ | + + + | |
| R90 | X | + + + |
Shaded rows indicate residues residing in the designated helix. αA, α-helix A.
Residue numbering and identity are based on the S. cerevisiae protein. The residue was substituted with alanine except where indicated.
Conservation was evaluated for 32 fungal species listed in the alignment in Fig. 2. The amino acids conserved in fungi are shown. The percent conservation is shown in parentheses. “X” indicates that the residue is not conserved (<25%), a check mark indicates the residue is completely conserved (100%), and a check mark next to a set of residues indicates there is more than one conserved residue at that position.
R1, R2, and R3 refer to the Sln1, Ssk1, and Skn7 receiver domains, respectively.
H64A was used in the interaction assays, and H64Q was used in the biochemical assays.
Several substitution mutants were tested for their effect on signaling. Point mutations in YPD1 might cause a partial decrease in phosphotransfer (complete inactivation is lethal). Alternatively, such mutations could cause an increase in phosphotransfer. The G74C mutation was isolated in a directed mutagenesis screen for mutants resistant to the fungicidal compound pradimicin A (69). Glycine 74 is located in a three-residue reverse turn that connects helix C and helix D and is postulated to be important for structural integrity of the protein. The G74 residue is highly conserved in Ypd1 orthologs (Table 3 and Fig. 2). Substitution of G74 to C was postulated to alter Ypd1 structure, causing resistance to pradimicin and sensitivity to osmotic stress (69). In vivo SLN1 pathway reporter gene assays using a SKN7-dependent lacZ reporter showed that this mutation increased signaling. Thus, the G74C mutation enhances Ypd1 phosphotransfer activity rather than diminishing it (J. S. Fassler and Y. Igarashi, unpublished results). The osmosensitivity of this YPD1 mutant is consistent with previous observations of osmosensitivity in SLN1 mutants (sln1*) that increase signaling (70) and is presumably due to reduction of the dephosphorylated form of Ssk1 required for activation of the Ssk2/22 MAPKKKs in the Hog1 MAPK pathway. Pradimicin resistance may likewise be a function of loss of Ssk1 signaling to Hog1 and to Ssk1-dependent cell death pathways.
Several additional ypd1 mutants were tested for their viability, pradimicin resistance and signaling phenotypes. Of these ypd1 mutants, the F27A and L73A mutants were viable and exhibited both pradimicin resistance and pathway activation. Other mutants tested (R48A, Q76A, and Q86A) were pradimicin sensitive and showed no pathway activation (J. S. Fassler, unpublished results). Interestingly, mutants exhibiting activation phenotypes were defective in all (three) receiver domain interactions. Hence, the observed changes in activity do not correlate well with two-hybrid experiments (66, 67). It will be of interest to structurally characterize the Ypd1 protein from one or more of the activated mutants.
PROSPECTS FOR Ypd1 AS AN ANTIFUNGAL DRUG TARGET
Previous screens for inhibitors of bacterial TCST pathways have been based on detection of the autophosphorylated HK protein or the phosphorylated RR protein (71–74). Of the inhibitors identified in this way, some failed to inhibit growth of microbial test strains (74), and others were found to be inadequate due to lack of specificity (75). Recent TCST-directed antibacterial efforts have focused more on the kinase sensory domain and the response regulator domain (76). In fungi, however, simpler high-throughput screens for HK inhibitors have been recently reported. One screen was based on the potential for growth inhibition due to activation of the HOG1 osmotic response MAPK pathway by inhibitors of group III HKs from C. albicans expressed heterologously in S. cerevisiae (77). In this study, several known antifungals were used successfully in proof-of-principle experiments (77). In a related screen, small molecules were tested for fungicidal activity against an S. cerevisiae reporter strain expressing a group III HK from Magnaporthe grisea. Here, compounds with broad antifungal activity were identified, but these compounds were ultimately shown to be HK independent in their effects (78).
