Abstract
In this work, we have compared the ability of Pandoraea pnomenusa B356 and of Burkholderia xenovorans LB400 to metabolize diphenylmethane and benzophenone, two biphenyl analogs in which the phenyl rings are bonded to a single carbon. Both chemicals are of environmental concern. P. pnomenusa B356 grew well on diphenylmethane. On the basis of growth kinetics analyses, diphenylmethane and biphenyl were shown to induce the same catabolic pathway. The profile of metabolites produced during growth of strain B356 on diphenylmethane was the same as the one produced by isolated enzymes of the biphenyl catabolic pathway acting individually or in coupled reactions. The biphenyl dioxygenase oxidizes diphenylmethane to 3-benzylcyclohexa-3,5-diene-1,2-diol very efficiently, and ultimately this metabolite is transformed to phenylacetic acid, which is further metabolized by a lower pathway. Strain B356 was also able to cometabolize benzophenone through its biphenyl pathway, although in this case, this substrate was unable to induce the biphenyl catabolic pathway and the degradation was incomplete, with accumulation of 2-hydroxy-6,7-dioxo-7-phenylheptanoic acid. Unlike strain B356, B. xenovorans LB400 did not grow on diphenylmethane. Its biphenyl pathway enzymes metabolized diphenylmethane, but they poorly metabolize benzophenone. The fact that the biphenyl catabolic pathway of strain B356 metabolized diphenylmethane and benzophenone more efficiently than that of strain LB400 brings us to postulate that in strain B356, this pathway evolved divergently to serve other functions not related to biphenyl degradation.
INTRODUCTION
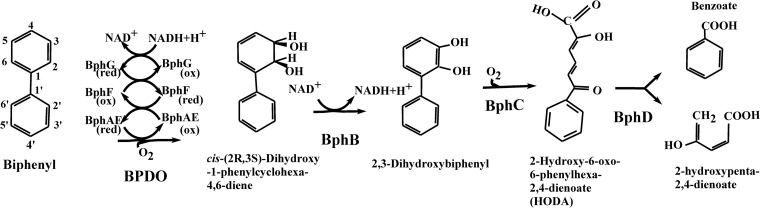
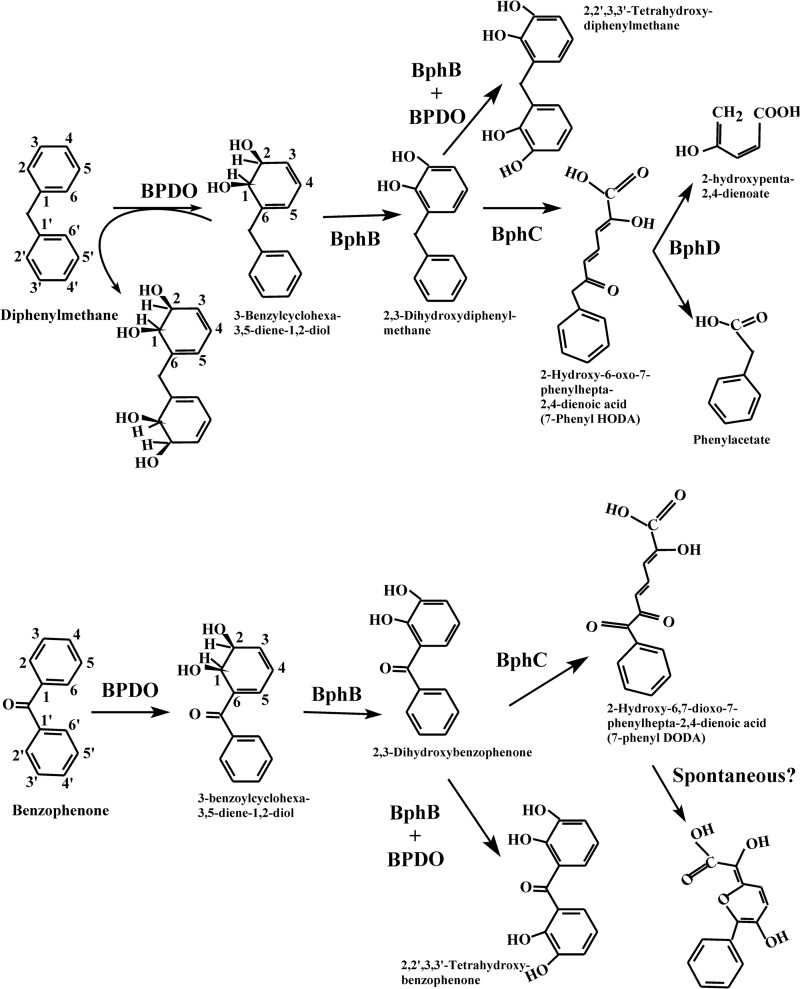
Many investigations have shown that the bacterial biphenyl catabolic pathway enzymes, especially biphenyl dioxygenase (BPDO), which initiates the degradation process, are very versatile (1). The biphenyl pathway, also called the upper pathway, comprises four enzymatic steps that transform biphenyl into benzoic acid, which is further metabolized by a lower pathway (Fig. 1).
