Abstract
The sirtuins are members of the NAD+-dependent histone deacetylase family that contribute to various cellular functions that affect aging, disease, and cancer development in metazoans. However, the physiological roles of the fungus-specific sirtuin family are still poorly understood. Here, we determined a novel function of the fungus-specific sirtuin HstD/Aspergillus oryzae Hst4 (AoHst4), which is a homolog of Hst4 in A. oryzae yeast. The deletion of all histone deacetylases in A. oryzae demonstrated that the fungus-specific sirtuin HstD/AoHst4 is required for the coordination of fungal development and secondary metabolite production. We also show that the expression of the laeA gene, which is the most studied fungus-specific coordinator for the regulation of secondary metabolism and fungal development, was induced in a ΔhstD strain. Genetic interaction analysis of hstD/Aohst4 and laeA clearly indicated that HstD/AoHst4 works upstream of LaeA to coordinate secondary metabolism and fungal development. The hstD/Aohst4 and laeA genes are fungus specific but conserved in the vast family of filamentous fungi. Thus, we conclude that the fungus-specific sirtuin HstD/AoHst4 coordinates fungal development and secondary metabolism via the regulation of LaeA in filamentous fungi.
INTRODUCTION
Histone acetylation plays key roles in the control of chromatin structure and function (1). The acetylation state is controlled by two histone modification enzymes with opposing actions, histone acetyltransferases (HATs) and deacetylases (HDACs). Acetylation is generally associated with transcriptional activation. In contrast, histone deacetylation is generally associated with transcriptional repression. These enzymes are highly conserved from yeasts to humans, and they are also conserved in filamentous fungi (2–5).
HDACs remove the acetyl moiety from the lysine residue of a histone tail. In addition to histones, these enzymes deacetylate many nonhistone substrates (6). Protein deacetylation affects diverse cellular processes, such us development, metabolism, and stress responses in eukaryotic cells (6). In mammalian and yeast cells, HDACs are divided into two major families called the sirtuins and the classical HDACs. The HDACs are phylogenetically classified into four classes. The sirtuins constitute class III, and the classical HDACs are grouped into classes I, II, and IV (4, 6). Except for mammal-specific class IV HDACs, these enzymes are also conserved in the genome of filamentous fungi (2, 5).
Recently, histone deacetylases in some filamentous fungi have been investigated for their role in the regulation of histone modification, developmental processes, stress resistance, pathogenesis, metabolism, and other such processes (2, 7–10). For example, the homolog of yeast Rpd3 is required for growth and conidiation in several filamentous fungi (11). The class II HDAC HdaA/Aspergillus fumigatus Hda1 (AfHda1), a homolog of yeast Hda1, is involved in germination and the oxidative stress response in Aspergillus fumigatus (12, 13). The yeast Hos2 homolog is required for conidial development, invasive growth, and the production of virulence factors in some plant-pathogenic filamentous fungi (7, 8, 14).
Filamentous fungi produce wide varieties of secondary metabolites (SMs), which are small bioactive molecules that include both beneficial medicines and cosmetics and toxins that are harmful for animals and plants (15, 16). Therefore, because of the importance of fungal SMs, there has been much research into the mechanisms that regulate their production. LaeA, a putative fungus-specific methyltransferase, is implicated in the global regulation of SM production (16, 17). LaeA also has an important role in coordinating fungal development and SM production (17). Recent studies have shown that histone modification plays key roles in the regulation of SM biosynthetic gene expression (18). In Aspergillus nidulans, the HDACs HdaA/Aspergillus nidulans Hda1 (AnHda1) and SirA/A. nidulans Sir2 (AnSir2) regulate carcinogenic sterigmatocystin and production of the antibiotic penicillin (19, 20). The loss of Hdf1/Fusarium graminearum Hos2 (FgHos2) reduces conidial development and the production of deoxynivalenol, which is the most characterized virulence factor in Fusarium graminearum (7).
However, the importance of fungal HDACs in the regulation of secondary metabolism and fungal development is still not known; studies of these HDACs are limited, even though several types of HDACs are found in fungal genomes (2). Moreover, the relationship between the global regulator laeA and histone modification is still poorly understood.
In this study, we examined the phenotypes caused by the disruption of all HDACs using Aspergillus oryzae, which is an important filamentous fungus in industry and has potential for the production of pharmaceutical and cosmetic SMs (21–23). Our observations indicated that the fungus-specific sirtuin HstD/Aspergillus oryzae Hst4 (AoHst4) regulates conidial development and production of kojic acid (KA), which is an important cosmetic material for preventing melanogenesis in skin, as well as production of the antimicrobial penicillin (24). We also performed microarray analysis of the ΔhstD strain to examine the global function of this sirtuin and found that the disruption of this gene affects the expression of many metabolite genes. As described above, laeA is an important coordinator for the regulation of secondary metabolism and development. In this context, we also analyzed the genetic interaction between hstD/Aohst4 and laeA and found that hstD/Aohst4 regulates laeA expression. We describe the function of the fungus-specific sirtuin HstD/AoHst4 first in SM production and then in conidial development through the regulation of laeA gene expression.
MATERIALS AND METHODS
Strains, media, and physiological tests.
The strains used in this study are listed in Table S1 in the supplemental material. Aspergillus oryzae RIB40 was used as the DNA donor. The A. oryzae NSR-ΔLD2 strain was used as the host for A. oryzae HDAC (AoHDAC) disruption (25). M+Met medium [2 g NH4Cl, 1 g (NH4)2SO4, 0.5 g KCl, 1 g KH2PO4, 0.5 g MgSO4·7H2O, 0.02 g FeSO4, 20 g glucose, 1.5 g l-methionine, pH 5.5, in 1 liter] or M+Ade medium (M+Met medium containing 0.5 g of adenine sulfate dihydrate instead of l-methionine) was used as the selection medium for A. oryzae adeA+ transformants and A. oryzae sC+ transformants, respectively (25). TS medium (6 g of NaNO2, 0.52 g of KCl, 1.52 g of KH2PO4, 0.52 g of MgSO4·7H2O, 10 g of glucose, 1 ml of trace elements, pH 6.5, in 1 liter) was used as the selectable medium for A. oryzae adeA+ sC+ transformants. M+Met, M+Ade, or TS medium with 0.8 M NaCl added was used for transformation. KAS medium (10 g of tryptone, 1.52 g of K2HPO4, 1.5 g of l-methionine, 0.5 g of MgSO4·7H2O, 1 ml of trace elements, pH 6.5, in 1 liter) was used for screening for HDAC-affected kojic acid productivity. KA medium (1 g of yeast extract, 0.5 g of KCl, 1 g of K2HPO4, 1.5 g of l-methionine, 0.5 g of MgSO4·7H2O, 100 g of glucose, 0.5 g of adenine sulfate dihydrate, pH 6.0, in 1 liter) was used to test for KA production and RNA preparation. N medium (3 g of l-glutamic acid, 0.52 g of KCl, 1.52 g of K2HPO4, 0.52 g of MgSO4·7H2O, 30 g of glucose, 1.5 g of l-methionine, 1 ml of trace elements, pH 6.5, in 1 liter) was used for morphological analysis. TSB medium (20 g of tryptic soy broth, 1.5 g of l-methionine, 3 g of l-glutamic acid, 0.5 g of adenine sulfate dihydrate, pH 7.5, in 1 liter) was used for the penicillin bioassay and RNA preparation.