Due to the dearth of safe and effective antifungal drugs, both natural and synthetic peptides have been proposed as new antifungal agents (79, 80). Peptides are a promising class of antifungal agent because they work rapidly with high specificity and can be used in combination with other therapeutic agents. Naturally occurring peptides exhibiting anticryptococcal activity include the membrane active class of antimicrobial peptides (AMP) (81, 82), the human salivary MUC7 mucin peptides (83, 84), and the cationic antimicrobial peptides (85, 86). Synthetic peptides with anticryptococcal activity have also been reported (87, 88). Several examples of protein interaction surfaces that have been specifically targeted with inhibitory peptides are described in recent reviews (89, 90). In each case, the inhibitors were short peptides derived from one of the binding partners.
Structural information and data from the in vivo and in vitro characterization of mutant proteins could be used to rationally design biologically relevant peptides that would inhibit phosphorelay pathways in fungal pathogens like C. neoformans. The interactions between the Ypd1 HPt and receiver domain-containing proteins in the SLN1 pathway are essential for viability, and inhibitors that disturb these interactions are predicted to have potential as antifungal drug leads. Cryptococcus Ypd1 is an excellent target for antifungal drug design because Ypd1 is a central molecule in fungal TCST pathways and because reduction in Ypd1 activity is expected to compromise fungal fitness, virulence, and viability.
CONCLUSIONS
The structurally and genetically well-characterized HPt from S. cerevisiae exhibits many attributes of a useful antifungal drug target. It is essential in at least some fungal pathogens, it is a unique, nonredundant protein in all fungal TCST pathways, and it plays an important role in fungal pathogenesis. While the HPt is nonenzymatic, it is nonetheless possible to interfere with the protein interactions that are required for its activity. In S. cerevisiae and in other fungi for which TCST control of the HOG1 MAPK osmotic response pathway is known, Ypd1 interactions with the upstream HK receiver domain from which it receives a phosphoryl group and with the downstream Ssk1 RR to which it donates a phosphoryl group will be essential. The existing cocrystal structures of Ypd1-receiver domain complexes (91, 92) could facilitate the design of such inhibitors.
ACKNOWLEDGMENTS
A.H.W. gratefully acknowledges support from the NSF (MCB1158319) and the Oklahoma Center for the Advancement of Science and Technology (HR12-059).
Footnotes
Published ahead of print 14 June 2013
REFERENCES
- 1.MacRitchie DM, Buelow DR, Price NL, Raivio TL. 2008. Two-component signaling and gram negative envelope stress response systems. Adv. Exp. Med. Biol. 631:80–110 [DOI] [PubMed] [Google Scholar]
- 2.Whistler CA, Corbell NA, Sarniguet A, Ream W, Loper JE. 1998. The two-component regulators GacS and GacA influence accumulation of the stationary-phase sigma factor sigmaS and the stress response in Pseudomonas fluorescens Pf-5. J. Bacteriol. 180:6635–6641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Martinez-Wilson HF, Tamayo R, Tischler AD, Lazinski DW, Camilli A. 2008. The Vibrio cholerae hybrid sensor kinase VieS contributes to motility and biofilm regulation by altering the cyclic diguanylate level. J. Bacteriol. 190:6439–6447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hickman JW, Tifrea DF, Harwood CS. 2005. A chemosensory system that regulates biofilm formation through modulation of cyclic diguanylate levels. Proc. Natl. Acad. Sci. U. S. A. 102:14422–14427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cotter PA, Stibitz S. 2007. c-di-GMP-mediated regulation of virulence and biofilm formation. Curr. Opin. Microbiol. 10:17–23 [DOI] [PubMed] [Google Scholar]