Fig 1.
Biphenyl catabolic pathway enzymes and metabolites.
Aside from its ability to metabolize polychlorinated biphenyls (PCBs) (1), BPDO metabolizes many biphenyl analogs (2–7) to generate hydroxylated aromatics. BPDO is composed of three components (Fig. 1). The catalytic component, which is a Rieske-type dioxygenase (RO) (BphAE), is a heterohexamer made up of three α (BphA) and three β (BphE) subunits. The other two components are ferredoxin (BphF) and ferredoxin reductase (BphG), both of which are involved in electron transfer from NADH to BphAE. The catalytic center of the enzyme is located on the C-terminal portion of the BphAE α subunit, which also carries the major structural determinants for substrate specificity (8). There are three phylogenetically distinct clusters of BphAEs (9–11), and the structure of a representative BphAE (also called BphA1A2) from each of these three clusters has now been elucidated. Thus, the Protein Data Bank (PDB) coordinate file for Burkholderia xenovorans LB400 BphAE (BphAELB400) is available (8), as are those for Pandoraea pnomenusa B356 BphAE (BphAEB356) (12) and Rhodococcus jostii RHA1 BphA1A2 (BphA1A2RHA1) (13).
BphAELB400 has been thoroughly investigated, because B. xenovorans LB400 is considered one of the best PCB degraders (8). However, recent studies have shown that BphAEB356 metabolizes flavone, isoflavone, and flavanone (14), as well as 2,6-dichlorobiphenyl (15) and 1,1,1-trichloro-2,2-bis(4-chlorophenyl)ethane (DDT) (16), significantly more efficiently than BphAELB400. In this work, we compared the abilities of strain LB400 and B356 BPDOs and of further enzymes of their biphenyl catabolic pathway to metabolize two biphenyl analogs (diphenylmethane and benzophenone) in which two phenyl rings are bonded to a single carbon. Both are chemicals of environmental importance. According to the U.S. Environmental Protection Agency, in 2003, benzophenone was classified as a high-volume chemical, with an annual production exceeding 450,000 kg (http://toxnet.nlm.nih.gov/). Benzophenone is widely used as a photoinitiator (17). Hydroxybenzophenones are useful building blocks for chemical syntheses, and they are also used as photosensitizers (17). Benzophenones and their xanthone analogs are common plant metabolites with medicinal properties (18), but because of their high demand, they are synthesized industrially. A major synthetic process is through atmospheric oxidation of diphenylmethane in the presence of metal catalysts (17). Aside from being a precursor for benzophenones, diphenylmethane and many of its analogs are used in various other industrial applications. The benzhydryl motif is a fundamental component in antiallergenic agents. It is also a component of hexachlorophene and DDT, and diphenylmethane diisocyanate is a major component of polyurethane. However, very few investigations have addressed the bacterial degradation of diphenylmethane (19, 20) or benzophenone (21). Focht and Alexander (22) have described a Hydrogenomonas isolate that grew on diphenylmethane and was able to cometabolize benzophenone and several related chlorinated analogs. However, the ability of this isolate to metabolize biphenyl has not been examined. More recently, Misawa et al. (19) have shown that Pseudomonas alcaligenes KF707 BPDO and variants derived from it were able to metabolize diphenylmethane. However, the metabolites produced have not been identified, and the steady-state kinetics of these BPDOs toward diphenylmethane were not determined. On the other hand, the ability of BPDO to metabolize benzophenone has never been examined.
While examining the ability of the biphenyl catabolic enzymes of P. pnomenusa B356 and of B. xenovorans LB400 to metabolize these two chemicals, we unexpectedly found that strain B356 grows well on diphenylmethane. In this context, we further investigated diphenylmethane metabolism by strain B356, and we obtained evidence that during growth of the strain on either biphenyl or diphenylmethane, both substrates are metabolized by the same catabolic pathway. This led us to postulate that in strain B356, the biphenyl catabolic pathway evolved to serve other functions not related to biphenyl degradation.
MATERIALS AND METHODS
Bacterial strains, plasmids, chemicals, and general protocols.
Escherichia coli DH11S (23) and E. coli C41(DE3) (24) were used in this study. Wild-type strains P. pnomenusa B356 and B. xenovorans LB400 were described previously (25, 26). BphAEp4 is a mutant of BphAELB400, described previously (8), which was obtained by substitution at two residues, Thr335Ala and Phe336Met. This mutant exhibits an expanded substrate range compared to that of the parent enzyme. Most plasmids used in this study were described previously and are listed in Table 1. Plasmids pET14b[B356-bphF] and pET14b[B356-bphG] carry the genes encoding BphFB356 and BphGB356, respectively, and they were prepared by subcloning the respective genes from pQE31[B356-bphF] and pQE31[B356-bphG] (27) into pET14b. The culture media used were Luria-Bertani (LB) broth (28), basal medium M9 (28), or minimal mineral medium 30 (MM30) (29) amended with various sources of carbon and antibiotics, depending on the experiment. DNA general protocols were done according to Sambrook et al. (28). Diphenylmethane and benzophenone (99% pure) were from Sigma-Aldrich.