Morphological analysis was performed in 20 ml of N 2% agar medium or 100 ml of N liquid medium. Three independent disruptants were used for each experiment. For the spore count, suspensions of conidia of each strain were point inoculated (1 × 105 conidia) on the center of each plate, and the strain was grown for 5 days at 30°C. Colony diameters were measured at this time, and the spores were harvested in suspension solution (0.025% Tween 80, 0.5% NaCl), vortexed vigorously, and counted using a TC10 automated cell counter (Bio-Rad). The conidiation rate was calculated by the conidium number/radial growth area (cm2). For biomass analysis in N liquid medium, 4 cm2 of full-growth colonies in plate cultures of each strain were cut out, homogenized in 1 ml of suspension solution, and then used to inoculate each flask. Flasks were incubated for 2 days at 30°C with shaking at 100 rpm. Then, mycelia were harvested, dried at 105°C for 2 h, and weighed.
Protein identification, domain prediction, and phylogenetic analysis.
HDAC sequences of Saccharomyces cerevisiae were obtained from the Saccharomyces genome database (http://www.yeastgenome.org/). HDAC sequences of Homo sapiens, Neurospora crassa, and A. nidulans were obtained from the NCBI proteins database (http://www.ncbi.nlm.nih.gov/guide/proteins/), the Neurospora crassa database (http://www.broadinstitute.org/annotation/genome/neurospora/MultiHome.html), and AspGD (http://www.aspgd.org/), respectively. The HDAC genes of A. oryzae were identified from the Comparative Fungal Genome Database (CFGD; http://nribf2.nrib.go.jp/) by BLAST searching using the HDAC sequences of S. cerevisiae and H. sapiens as the query. The sequences of these HDACs in A. oryzae were verified by RNA sequencing using the SOLiD3 system (Applied Biosystems). The gene structures of all AoHDACs were confirmed from the RNA sequence data (details of our RNA sequence data are available in AspGD [accession no. ASPL0000367586; http://www.aspergillusgenome.org/download/large_scale_data/Iwashita_2012/]) (26). The mapping data for all reads are also available in CFGD. For recognizable domains, the protein sequence of the HDAC homolog in A. oryzae was analyzed using the InterProScan tool (27). The protein sequence of the HstD homolog in filamentous fungi was identified using an NCBI blast search with Pezizomycotina genomes (http://www.ncbi.nlm.nih.gov/sutils/genom_tree.cgi) and the amino acid sequence of HstD as a query.
For the classification of HDACs in A. oryzae, protein sequences of HDACs in S. cerevisiae, N. crassa, A. nidulans, and A. oryzae were aligned with the ClustalW software in the Molecular Evolutionary Genetic Analysis, v.5 (MEGA5), program (28). Phylogenetic analysis was carried out by the MEGA5 program using the neighbor-joining method with 1,000 bootstrap replicates. For the classification of HstD in filamentous fungi, alignment and phylogenetic analysis were performed as described above. A list of sequence accession numbers used for AoHDAC analysis is provided in Table S2 in the supplemental material, and the sequence accession numbers of each HstD homolog are provided in Fig. S3 in the supplemental material.
RNA preparation.
KA culture was performed in 20 ml of KA liquid medium inoculated with 200 μl of 1 × 108 conidia/ml suspension, and incubation was at 30°C for 4 or 7 days with shaking at 130 rpm. TSB culture was performed in 40 ml of TSB liquid medium inoculated with 400 μl of 1 × 108 conidia/ml suspension, and incubation was at 30°C for 1 day with shaking at 200 rpm. After cultivation, mycelia were harvested using Miracloth (Merck). Then, mycelia were immediately frozen in liquid N2 and ground to a fine powder. Total RNA was isolated from mycelia from KA or TSB liquid medium using the Isogen reagent (Nippon Gene) according to the manufacturer's instructions.
Northern hybridization.
Denatured total RNA (20 μg) was electrophoresed on a formaldehyde-agarose gel and transferred in 20× SSC (1× SSC is 0.15 M NaCl plus 0.015M sodium citrate) onto a Hybond N+ membrane. Northern analysis was performed with a Detection starter kit II (Roche) according to the manufacturer's instructions. Digoxigenin-labeled probes were prepared using a PCR digoxigenin probe synthesis kit (Roche) with genomic A. oryzae RIB40 DNA as the template and the primers X-probe-F and X-probe-R. The letter X means the respective gene used for Northern analysis. Each blot was imaged using an LAS1000plus luminescent image analyzer (Fujifilm). A list of the primers used for these PCRs is shown in Table S3 in the supplemental material.
Construction of the disruption cassette.
Each disruption cassette was constructed by fusion PCR of three mutually primed DNA fragments, the 5′ and 3′ flanking regions of the target genes and the adeA fragment (29). About 1 kb of the 5′ and 3′ flanking regions of the target genes and the adeA gene was amplified from genomic A. oryzae RIB40 DNA with primers X-A and X-B, X-C and X-D, and adeA-F and adeA-R, respectively. Only for the construction of the ΔhstD and ΔlaeA genes and the A. nidulans sC gene, amplified from pUSA with primers sC-F and sC-R, was the gene fused to the flanking region of laeA (30). The letter X in the primer names represents the name of each target gene. Each region was amplified by KOD Plus DNA polymerase (Toyobo). These fragments were combined by a second PCR with KOD Plus DNA polymerase and the primers X-A and X-D or X-A2 and X-D2. The amplified fragment was purified by a QIAquick PCR purification kit (Qiagen) and then used as a disruption cassette. A list of the primers used for these PCRs is shown in Table S3 in the supplemental material.
Complementation of hstD.
To recover the native locus of hstD, we first amplified the same 5′ flanking region of the disruption construct of the hstD and adeA fragments from RIB40 DNA with primers hstD-A and hstD-compB or adeA fusion primers sC-F and adeA-R, respectively. We also amplified the A. nidulans sC gene from pUSA with sC-F and sC-R as autotrophic markers (30). Then, these fragments were combined by fusion PCR using nested primers adeA-R and hstD-A2 (29). The amplified fragment was purified by a QIAquick PCR purification kit and used as a complementation cassette. A list of the primers used for these PCRs is shown in Table S3 in the supplemental material.
Construction of overexpression plasmids.
The open reading frames (ORFs) of laeA or hstD/Aohst4 were amplified from the genomic DNA of A. oryzae RIB40 with Fusion-laeA-F and Fusion-laeA-R or Fusion-hstD-F and Fusion-hstD-R, respectively. The resulting fragments were fused into SmaI-cut pUSA using an In-Fusion HD cloning kit (TaKaRa) (30). The amyB promoter of pUSA was used to drive the overexpression of laeA and hstD/Aohst4, respectively. The resulting plasmids, pUSlaeA and pUShstD, were linearized with one cut of restriction enzyme BglII and EcoT22I on the laeA ORF and hstD/Aohst4 ORF, respectively. The resulting linearized fragments were used as overexpression cassettes. A list of the primers used for these PCRs is shown in Table S3 in the supplemental material.
Transformation of A. oryzae.
Transformation of A. oryzae strains was performed using the protoplast-polyethylene glycol method (31). To verify the disruption of the target gene, direct colony PCR was performed using primers (X-F and X-G, X-A and X-D), KOD-FX (Toyobo), and a crude DNA sample of each transformant. Primers X-F and X-G were designed at the region of each target gene. The crude DNA sample was prepared as follows: conidia and hyphae from each transformant culture were suspended in 100 μl buffer A (100 mM Tris-HCl [pH 9.5], 1 M KCl, 10 mM EDTA). This mycelial suspension was vigorously vortexed and incubated at 95°C for 10 min. Immediately thereafter, this hot solution was vigorously vortexed and centrifuged at 5,000 rpm for 1 min. A total of 1 μl of supernatant was used as the crude DNA sample. A list of the primers used for these assays is shown in Table S3 in the supplemental material.