- 6.Ohta N, Lane T, Ninfa EG, Sommer JM, Newton A. 1992. A histidine protein kinase homologue required for regulation of bacterial cell division and differentiation. Proc. Natl. Acad. Sci. U. S. A. 89:10297–10301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hoch JA. 1993. Regulation of the phosphorelay and the initiation of sporulation in Bacillus subtilis. Annu. Rev. Microbiol. 47:441–465 [DOI] [PubMed] [Google Scholar]
- 8.Ulrich LE, Zhulin IB. 2010. The MiST2 database: a comprehensive genomics resource on microbial signal transduction. Nucleic Acids Res. 38:D401–D407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ninfa EG, Atkinson MR, Kamberov ES, Ninfa AJ. 1993. Mechanism of autophosphorylation of Escherichia coli nitrogen regulator II (NRII or NtrB): trans-phosphorylation between subunits. J. Bacteriol. 175:7024–7032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Swanson RV, Bourret RB, Simon MI. 1993. Intermolecular complementation of the kinase activity of CheA. Mol. Microbiol. 8:435–441 [DOI] [PubMed] [Google Scholar]
- 11.Wolfe AJ, Stewart RC. 1993. The short form of the CheA protein restores kinase activity and chemotactic ability to kinase-deficient mutants. Proc. Natl. Acad. Sci. U. S. A. 90:1518–1522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yang Y, Inouye M. 1993. Requirement of both kinase and phosphatase activities of an Escherichia coli receptor (Taz1) for ligand-dependent signal transduction. J. Mol. Biol. 231:335–342 [DOI] [PubMed] [Google Scholar]
- 13.McEvoy MM, Dahlquist FW. 1997. Phosphohistidines in bacterial signaling. Curr. Opin. Struct. Biol. 7:793–797 [DOI] [PubMed] [Google Scholar]
- 14.Alex LA, Simon MI. 1994. Protein histidine kinases and signal transduction in prokaryotes and eukaryotes. Trends Genet. 10:133–138 [DOI] [PubMed] [Google Scholar]
- 15.Stock AM, Robinson VL, Goudreau PN. 2000. Two-component signal transduction. Annu. Rev. Biochem. 69:183–215 [DOI] [PubMed] [Google Scholar]
- 16.Tsuzuki M, Ishige K, Mizuno T. 1995. Phosphotransfer circuitry of the putative multi-signal transducer, ArcB, of Escherichia coli: in vitro studies with mutants. Mol. Microbiol. 18:953–962 [DOI] [PubMed] [Google Scholar]
- 17.Santos JL, Shiozaki K. 2001. Fungal histidine kinases. Sci. STKE 2001(98):re1. [DOI] [PubMed] [Google Scholar]
- 18.Schaller GE, Shiu SH, Armitage JP. 2011. Two-component systems and their co-option for eukaryotic signal transduction. Curr. Biol. 21:R320–R330 [DOI] [PubMed] [Google Scholar]
- 19.Posas F, Wurgler-Murphy SM, Maeda T, Witten EA, Thai TC, Saito H. 1996. Yeast HOG1 MAP kinase cascade is regulated by a multistep phosphorelay mechanism in the SLN1-YPD1-SSK1 “two-component” osmosensor. Cell 86:865–875 [DOI] [PubMed] [Google Scholar]
- 20.Li D, Agrellos OA, Calderone R. 2010. Histidine kinases keep fungi safe and vigorous. Curr. Opin. Microbiol. 13:424–430 [DOI] [PubMed] [Google Scholar]
- 21.Selitrennikoff CP, Alex L, Miller TK, Clemons KV, Simon MI, Stevens DA. 2001. COS-l, a putative two-component histidine kinase of Candida albicans, is an in vivo virulence factor. Med. Mycol. 39:69–74 [DOI] [PubMed] [Google Scholar]
- 22.Kruppa M, Krom BP, Chauhan N, Bambach AV, Cihlar RL, Calderone RA. 2004. The two-component signal transduction protein Chk1p regulates quorum sensing in Candida albicans. Eukaryot. Cell 3:1062–1065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bahn YS, Kojima K, Cox GM, Heitman J. 2006. A unique fungal two-component system regulates stress responses, drug sensitivity, sexual development, and virulence of Cryptococcus neoformans. Mol. Biol. Cell 17:3122–3135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Boyce KJ, Schreider L, Kirszenblat L, Andrianopoulos A. 2011. The two-component histidine kinases DrkA and SlnA are required for in vivo growth in the human pathogen Penicillium marneffei. Mol. Microbiol. 82:1164–1184 [DOI] [PubMed] [Google Scholar]
- 25.Wang F, Tao J, Qian Z, You S, Dong H, Shen H, Chen X, Tang S, Ren S. 2009. A histidine kinase PmHHK1 regulates polar growth, sporulation and cell wall composition in the dimorphic fungus Penicillium marneffei. Mycol. Res. 113:915–923 [DOI] [PubMed] [Google Scholar]
- 26.Nemecek JC, Wuthrich M, Klein BS. 2006. Global control of dimorphism and virulence in fungi. Science 312:583–588 [DOI] [PubMed] [Google Scholar]
- 27.Ochiai N, Tokai T, Nishiuchi T, Takahashi-Ando N, Fujimura M, Kimura M. 2007. Involvement of the osmosensor histidine kinase and osmotic stress-activated protein kinases in the regulation of secondary metabolism in Fusarium graminearum. Biochem. Biophys. Res. Commun. 363:639–644 [DOI] [PubMed] [Google Scholar]
- 28.Rispail N, Di Pietro A. 2010. The two-component histidine kinase Fhk1 controls stress adaptation and virulence of Fusarium oxysporum. Mol. Plant Pathol. 11:395–407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ma Z, Luo Y, Michailides T. 2006. Molecular characterization of the two-component histidine kinase gene from Monilinia fructicola. Pest Manag. Sci. 62:991–998 [DOI] [PubMed] [Google Scholar]
- 30.Cui W, Beever RE, Parkes SL, Weeds PL, Templeton MD. 2002. An osmosensing histidine kinase mediates dicarboximide fungicide resistance in Botryotinia fuckeliana (Botrytis cinerea). Fungal Genet. Biol. 36:187–198 [DOI] [PubMed] [Google Scholar]
- 31.Oshima M, Fujimura M, Banno S, Hashimoto C, Motoyama T, Ichiishi A, Yamaguchi I. 2002. A point mutation in the two-component histidine kinase BcOS-1 gene confers dicarboximide resistance in field isolates of Botrytis cinerea. Phytopathology 92:75–80 [DOI] [PubMed] [Google Scholar]
- 32.Viaud M, Fillinger S, Liu W, Polepalli JS, Le Pecheur P, Kunduru AR, Leroux P, Legendre L. 2006. A class III histidine kinase acts as a novel virulence factor in Botrytis cinerea. Mol. Plant Microbe Interact. 19:1042–1050 [DOI] [PubMed] [Google Scholar]
- 33.Cho Y, Kim KH, La Rota M, Scott D, Santopietro G, Callihan M, Mitchell TK, Lawrence CB. 2009. Identification of novel virulence factors associated with signal transduction pathways in Alternaria brassicicola. Mol. Microbiol. 72:1316–1333 [DOI] [PubMed] [Google Scholar]
- 34.Dry IB, Yuan KH, Hutton DG. 2004. Dicarboximide resistance in field isolates of Alternaria alternata is mediated by a mutation in a two-component histidine kinase gene. Fungal Genet. Biol. 41:102–108 [DOI] [PubMed] [Google Scholar]
- 35.Oide S, Liu J, Yun SH, Wu D, Michev A, Choi MY, Horwitz BA, Turgeon BG. 2010. Histidine kinase two-component response regulator proteins regulate reproductive development, virulence, and stress responses of the fungal cereal pathogens Cochliobolus heterostrophus and Gibberella zeae. Eukaryot. Cell 9:1867–1880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lu JM, Deschenes RJ, Fassler JS. 2003. Saccharomyces cerevisiae histidine phosphotransferase Ypd1p shuttles between the nucleus and cytoplasm for SLN1-dependent phosphorylation of Ssk1p and Skn7p. Eukaryot. Cell 2:1304–1314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Geer LY, Domrachev M, Lipman DJ, Bryant SH. 2002. CDART: protein homology