Time-lapse imaging.
For time-lapse imaging, conidiophores were germinated in 1.5% agar N medium in 35-mm glass-bottom dishes. Cells were imaged using a real-time culture cell monitoring system (Astec) controlled by CCM software. Differential interference contrast images of each disruptant were taken every 20 min for approximately 84 h. All imaging was carried out at 30°C. Pictures and movies were edited with CCM software (Astec).
Secondary metabolite analysis.
For the plate assay of kojic acid production, KAS medium and KA medium containing 5 mM FeCl3 were used for screening and genetic interaction analysis, respectively. Suspensions of conidia of each strain were point inoculated (1 × 105 conidia) on the center of the 2% agar medium and grown for 5 days at 30°C. Then, a red halo, which indicates the existence of kojic acid, was observed.
For the quantification of kojic acid, 20 ml of KA liquid medium inoculated with 200 μl of 1 × 108 conidia/ml suspension was incubated at 30°C with shaking at 130 rpm. After cultivation for the appropriate period, mycelia were filtered through Miracloth (Merck), and then the filtrate was collected. Harvested mycelia were dried at 105°C for 1 h and weighed. The collected KA medium was filtered through a MillexHV filter (Millipore), and then the kojic acid concentration was quantified by colorimetric methods.
Microarray analysis.
The A. oryzae GeneChip microarray (AoDNAChip; NCBI Gene Expression Omnibus [GEO] platform GPL16184) was designed by Affymetrix to refer to the entire genome sequence of A. oryzae, and predicted ORFs are published at the Comparative Fungal Genome Database (http://nribf2.nrib.go.jp/, accession number A. oryzae RIB40 Ace33v2). The AoDNAChip covered 13,765 ORFs and 6,143 promoters of A. oryzae. In this study, we performed transcriptome analysis using probes of 13,765 ORFs set on this microarray.
Total RNA was purified for microarray analysis by using an RNeasy minikit (Qiagen). RNA quality was determined by using a BioAnalyzer 2100 system (Agilent Technology), and the quantity was determined by using an Ultrospec 3300 pro spectrophotometer (Amersham Pharmacia Biotech). Fragmented biotin-labeled cRNA was prepared by using a GeneChip one-cycle target labeling and control reagent kit (Affymetrix) according to the manufacturer's instructions. The fragmented cRNA was hybridized to an AoDNAChip. Then, this GeneChip was washed, stained, and scanned by using a GeneChip FS-450 fluidics station (fluidics protocol FS450_001) and a GeneChip 3000 scanner.
Scanned probe array images were converted into CEL files and normalized by using the GCOS v.1.4 program (Affymetrix). Calculations of signal intensity and detection P values were also performed using GCOS v.1.4. The trimmed mean signal of the array was scaled to the target signal of 500 with the all probe sets scaling option. The detection call was used for detection of a particular transcript, with a detection P value of <0.04 considered present (P), 0.04 ≤ P < 0.06 considered marginal (M), and a P value of ≥0.06 considered absent (A). These calculation data were exported as CHP files. For microarray data analysis, CHP files were imported into GeneSpring v.7.3 (Agilent Technologies). Expression data were normalized per chip to the 50th percentile. In this study, we analyzed genes detected as P or M flags. Genes with statistically significant changes in transcript abundance were identified using a cutoff value of 2-fold and a Welch's t test value of less than 5%. FungiFun software was used for functional catalogue (FunCat) categorization (32, 33). Significantly enriched FunCat categories were extracted using FungiFun software (cutoff P value, <0.05; the P value indicates the significance of the number of hits for each category in the data set, taking the number of hits for the whole genome of A. oryzae as the background). The calculation is based on a two-tailed Fisher's exact test). The distribution of whole A. oryzae genes is indicated in Fig. S4 and S5 in the supplemental material and was used as a reference for FunCat enrichment analysis. Two biological replicates were used for the microarray analysis.
Accession numbers.
The microarray data have been deposited in the NCBI Gene Expression Omnibus (GEO) database (http://www.ncbi.nlm.nih.gov/geo/) and are accessible through GEO series accession number GSE41612. The coding sequences of hdaB and hstD were deposited in DDBJ under accession no. AB744040 and AB744041, respectively.
RESULTS
Phylogeny and morphology of AoHDACs.
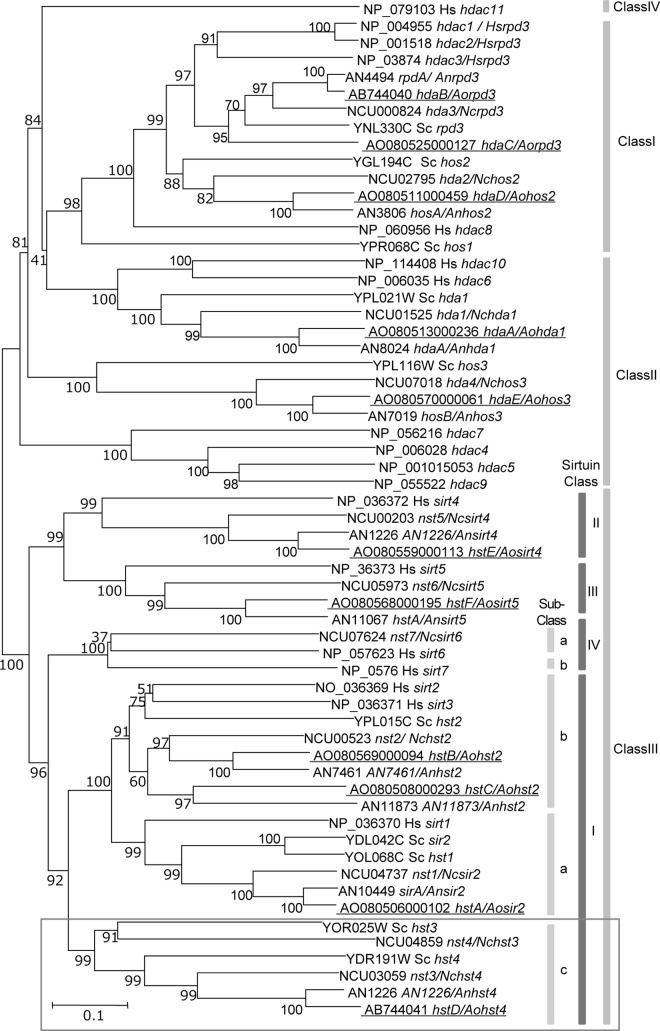
A total of 11 AoHDACs were found in the A. oryzae genome on the basis of a BLAST analysis. We classified the AoHDACs according to a previous phylogenetic study of HDACs. These AoHDACs were phylogenetically divided into class I to III HDACs, but the mammal-specific class IV HDACs were not found in the genome. Class III HDACs are generally described as sirtuin-type HDACs, and 6 AoHDACs belonged to this class. The A. oryzae sirtuins (AoSirtuins) were classified into classes I to III, but class IV sirtuins were not found in the genome. The class I sirtuins were further categorized into three subclasses, including the fungus-specific HDACs of sirtuin subclass C (Fig. 1). We attempted to disrupt these 11 AoHDACs and succeeded for 10 of the AoHDACs. However, only heterokaryon transformants were obtained in the case of hdaB/A. oryzae rpd3 (Aorpd3) over several trials (data not shown). This result suggests that hdaB/Aorpd3 is essential in A. oryzae, but further experiments are required to confirm this hypothesis. Thus, in this report, the heterokaryon disruptant of hdaB/Aorpd3 and AoHDAC disruptants were used for subsequent experiments.
Fig 1.
Phylogenetic analysis of histone deacetylase in A. oryzae. Accession numbers and HDAC names are indicated for each branch. The HDAC names of S. cerevisiae or H. sapiens with the species name indicated are followed by a slash. The numbers at the nodes are bootstrap values obtained from 1,000 replicates and are indicated as percentages. Scale bar, a distance corresponding to 0.2 amino acid substitution per site. The class or subclass of HDACs is shown on the right. These classes of HDACs are referred to in previous phylogenetic studies (4, 6, 34). AoHDACs are indicated by underlines. Abbreviations of AoHDAC gene names are as follows: hda, histone deacetylase; hst, homolog of sirtuin. The class to which hstD belongs is surrounded by a gray border. The gene names and their accession numbers are identified in Table S2 in the supplemental material. Sc, Saccharomyces cerevisiae; An, Aspergillus nidulans; Nc, Neurospora crassa; Hs, Homo sapiens; Ao, Aspergillus oryzae.
We first observed the growth and generation of conidia of AoHDAC disruptants and hdaB/Aorpd3 heterokaryon transformants on the plate culture (see Fig. S1 and S2B and C in the supplemental material). Observation of the growth of AoHDAC disruptants revealed an obvious defect of the ΔhstD and ΔhdaD strains in morphogenesis (Fig. 2A). The conidial formation of these disruptants was significantly decreased, and a slight growth effect was observed in plate cultivation (Fig. 2B and C). We additionally observed the growth of AoHDAC disruptants and the hdaB/Aorpd3 heterokaryon transformant in liquid culture (see Fig. S2D in the supplemental material). A growth defect of the ΔhdaD strain was observed in submerged cultivation (see Fig. S2D in the supplemental material). Thus, hdaD/Aohos2 is required for the growth integrity of A. oryzae. These two disruptants were further examined using time-lapse imaging for more detailed observation. As expected, the ΔhdaD strain showed slow growth but exhibited more crowded aerial hyphae than the wild-type strain (see Fig. S2A in the supplemental material). However, a few invasive hyphae were detected in the ΔhstD strain but not in the ΔhdaD strain. In this time-lapse imaging analysis, a significant defect of conidial development was also found in both the ΔhdaD and ΔhstD strains.
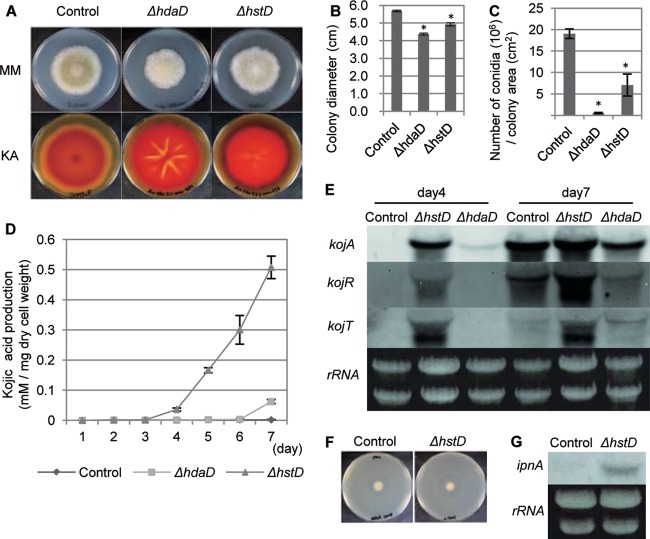
Fig 2.
hstD/Aohst4 and hdaD/Aohos2 regulate SM production and development. (A) The morphological phenotype on N agar medium (MM) and results of the kojic acid production plate assay (KA) are provided for the indicated strains. (B, C) Radial growth and conidiation of the indicated strains on N agar medium, respectively. (D) Time course characteristics of kojic acid production of the indicated disruptants. (E) Expression profiles of kojic acid cluster genes represented by Northern hybridization. The culture times of the indicated strains are shown at the top of the panel. The analyzed gene is indicated on the left side of each blot. The results for rRNA, used as the loading control, are shown. (F) Bioassay of penicillin production of the ΔhstD strain. (G) Northern hybridization of the penicillin biosynthetic gene ipnA in the ΔhstD strain. The results for rRNA, used as the loading control, are shown. The adeA+ strain was used as a control in the experiment whose results are presented in this figure. All data are represented as means ± SDs (n = 3); *, P < 0.01, t test.
These results indicate that both HdaD/AoHos2 and HstD/AoHst4 play an important role in the growth and development of A. oryzae, especially in asexual development.
Fungus-specific sirtuin regulates SM production.
We further examined SM production of AoHDAC disruptants and the hdaB/Aorpd3 heterokaryon transformant using a plate assay for kojic acid productivity (see Fig. S1 in the supplemental material). High levels of production of kojic acid were observed in the ΔhdaB and ΔhstA strains (see Fig. S1 in the supplemental material), and significant overproduction was observed in the ΔhdaD and ΔhstD strains (Fig. 2A). We quantified the kojic acid production of these two HDAC disruptants, and the ΔhstD strain showed a 200-fold increased productivity in a 7-day culture (Fig. 2D). In contrast, the ΔhdaD strain showed a 30-fold overproduction compared with the level of production for the control strain (Fig. 2D). The ΔhstD strain started to produce kojic acid by 4 days in culture, while kojic acid was not detected in the wild-type strain culture.
We further analyzed kojic acid production at the gene expression level by examining three key genes in the kojic acid gene cluster (24). Northern analysis of 4-day-old cultures showed extremely high levels of expression of these genes in the ΔhstD strain, but no expression was observed in the wild-type strain (Fig. 2E). The kojA gene was also expressed in the ΔhdaD strain, but the expression was not as high as that in the ΔhstD strain. This earlier and higher expression in both disruptants is consistent with the earlier and higher production of kojic acid.
These results indicate the importance of ΔhdaD and ΔhstD in the regulation of kojic acid production. On the basis of phylogenetic classification, HstD/AoHst4 belongs to the fungus-specific class of sirtuins, and this protein is widely conserved in filamentous fungi (34) (see Fig. S3 in the supplemental material). Interestingly, nst3/N. crassa hst4 (Nchst4) is involved in the silencing mechanism of N. crassa (35). Moreover, the phenotype of the ΔhstD strain was reminiscent of the global use of HstD/AoHst4 for the production of various SMs in A. oryzae.
In this context, we examined the regulation of penicillin biosynthesis in the ΔhstD strain. As expected, a higher level of production of penicillin was found in the ΔhstD strain, and a higher level of expression of the penicillin biosynthetic gene was confirmed (Fig. 2F and G). Along with the morphogenetic defect and kojic acid production, these phenotypes were rescued by the complementation of hstD/Aohst4 (Fig. 3).