by domain architecture. Genome Res. 12:1619–1623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Horie T, Tatebayashi K, Yamada R, Saito H. 2008. Phosphorylated Ssk1 prevents unphosphorylated Ssk1 from activating the Ssk2 mitogen-activated protein kinase kinase kinase in the yeast high-osmolarity glycerol osmoregulatory pathway. Mol. Cell. Biol. 28:5172–5183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Posas F, Saito H. 1998. Activation of the yeast SSK2 MAP kinase kinase kinase by the SSK1 two-component response regulator. EMBO J. 17:1385–1394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Brown JL, Bussey H, Stewart RC. 1994. Yeast Skn7p functions in a eukaryotic two-component regulatory pathway. EMBO J. 13:5186–5194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Raitt DC, Johnson AL, Erkine AM, Makino K, Morgan B, Gross DS, Johnston LH. 2000. The Skn7 response regulator of Saccharomyces cerevisiae interacts with Hsf1 in vivo and is required for the induction of heat shock genes by oxidative stress. Mol. Biol. Cell 11:2335–2347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hwang I, Sheen J. 2001. Two-component circuitry in Arabidopsis cytokinin signal transduction. Nature 413:383–389 [DOI] [PubMed] [Google Scholar]
- 43.Tanaka Y, Suzuki T, Yamashino T, Mizuno T. 2004. Comparative studies of the AHP histidine-containing phosphotransmitters implicated in His-to-Asp phosphorelay in Arabidopsis thaliana. Biosci. Biotechnol. Biochem. 68:462–465 [DOI] [PubMed] [Google Scholar]
- 44.Punwani JA, Hutchison CE, Schaller GE, Kieber JJ. 2010. The subcellular distribution of the Arabidopsis histidine phosphotransfer proteins is independent of cytokinin signaling. Plant J. 62:473–482 [DOI] [PubMed] [Google Scholar]
- 45.Makino S, Kiba T, Imamura A, Hanaki N, Nakamura A, Suzuki T, Taniguchi M, Ueguchi C, Sugiyama T, Mizuno T. 2000. Genes encoding pseudo-response regulators: insight into His-to-Asp phosphorelay and circadian rhythm in Arabidopsis thaliana. Plant Cell Physiol. 41:791–803 [DOI] [PubMed] [Google Scholar]
- 46.Wuichet K, Cantwell BJ, Zhulin IB. 2010. Evolution and phyletic distribution of two-component signal transduction systems. Curr. Opin. Microbiol. 13:219–225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Keeling PJ, Palmer JD. 2008. Horizontal gene transfer in eukaryotic evolution. Nat. Rev. Genet. 9:605–618 [DOI] [PubMed] [Google Scholar]
- 48.Maeda T, Wurgler-Murphy SM, Saito H. 1994. A two-component system that regulates an osmosensing MAP kinase cascade in yeast. Nature 369:242–245 [DOI] [PubMed] [Google Scholar]
- 49.Brown JL, North S, Bussey H. 1993. SKN7, a yeast multicopy suppressor of a mutation affecting cell wall beta-glucan assembly, encodes a product with domains homologous to prokaryotic two-component regulators and to heat shock transcription factors. J. Bacteriol. 175:6908–6915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Aoyama K, Mitsubayashi Y, Aiba H, Mizuno T. 2000. Spy1, a histidine-containing phosphotransfer signaling protein, regulates the fission yeast cell cycle through the Mcs4 response regulator. J. Bacteriol. 182:4868–4874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Calera JA, Herman D, Calderone R. 2000. Identification of YPD1, a gene of Candida albicans which encodes a two-component phosphohistidine intermediate protein. Yeast 16:1053–1059 [DOI] [PubMed] [Google Scholar]
- 52.Azuma N, Kanamaru K, Matsushika A, Yamashino T, Mizuno T, Kato M, Kobayashi T. 2007. In vitro analysis of His-Asp phosphorelays in Aspergillus nidulans: the first direct biochemical evidence for the existence of His-Asp phosphotransfer systems in filamentous fungi. Biosci. Biotechnol. Biochem. 71:2493–2502 [DOI] [PubMed] [Google Scholar]