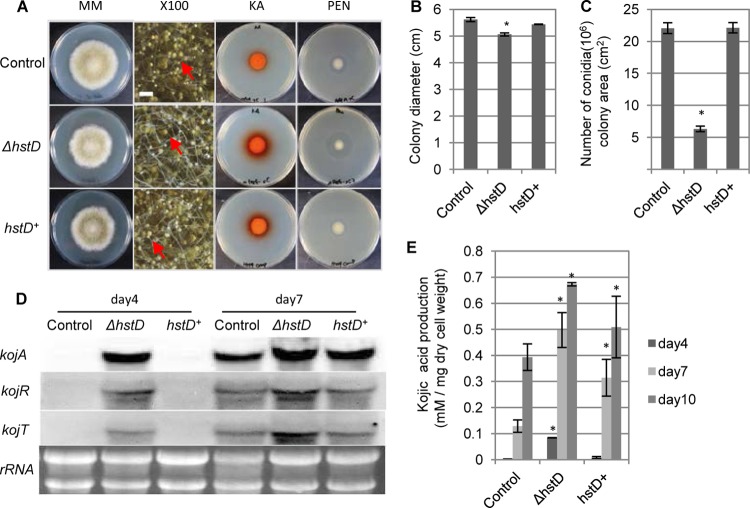
Fig 3.
Complementation analysis of hstD. (A) Analysis of the morphology and SM production of the ΔhstD and hstD+ strains. MM, morphological phenotype of the indicated strain on N agar medium; ×100, closeup stereomicroscopic images of the strains on N agar medium (magnification, ×100; bar, 500 μm; red arrows, examples of conidia); KA and PEN, plate assay or bioassay of kojic acid and penicillin, respectively. (B, C) Quantification of colony diameter and rate of conidiation of ΔhstD and hstD+ strains, respectively. (D) The expression profiles of the kojic acid cluster genes were determined by Northern hybridization. The culture time of the indicated strain is shown at the top. The analyzed gene is indicated on the left side of each blot. The results for rRNA, used as the loading control, are shown. (E) Quantification of kojic acid production. The adeA+ sC+ strain was used as the control, and the ΔhstD sC+ strain represents the ΔhstD strain. All data are represented as means ± SDs (n = 3); *, P < 0.01, t test.
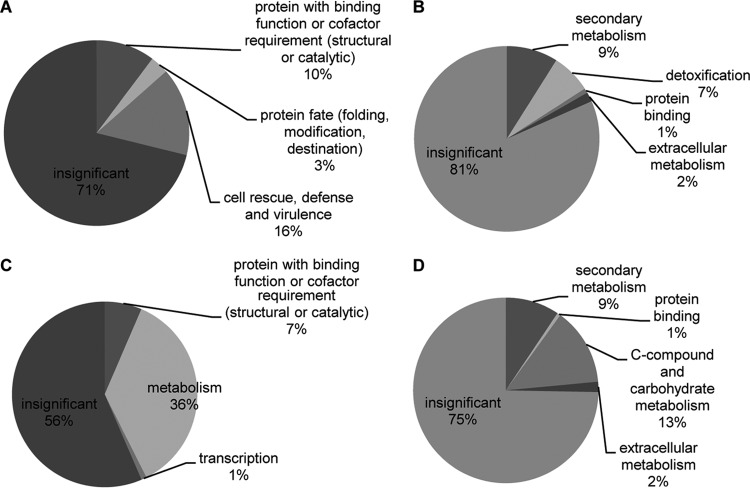
These results suggest that hstD/Aohst4 is required for the global regulation of SM biosynthesis. Thus, we examined the ΔhstD strain by microarray analysis to investigate the expression of other SM-related genes. The expression of 388 genes was significantly affected by hstD/Aohst4 deletion (absolute fold change, >2; P < 0.05) (see Table S4 in the supplemental material). These genes were spread across the whole genome; 299 of 388 genes were upregulated in the ΔhstD strain. To reveal the functional distribution of genes affected by hstD/Aohst4, FunCat enrichment analysis was carried out using FungiFun software (32, 33) (Fig. 4). The C-compound and carbohydrate metabolism and the secondary metabolism categories were significantly enriched in downregulated genes. Most of the genes categorized as secondary metabolism overlapped with the genes categorized as C-compound and carbohydrate metabolism. Genes categorized as C-compound and carbohydrate metabolism were mainly related to polysaccharide degradation or glycolysis. This result suggests that carbon source degradation-related genes are downregulated in the ΔhstD strain. The secondary metabolism and detoxification categories were significantly enriched in genes upregulated by hstD deletion. Genes categorized as secondary metabolism and detoxification were mainly constituted by cytochrome P450 (CYP) genes. Interestingly, three nonribosomal peptide synthetases (NRPSs) and one polyketide synthase (PKS) were also upregulated in the ΔhstD strain. One of three NRPSs encoded wykN (AO080501000008), which is involved in Wyk-1 production in A. oryzae (23). PKS and NRPS enzymes generate the general structural scaffolds of most secondary metabolites (36). Additionally, fungal CYPs have PKS- and NRPS-associated functions and generate structural variation of fungal SMs. Thus, hstD/Aohst4 affects many kinds of SM production (at least 6 different SM gene clusters) (37, 38).
Fig 4.
Enrichment analysis of the FunCat categorization of the microarray analysis. Significantly enriched FunCat level 1 and level 2 categories of genes upregulated (A, B) or downregulated (C, D) by hstD/Aohst4 deletion are shown. The FunCat is the organism-independent functional description of proteins (33). FunCat consists of 28 main functional categories (level 1). Level 1 is the most general one, whereas level 2 shows much more detail. The percentage indicated for each category contributes to the total mapping. Insignificant FunCat categories are indicated as insignificant in the pie charts. Significantly enriched categories were extracted by FungiFun software (cutoff P value, <0.05; Fisher's exact test) (32). Details of the enrichment analysis are available at the FungiFun website (https://sbi.hki-jena.de/FungiFun/FungiFunHelp.html).
hstD/Aohst4 regulates expression of laeA.
The LaeA complex coordinates the development and SM biosynthesis in A. nidulans and is conserved in numerous fungal genomes (17). Recently, it was reported that deleting laeA diminishes kojic acid production and gene expression in A. oryzae (39). Additionally, penicillin production in A. oryzae is diminished after deletion of veA, which is a member of the LaeA complex (17, 40). These reports suggest that the LaeA complex also plays a general role in the induction of the SM gene cluster in A. oryzae. Thus, the SM overproduction phenotype of the hstD/Aohst4 disruptant should be associated with the LaeA complex, and HstD/AoHst4 may play a role under the control of LaeA. According to this hypothesis, the expression of laeA should not be altered in the hstD/Aohst4 disruptant. Thus, we examined the expression of laeA in the hstD/Aohst4 disruptant. Surprisingly, laeA was highly expressed in the hstD/Aohst4 disruptant, even in the 4-day culture, but remained unexpressed in the wild-type strain (Fig. 5A). This result suggests that HstD/AoHst4 is involved in laeA gene repression. Thus, both SM overproduction phenotypes and the altered morphological phenotype of the ΔhstD strain are caused by the high level of expression of laeA.
Fig 5.
Genetic interaction between hstD and laeA. (A) Expression profile of laeA under the KA-producing condition. The adeA+ strain was used as a control. The culture time of the indicated strain is shown at the top of the panel. The results for rRNA, used as the loading control, are shown. (B) Analysis of morphology and SM production of the ΔhstD, ΔlaeA, and ΔhstD ΔlaeA strains. MM, morphological phenotype of the indicated strain on N agar medium; ×100, closeup stereomicroscopic images of the strains on N agar medium (magnification, ×100; bar, 500 μm; red arrows, examples of conidia); KA and PEN, plate assay or bioassay of kojic acid and penicillin, respectively. (C, D) Quantification of the conidiation rate and kojic acid production of the ΔhstD, ΔlaeA, and ΔhstD ΔlaeA strains, respectively. Except for panel A, the adeA+ sC+ strain was used as the control, and the ΔhstD sC+ and ΔlaeA sC+ strains represent the ΔhstD and ΔlaeA strains, respectively, in this figure. All data are represented as means ± SDs (n = 3); *, P < 0.01, t test.