- 53.Tan H, Janiak-Spens F, West AH. 2007. Functional characterization of the phosphorelay protein Mpr1p from Schizosaccharomyces pombe. FEMS Yeast Res. 7:912–921 [DOI] [PubMed] [Google Scholar]
- 54.Lee JW, Ko YJ, Kim SY, Bahn YS. 2011. Multiple roles of Ypd1 phosphotransfer protein in viability, stress response, and virulence factor regulation in Cryptococcus neoformans. Eukaryot. Cell 10:998–1002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Nguyen AN, Lee A, Place W, Shiozaki K. 2000. Multistep phosphorelay proteins transmit oxidative stress signals to the fission yeast stress-activated protein kinase. Mol. Biol. Cell 11:1169–1181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Chang WT, Thomason PA, Gross JD, Neweil PC. 1998. Evidence that the RdeA protein is a component of a multistep phosphorelay modulating rate of development in Dictyostelium. EMBO J. 17:2809–2816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ohmiya R, Kato C, Yamada H, Aiba H, Mizuno T. 1999. A fission yeast gene (prr1(+)) that encodes a response regulator implicated in oxidative stress response. J. Biochem. 125:1061–1066 [DOI] [PubMed] [Google Scholar]
- 58.Quinn J, Malakasi P, Smith DA, Cheetham J, Buck V, Millar JB, Morgan BA. 2011. Two-component mediated peroxide sensing and signal transduction in fission yeast. Antioxid. Redox Signal. 15:153–165 [DOI] [PubMed] [Google Scholar]
- 59.Banno S, Noguchi R, Yamashita K, Fukumori F, Kimura M, Yamaguchi I, Fujimura M. 2007. Roles of putative His-to-Asp signaling modules HPT-1 and RRG-2, on viability and sensitivity to osmotic and oxidative stresses in Neurospora crassa. Curr. Genet. 51:197–208 [DOI] [PubMed] [Google Scholar]
- 60.Vargas-Perez I, Sanchez O, Kawasaki L, Georgellis D, Aguirre J. 2007. Response regulators SrrA and SskA are central components of a phosphorelay system involved in stress signal transduction and asexual sporulation in Aspergillus nidulans. Eukaryot. Cell 6:1570–1583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Nemecek JC, Wüthrich M, Klein BS. 2007. Detection and measurement of two-component systems that control dimorphism and virulence in fungi. Methods Enzymol. 422:465–487 [DOI] [PubMed] [Google Scholar]
- 62.Xu Q, Nguyen V, West AH. 1999. Purification, crystallization and preliminary X-ray diffraction analysis of the yeast phosphorelay protein YPD1. Acta Crystallogr. D Biol. Crystallogr. 55:291–293 [DOI] [PubMed] [Google Scholar]
- 63.Janiak-Spens F, West AH. 2000. Functional roles of conserved amino acid residues surrounding the phosphorylatable histidine of the yeast phosphorelay protein YPD1. Mol. Microbiol. 37:136–144 [DOI] [PubMed] [Google Scholar]
- 64.Fassler JS, West AH. 2010. Genetic and biochemical analysis of the SLN1 pathway in Saccharomyces cerevisiae. Methods Enzymol. 471:291–317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Janiak-Spens F, Sparling DP, West AH. 2000. Novel role for an HPt domain in stabilizing the phosphorylated state of a response regulator domain. J. Bacteriol. 182:6673–6678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Porter SW, West AH. 2005. A common docking site for response regulators on the yeast phosphorelay protein YPD1. Biochim. Biophys. Acta 1748:138–145 [DOI] [PubMed] [Google Scholar]
- 67.Porter SW, Xu Q, West AH. 2003. Ssk1p response regulator binding surface on histidine-containing phosphotransfer protein Ypd1p. Eukaryot. Cell 2:27–33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Ault AD. 2001. Analysis of the molecular mechanisms of signaling by Sln1, part of a two-component phosphorelay pathway in Saccharomyces cerevisiae. University of Iowa, Iowa City, IA: [Google Scholar]
- 69.Hiramoto F, Nomura N, Furumai T, Igarashi Y, Oki T. 2003. Pradimicin-resistance of yeast is caused by a point mutation of the histidine-containing phosphotransfer protein Ypd1. J. Antibiot. (Tokyo) 56:1053–1057 [DOI] [PubMed] [Google Scholar]
- 70.Fassler JS, Gray WM, Malone CL, Tao W, Lin H, Deschenes RJ. 1997. Activated alleles of yeast SLN1 increase Mcm1-dependent reporter gene expression and diminish signaling through the Hog1 osmosensing pathway. J. Biol. Chem. 272:13365–13371 [DOI] [PubMed] [Google Scholar]
- 71.Barrett JF, Goldschmidt RM, Lawrence LE, Foleno B, Chen R, Demers JP, Johnson S, Kanojia R, Fernandez J, Bernstein J, Licata L, Donetz A, Huang S, Hlasta DJ, Macielag MJ, Ohemeng K, Frechette R, Frosco MB, Klaubert DH, Whiteley JM, Wang L, Hoch JA. 1998. Antibacterial agents that inhibit two-component signal transduction systems. Proc. Natl. Acad. Sci. U. S. A. 95:5317–5322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Barrett JF, Hoch JA. 1998. Two-component signal transduction as a target for microbial anti-infective therapy. Antimicrob. Agents Chemother. 42:1529–1536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Foster JE, Sheng Q, McClain JR, Bures M, Nicas TI, Henry K, Winkler ME, Gilmour R. 2004. Kinetic and mechanistic analyses of new classes of inhibitors of two-component signal transduction systems using a coupled assay containing HpkA-DrrA from Thermotoga maritima. Microbiology 150:885–896 [DOI] [PubMed] [Google Scholar]
- 74.Sugahara H, Kondo T, Okada M, Ikeda Y, Kaida K, Fudou R, Mizuno T, Sakagami Y. 2008. Articulospora sp. produces Art1, an inhibitor of bacterial histidine kinase. Biosci. Biotechnol. Biochem. 72:2521–2525 [DOI] [PubMed] [Google Scholar]
- 75.Deschenes RJ, Lin H, Ault AD, Fassler JS. 1999. Antifungal properties and target evaluation of three putative bacterial histidine kinase inhibitors. Antimicrob. Agents Chemother. 43:1700–1703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Gotoh Y, Eguchi Y, Watanabe T, Okamoto S, Doi A, Utsumi R. 2010. Two-component signal transduction as potential drug targets in pathogenic bacteria. Curr. Opin. Microbiol. 13:232–239 [DOI] [PubMed] [Google Scholar]
- 77.Buschart A, Gremmer K, El-Mowafy M, van den Heuvel J, Mueller PP, Bilitewski U. 2012. A novel functional assay for fungal histidine kinases group III reveals the role of HAMP domains for fungicide sensitivity. J. Biotechnol. 157:268–277 [DOI] [PubMed] [Google Scholar]
- 78.Tebbets B, Stewart D, Lawry S, Nett J, Nantel A, Andes D, Klein BS. 2012. Identification and characterization of antifungal compounds using a Saccharomyces cerevisiae reporter bioassay. PLoS One 7:e36021. 10.1371/journal.pone.0036021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Epand RM, Vogel HJ. 1999. Diversity of antimicrobial peptides and their mechanisms of action. Biochim. Biophys. Acta 1462:11–28 [DOI] [PubMed] [Google Scholar]
- 80.Tossi A, Sandri L, Giangaspero A. 2000. Amphipathic, alpha-helical antimicrobial peptides. Biopolymers 55:4–30 [DOI] [PubMed] [Google Scholar]
- 81.Hancock RE, Lehrer R. 1998. Cationic peptides: a new source of antibiotics. Trends Biotechnol. 16:82–88 [DOI] [PubMed] [Google Scholar]
- 82.Zasloff M. 2002. Antimicrobial peptides of multicellular organisms. Nature 415:389–395 [DOI] [PubMed] [Google Scholar]