Genetic interaction of hstD and laeA.
To confirm the hypothesis presented above, we prepared a ΔlaeA strain and a ΔhstD ΔlaeA double disruptant and then examined the strains for SM production and conidial development. At first, these strains exhibited the phenotype of the ΔlaeA strain, including a lack of kojic acid production (Fig. 5B and D), similar to previous reports (39). The production of penicillin and conidial development were also lost in the ΔlaeA strain (Fig. 5B and C). Next, we observed the effect of laeA disruption in the ΔhstD strain background and found that the SM overproduction phenotype of the ΔhstD strain was abolished by laeA disruption (Fig. 5B and D). We further observed conidial development in the double disruptant, which exhibited a ΔlaeA strain-like fluffy phenotype (Fig. 5B and C). These results clearly indicate a genetic interaction between hstD/Aohst4 and laeA and that HstD/AoHst4 plays a role upstream of LaeA.
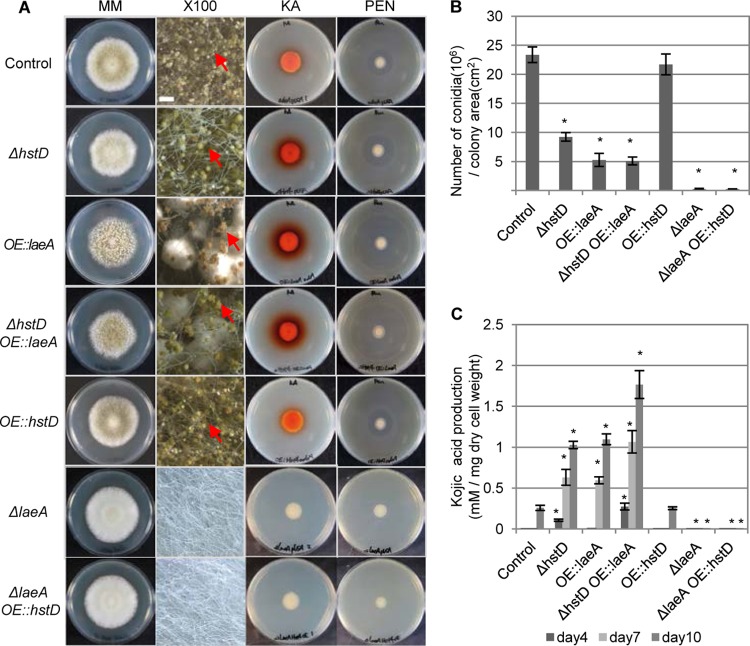
To confirm this epistatic relationship, we examined the effect of laeA and hstD/Aohst4 overexpression using the amyB promoter. The strain overexpressing laeA exhibited a ΔhstD strain-like phenotype, such as SM overproduction and low levels of conidial formation (Fig. 6). The effects of laeA overexpression were also observed in the ΔhstD strain background (Fig. 6). Additionally, both the OE::laeA strain and ΔhstD OE::laeA strain showed high levels of expression of laeA, kojA, and ipnA (see Fig. S6 in the supplemental material). These results indicate that laeA is downstream of hstD/Aohst4. Therefore, we overexpressed hstD/Aohst4 in a laeA disruption background. As expected, the overexpression of hstD/Aohst4 resulted in a ΔlaeA strain-like phenotype, such as no SM production and a fluffy morphology, in the ΔlaeA background (Fig. 6). Overexpression of hstD/Aohst4 in the wild-type laeA background had no measurable effect on SM production and development (Fig. 6).
Fig 6.
Epistatic relationship between hstD and laeA. (A) Analysis of the morphology and SM production of the ΔhstD OE::laeA and ΔlaeA OE::hstD strains. MM, morphological phenotype of the indicated strain on N agar medium; ×100, closeup stereomicroscopic images of the strains on N agar medium (magnification, ×100; bar, 500 μm; red arrows, examples of conidia); KA and PEN, plate assay or bioassay of kojic acid and penicillin, respectively. (B, C) Quantification of the conidiation rate and kojic acid production of the ΔhstD OE::laeA and ΔlaeA OE::hstD strains, respectively. The adeA+ pUSA+ strain was used as the control, and the ΔhstD pUSA+, OE::laeA adeA+, ΔlaeA pUSA+, and OE::hstD adeA+ strains represent the ΔhstD, OE::laeA, ΔlaeA, and OE::hstD strains, respectively, in this figure. The amyB promoter was used to drive overexpression of laeA and hstD. All data are represented as means ± SDs (n = 3); *, P < 0.01, t test.
From these results, we suggest that the fungus-specific sirtuin HstD/AoHst4 controls SM production and fungal development through the regulation of laeA gene expression.
DISCUSSION
In the past decade, the importance of the role of classical HDACs in filamentous fungi in the regulation of some fungal phenotypes has been revealed (2). For example, F. graminearum hdf1/F. graminearum hos2 is important for conidial development and SM production (7). In this study, we also determined the importance of hdaD/Aohos2 in the regulation of hyphal growth, conidiation, and SM production. Compared with the study of classical HDACs, only a few reports have been published for sirtuin-type HDACs in filamentous fungi (20, 35). Recently, sirA/A. nidulans sir2 (Ansir2), which is a homolog of yeast sir2 and mammalian sirt1, was reported to function in secondary metabolism regulation, but no effects on growth, conidiation, or morphogenesis were reported (20).
The fungus-specific putative methyltransferase LaeA coordinates fungal development and SMs in several filamentous fungi (17). It was previously reported that kojic acid production is regulated by LaeA, but we found that laeA coordinates both SM production and conidial development (39). In general, the production of fungal SMs is coordinated with fungal development, and the LaeA complex coordinates these (17). In higher eukaryotes, sirtuin affects various physiological functions, such as differentiation, metabolism, and the stress response (41–43). In this study, we found that the fungus-specific sirtuin HstD/AoHst4 affects both fungal development and SM production. Furthermore, the epistatic study revealed that HstD/AoHst4 is involved in the coordination of fungal development and SM production via LaeA expression. These results indicate that HstD/AoHst4 plays a Sirt1-like central role that coordinates the developmental state and metabolism in filamentous fungi.
The yeast Hst4, which is a homolog of hstD/Aohst4 of A. oryzae, is a NAD+-dependent histone H3K56 deacetylase (44). In the Northern analysis, deletion of hstD/Aohst4 affected the level of laeA gene expression. Therefore, HstD/AoHstD may be involved in the epigenetic regulation of laeA gene expression. However, the overexpression of hstD/Aohst4 did not affect SM production or conidial development. In general, histone acetyltransferase (HAT) is required for activation of silenced genes. In budding yeast, it has been reported that the fungus-specific HAT of RTT109 catalyzes H3K56 acetylation and restores the silencing defects of the Δhst3 Δhst4 mutant (44, 45). We found one rtt109 homologue of AO090020000581 (A. oryzae rtt109 [Aortt109]) in the A. oryzae genome. This suggests that Aortt109 is required for the expression of the silenced laeA by H3K56 deacetylation and overcomes even hstD/Aohst4 overexpression. In this context, we will investigate histone H3K56 acetylation of the laeA locus and the effect of Aortt109 on the acetylation and expression of laeA in future studies.