- 83.Bobek LA, Situ H. 2003. MUC7 20-mer: investigation of antimicrobial activity, secondary structure, and possible mechanism of antifungal action. Antimicrob. Agents Chemother. 47:643–652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Satyanarayana J, Situ H, Narasimhamurthy S, Bhayani N, Bobek LA, Levine MJ. 2000. Divergent solid-phase synthesis and candidacidal activity of MUC7 D1, a 51-residue histidine-rich N-terminal domain of human salivary mucin MUC7. J. Pept. Res. 56:275–282 [DOI] [PubMed] [Google Scholar]
- 85.Hancock RE. 1997. Peptide antibiotics. Lancet 349:418–422 [DOI] [PubMed] [Google Scholar]
- 86.Hancock RE, Rozek A. 2002. Role of membranes in the activities of antimicrobial cationic peptides. FEMS Microbiol. Lett. 206:143–149 [DOI] [PubMed] [Google Scholar]
- 87.Garibotto FM, Garro AD, Masman MF, Rodriguez AM, Luiten PG, Raimondi M, Zacchino SA, Somlai C, Penke B, Enriz RD. 2010. New small-size peptides possessing antifungal activity. Bioorg. Med. Chem. 18:158–167 [DOI] [PubMed] [Google Scholar]
- 88.Grimaldi M, De Rosa M, Di Marino S, Scrima M, Posteraro B, Sanguinetti M, Fadda G, Soriente A, D'Ursi AM. 2010. Synthesis of new antifungal peptides selective against Cryptococcus neoformans. Bioorg. Med. Chem. 18:7985–7990 [DOI] [PubMed] [Google Scholar]
- 89.Arkin MR, Whitty A. 2009. The road less traveled: modulating signal transduction enzymes by inhibiting their protein-protein interactions. Curr. Opin. Chem. Biol. 13:284–290 [DOI] [PubMed] [Google Scholar]
- 90.Neduva V, Russell RB. 2006. Peptides mediating interaction networks: new leads at last. Curr. Opin. Biotechnol. 17:465–471 [DOI] [PubMed] [Google Scholar]
- 91.Xu Q, Porter SW, West AH. 2003. The yeast YPD1/SLN1 complex: insights into molecular recognition in two-component signaling systems. Structure 11:1569–1581 [DOI] [PubMed] [Google Scholar]
- 92.Zhao X, Copeland DM, Soares AS, West AH. 2008. Crystal structure of a complex between the phosphorelay protein YPD1 and the response regulator domain of SLN1 bound to a phosphoryl analog. J. Mol. Biol. 375:1141–1151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Edgar RC. 2004. Local homology recognition and distance measures in linear time using compressed amino acid alphabets. Nucleic Acids Res. 32:380–385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Nicholas KB, Nicholas HBJ, Deerfield DWI. 1997. GeneDoc: analysis and visualization of genetic variation. EMBNEW.NEWS 4:14 [Google Scholar]
- 95.Song HK, Lee JY, Lee MG, Moon J, Min K, Yang JK, Suh SW. 1999. Insights into eukaryotic multistep phosphorelay signal transduction revealed by the crystal structure of Ypd1p from Saccharomyces cerevisiae. J. Mol. Biol. 293:753–761 [DOI] [PubMed] [Google Scholar]
- 96.Xu Q, West AH. 1999. Conservation of structure and function among histidine-containing phosphotransfer (HPt) domains as revealed by the crystal structure of YPD1. J. Mol. Biol. 292:1039–1050 [DOI] [PubMed] [Google Scholar]
- 97.Ochiai N, Fujimura M, Motoyama T, Ichiishi A, Usami R, Horikoshi K, Yamaguchi I. 2001. Characterization of mutations in the two-component histidine kinase gene that confer fludioxonil resistance and osmotic sensitivity in the os-1 mutants of Neurospora crassa. Pest Manag. Sci. 57:437–442 [DOI] [PubMed] [Google Scholar]
- 98.Xu D, Jiang B, Ketela T, Lemieux S, Veillette K, Martel N, Davison J, Sillaots S, Trosok S, Bachewich C, Bussey H, Youngman P, Roemer T. 2007. Genome-wide fitness test and mechanism-of-action studies of inhibitory compounds in Candida albicans. PLoS Pathog. 3:e92. 10.1371/journal.ppat.0030092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Furukawa K, Hoshi Y, Maeda T, Nakajima T, Abe K. 2005. Aspergillus nidulans HOG pathway is activated only by two-component signalling pathway in response to osmotic stress. Mol. Microbiol. 56:1246–1261 [DOI] [PubMed] [Google Scholar]