In the overexpression analysis of laeA, several different phenotypes of kojic acid production were observed compared with the phenotype of the hstD/Aohst4 mutant. The ΔhstD strain showed high levels of kojic acid production in a 4-day liquid culture, while the same phenotype was not observed in the OE::laeA strain. Compared with the OE::laeA strain, interestingly, higher levels of kojic acid production and higher levels of expression of kojA, ipnA, and laeA were observed in the ΔhstD OE::laeA strain. These results suggested that HstD/AoHst4 has some LaeA-independent role in the regulation of SM production.
We developed a model of the regulatory system of HstD/AoHst4 (Fig. 7). The expression of hstD/Aohst4 may be induced or suppressed by unknown signals. As described above, an unknown factor like AoRtt109 may compete with HstD/AoHst4 activity under hstD/Aohst4-inducing conditions. Under the hstD/Aohst4-suppressed condition, laeA expression was induced. This in turn led to conidiation and induction of secondary metabolite production. However, it is possible that HstD/AoHst4 directly regulates fungal development and secondary metabolism independently of LaeA.
Fig 7.
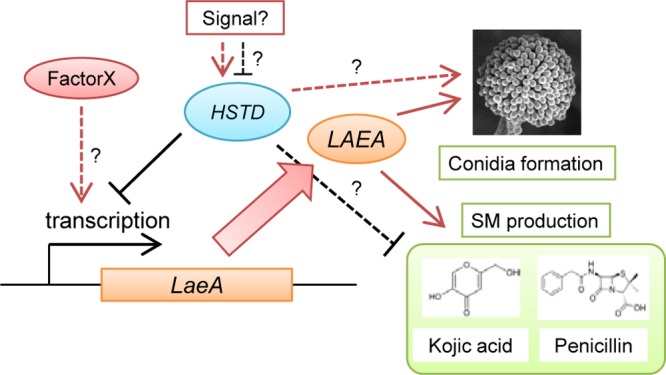
Schematic model of the regulation of SM production and development by HstD/AoHst4 through LaeA. An unknown signal induces or suppresses the function of HstD/AoHst4. Suppression of HstD/AoHst4 leads to expression of LaeA. This activation stimulates fungal development and secondary metabolite production. However, there is the possibility of an HstD/AoHst4 competitive mechanism by an unknown factor (described as factor X in this figure).
The hstD/Aohst4 gene is fungus specific but is conserved in the vast family of filamentous fungi (see Fig. S3 in the supplemental material). Furthermore, this gene plays a role in the coordination of fungal development and SM production. These results indicate that HstD/AoHst4 has great potential as a target to improve the productivity of useful SMs. It will also be important in the development of an attractive host for the production of several heterogeneous metabolites.
Supplementary Material
ACKNOWLEDGMENTS
We thank Ken Oda for great discussions, Katsuhiko Kitamoto for the gift of the A. oryzae NSR-ΔLD2 strain, Ryoko Hamada for technical help with the microarray analysis, and Kanae Koike (Electron Microscopy Service, Center for Gene Science, Hiroshima University) for technical help with electron microscopic analysis.
Footnotes
Published ahead of print 31 May 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/EC.00003-13.
REFERENCES
- 1.Shahbazian MD, Grunstein M. 2007. Functions of site-specific histone acetylation and deacetylation. Annu. Rev. Biochem. 76:75–100 [DOI] [PubMed] [Google Scholar]
- 2.Brosch G, Loidl P, Graessle S. 2008. Histone modifications and chromatin dynamics: a focus on filamentous fungi. FEMS Microbiol. Rev. 32:409–439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nishida H. 2009. Evolutionary conservation levels of subunits of histone-modifying protein complexes in fungi. Comp. Funct. Genomics 2009:379317. 10.1155/2009/379317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ekwall K. 2005. Genome-wide analysis of HDAC function. Trends Genet. 21:608–615 [DOI] [PubMed] [Google Scholar]
- 5.Borkovich KA, Alex LA, Yarden O, Freitag M, Turner GE, Read ND, Seiler S, Bell-Pedersen D, Paietta J, Plesofsky N, Plamann M, Goodrich-Tanrikulu M, Schulte U, Mannhaupt G, Nargang FE, Radford A, Selitrennikoff C, Galagan JE, Dunlap JC, Loros JJ, Catcheside D, Inoue H, Aramayo R, Polymenis M, Selker EU, Sachs MS, Marzluf GA, Paulsen I, Davis R, Ebbole DJ, Zelter A, Kalkman ER, O'Rourke R, Bowring F, Yeadon J, Ishii C, Suzuki K, Sakai W, Pratt R. 2004. Lessons from the genome sequence of Neurospora crassa: tracing the path from genomic blueprint to multicellular organism. Microbiol. Mol. Biol. Rev. 68:1–108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yang X-J, Seto E. 2008. The Rpd3/Hda1 family of lysine deacetylases: from bacteria and yeast to mice and men. Nat. Rev. Mol. Cell Biol. 9:206–218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Li Y, Wang C, Liu W, Wang G, Kang Z, Kistler HC, Xu J-R. 2011. The HDF1 histone deacetylase gene is important for conidiation, sexual reproduction, and pathogenesis in Fusarium graminearum. Mol. Plant Microbe Interact. 24:487–496 [DOI] [PubMed] [Google Scholar]
- 8.Ding S-L, Liu W, Iliuk A, Ribot C, Vallet J, Tao A, Wang Y, Lebrun M-H, Xu J-R. 2010. The tig1 histone deacetylase complex regulates infectious growth in the rice blast fungus Magnaporthe oryzae. Plant Cell 22:2495–2508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Izawa M, Takekawa O, Arie T, Teraoka T, Yoshida M, Kimura M, Kamakura T. 2009. Inhibition of histone deacetylase causes reduction of appressorium formation in the rice blast fungus Magnaporthe oryzae. J. Gen. Appl. Microbiol. 55:489–498 [DOI] [PubMed] [Google Scholar]
- 10.Smith K, Kothe G, Matsen C, Khlafallah T, Adhvaryu K, Hemphill M, Freitag M, Motamedi M, Selker E. 2008. The fungus Neurospora crassa displays telomeric silencing mediated by multiple sirtuins and by methylation of histone H3 lysine 9. Epigenetics Chromatin 1:5. 10.1186/1756-8935-1-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tribus M, Bauer I, Galehr J, Rieser G, Trojer P, Brosch G, Loidl P, Haas H, Graessle S. 2010. A novel motif in fungal class 1 histone deacetylases is essential for growth and development of Aspergillus. Mol. Biol. Cell 21:345–353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tribus M, Galehr J, Trojer P, Brosch G, Loidl P, Marx F, Haas H, Graessle S. 2005. HdaA, a major class 2 histone deacetylase of Aspergillus nidulans, affects growth under conditions of oxidative stress. Eukaryot. Cell 4:1736–1745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lee I, Oh J-H, Keats Shwab E, Dagenais TRT, Andes D, Keller NP. 2009. HdaA, a class 2 histone deacetylase of Aspergillus fumigatus, affects germination and secondary metabolite production. Fungal Genet. Biol. 46:782–790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Baidyaroy D, Brosch G, Ahn J-H, Graessle S, Wegener S, Tonukari NJ, Caballero O, Loidl P, Walton JD. 2001. A gene related to yeast HOS2 histone deacetylase affects extracellular depolymerase expression and virulence in a plant pathogenic fungus. Plant Cell 13:1609–1624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hoffmeister D, Keller NP. 2007. Natural products of filamentous fungi: enzymes, genes, and their regulation. Nat. Prod. Rep. 24:393–416 [DOI] [PubMed] [Google Scholar]
- 16.Sanchez JF, Somoza AD, Keller NP, Wang CCC. 2012. Advances in Aspergillus secondary metabolite research in the post-genomic era. Nat. Prod. Rep. 29:351–371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bayram Ö, Braus GH. 2012. Coordination of secondary metabolism and development in fungi: the velvet family of regulatory proteins. FEMS Microbiol. Rev. 36:1–24 [DOI] [PubMed] [Google Scholar]
- 18.Gacek A, Strauss J. 2012. The chromatin code of fungal secondary metabolite gene clusters. Appl. Microbiol. Biotechnol. 95:1389–1404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shwab EK, Bok JW, Tribus M, Galehr J, Graessle S, Keller NP. 2007. Histone deacetylase activity regulates chemical diversity in Aspergillus. Eukaryot. Cell 6:1656–1664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shimizu M, Masuo S, Fujita T, Doi Y, Kamimura Y, Takaya N. 2012. Hydrolase controls cellular NAD, sirtuin, and secondary metabolites. Mol. Cell. Biol. 32:3743–3755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Machida M, Yamada O, Gomi K. 2008. Genomics of Aspergillus oryzae: learning from the history of koji mold and exploration of its future. DNA Res. 15:173–183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Abe K, Gomi K, Hasegawa F, Machida M. 2006. Impact of Aspergillus oryzae genomics on industrial production of metabolites. Mycopathologia 162:143–153 [DOI] [PubMed] [Google Scholar]
- 23.Imamura K, Tsuyama Y, Hirata T, Shiraishi S, Sakamoto K, Yamada O, Akita O, Shimoi H. 2012. Identification of a gene involved in the synthesis of a dipeptidyl peptidase IV inhibitor in Aspergillus oryzae. Appl. Environ. Microbiol. 78:6996–7002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Terabayashi Y, Sano M, Yamane N, Marui J, Tamano K, Sagara J, Dohmoto M, Oda K, Ohshima E, Tachibana K, Higa Y, Ohashi S, Koike H, Machida M. 2010. Identification and characterization of genes responsible for biosynthesis of kojic acid, an industrially important compound from Aspergillus oryzae. Fungal Genet. Biol. 47:953–961 [DOI] [PubMed] [Google Scholar]
- 25.Maruyama J-I, Kitamoto K. 2008. Multiple gene disruptions by marker recycling with highly efficient gene-targeting background (ΔligD) in Aspergillus oryzae. Biotechnol. Lett. 30:1811–1817 [DOI] [PubMed] [Google Scholar]
- 26.Arnaud MB, Chibucos MC, Costanzo MC, Crabtree J, Inglis DO, Lotia A, Orvis J, Shah P, Skrzypek MS, Binkley G, Miyasato SR, Wortman JR, Sherlock G. 2010. The Aspergillus Genome Database, a curated comparative genomics resource for gene, protein and sequence information for the Aspergillus research community. Nucleic Acids Res. 38:D420–D427. 10.1093/nar/gkp751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Quevillon E, Silventoinen V, Pillai S, Harte N, Mulder N, Apweiler R, Lopez R. 2005. InterProScan: protein domains identifier. Nucleic Acids Res. 33:W116–W120. 10.1093/nar/gki442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. 2011. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 28:2731–2739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Szewczyk E, Nayak T, Oakley CE, Edgerton H, Xiong Y, Taheri-Talesh N, Osmani SA, Oakley BR. 2007. Fusion PCR and gene targeting in Aspergillus nidulans. Nat. Protoc. 1:3111–3120 [DOI] [PubMed] [Google Scholar]
- 30.Yamada O, Na Nan S, Akao T, Tominaga M, Watanabe H, Satoh T, Enei H, Akita O. 2003. dffA gene from Aspergillus oryzae encodes l-ornithine N5-oxygenase and is indispensable for deferriferrichrysin biosynthesis. J. Biosci. Bioeng. 95:82–88 [DOI] [PubMed] [Google Scholar]
- 31.Kitamoto K. 2002. Molecular biology of the koji molds. Adv. Appl. Microbiol. 51:129–153 [DOI] [PubMed] [Google Scholar]
- 32.Priebe S, Linde J, Albrecht D, Guthke R, Brakhage AA. 2011. FungiFun: a web-based application for functional categorization of fungal genes and proteins. Fungal Genet. Biol. 48:353–358 [DOI] [PubMed] [Google Scholar]
- 33.Ruepp A, Zollner A, Maier D, Albermann K, Hani J, Mokrejs M, Tetko I, Güldener U, Mannhaupt G, Münsterkötter M, Mewes HW. 2004. The FunCat, a functional annotation scheme for systematic classification of proteins from whole genomes. Nucleic Acids Res. 32:5539–5545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Frye RA. 2000. Phylogenetic classification of prokaryotic and eukaryotic Sir2-like proteins. Biochem. Biophys. Res. Commun. 273:793–798 [DOI] [PubMed] [Google Scholar]
- 35.Smith KM, Dobosy JR, Reifsnyder JE, Rountree MR, Anderson DC, Green GR, Selker EU. 2010. H2B- and H3-specific histone deacetylases are required for DNA methylation in Neurospora crassa. Genetics 186:1207–1216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Brakhage AA. 2013. Regulation of fungal secondary metabolism. Nat. Rev. Microbiol. 11:21–32 [DOI] [PubMed] [Google Scholar]
- 37.Podust LM, Sherman DH. 2012. Diversity of P450 enzymes in the biosynthesis of natural products. Nat. Prod. Rep. 29:1251–1266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kelly DE, Kraševec N, Mullins J, Nelson DR. 2009. The CYPome (cytochrome P450 complement) of Aspergillus nidulans. Fungal Genet. Biol. 46:S53–S61 [DOI] [PubMed] [Google Scholar]
- 39.Oda K, Kobayashi A, Ohashi S, Sano M. 2011. Aspergillus oryzae laeA regulates kojic acid synthesis genes. Biosci. Biotechnol. Biochem. 75:1832–1834 [DOI] [PubMed] [Google Scholar]
- 40.Marui J, Ohashi-Kunihiro S, Ando T, Nishimura M, Koike H, Machida M. 2010. Penicillin biosynthesis in Aspergillus oryzae and its overproduction by genetic engineering. J. Biosci. Bioeng. 110:8–11 [DOI] [PubMed] [Google Scholar]
- 41.Zhang T, Kraus WL. 2010. SIRT1-dependent regulation of chromatin and transcription: linking NAD+ metabolism and signaling to the control of cellular functions. Biochim. Biophys. Acta 1804:1666–1675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Imai S-I, Guarente L. 2010. Ten years of NAD-dependent SIR2 family deacetylases: implications for metabolic diseases. Trends Pharmacol. Sci. 31:212–220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Horio Y, Hayashi T, Kuno A, Kunimoto R. 2011. Cellular and molecular effects of sirtuins in health and disease. Clin. Sci. 121:191–203 [DOI] [PubMed] [Google Scholar]
- 44.Yang B, Miller A, Kirchmaier AL. 2008. HST3/HST4-dependent deacetylation of lysine 56 of histone H3 in silent chromatin. Mol. Biol. Cell 19:4993–5005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.D'Arcy S, Luger K. 2011. Understanding histone acetyltransferase Rtt109 structure and function: how many chaperones does it take? Curr. Opin. Struct. Biol. 21:728–734 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.