Table 1.
Plasmids used in the study
| Plasmid | Protein(s) expressed | Reference or source |
|---|---|---|
| pET14b[LB400-bphAE] | BphAELB400 | 8 |
| pET14b[p4-bphAE] | BphAEp4 | 8 |
| pET14b[B356-bphAE] | BphAEB356 | 16 |
| pQE31[B356-bphAE] | BphAEB356 | 16 |
| pDB31[LB400-bphFG] | BphFLB400, BphGLB400 | 47 |
| pYH31[LB400-bphFGBC] | BphFLB400, BphGLB400, BphBLB400, BphCLB400 | 48 |
| pET14b[B356-bphF] | BphFB356 | This study |
| pET14b[B356-bphG] | BphGB356 | This study |
| pET14b[B356-bphB] | BphBB356 | 49 |
| pQE31[B356-bphC] | BphCB356 | 35 |
Assays to assess ability of diphenylmethane and benzophenone to support growth of strains B356 and LB400 and to induce their biphenyl catabolic pathway.
Induction of the biphenyl catabolic pathway of strains B356 and LB400 by diphenylmethane and benzophenone was assessed by monitoring the amount of 4-chlorobenzoate produced from 4-chorobiphenyl as described previously (14). We also evaluated the ability of wild-type strains B356 and LB400 to grow on diphenylmethane or benzophenone as the sole growth substrate. Cells grown overnight in LB broth were washed twice in MM30 and suspended in MM30 basal medium to an optical density at 600 nm (OD600) of 0.5. This cell suspension was used (100 μl) to inoculate 20 ml of MM30 containing 2 mM biphenyl or diphenylmethane. The cultures were incubated with shaking at 28°C. Cell growth was monitored by determining the CFU. We also used the Bioscreen C system (Growth Curves USA, Piscataway, NJ) to compare the growth kinetics of strain B356 according to the substrate diphenylmethane or biphenyl. In order to prepare the inocula for the Bioscreen C experiments, cells were grown on 1 mM diphenylmethane or biphenyl or on 30 mM sodium acetate for 16 to 18 h, they were washed twice in MM30, and the cells were suspended in the same medium to an OD600 of 0.08. Each well of the Bioscreen C microplate contained 237.5 μl of MM30 supplemented with 1 mM biphenyl or diphenylmethane added in 30 μl dimethyl sulfoxide (DMSO) or 30 mM sodium acetate added in 30 μl water, and they were inoculated with 12.5 μl of the suspension described above. A series of cultures also contained 3-chlorobenzoic acid (2 mM) or 3-chlorocatechol (0.2 mM), each added in 5 μl DMSO. Control cultures with no substrate and uninoculated cultures were also run in the experiments. The cultures were incubated at 28°C and set at low revolution. Both biphenyl and diphenylmethane are poorly soluble in water, but when added at a concentration of 1 mM, the nonsoluble portion of the substrate did not interfere with the OD readings during the Bioscreen C experiments. Each set of cultures was run in triplicate. Growth was also monitored by determining the CFU at various intervals of time during the Bioscreen C experiments.
Analysis of the metabolites produced from diphenylmethane and benzophenone by strains B356 and LB400 and by enzymes of their biphenyl catabolic pathway.
The metabolites produced during growth of strain B356 on 2 mM diphenylmethane in 20 ml MM30 were extracted with ethyl acetate at neutral pH and at pH 4 from the supernatant of 22-h-old cultures. They were then treated with butylboronate (nBuB) or N,O-bis-trimethylsilyl trifluoroacetamide (TMS) for gas chromatography-mass spectrometry (GC-MS) analyses according to previously described protocols (30). A similar protocol was used to examine the metabolites produced from benzophenone by biphenyl-induced cells of strain B356. However, in this case, the cells were grown on biphenyl to reach log phase, and then they were suspended at an OD600 of 3.0 in M9 medium and incubated at 28°C and 100 rpm for 60 min in the presence of 0.2 mM benzophenone.
Metabolites from diphenylmethane and benzophenone also were analyzed from suspensions of isopropyl β-d-1-thiogalactopyranoside (IPTG)-induced whole cells of E. coli[pDB31 B356-bphFG] or E. coli[pYH31 LB400-bphFGBC] also harboring pQE31[B356-bphAE] or pQE31[LB400-bphAE] according to a previously published protocol (30). The induced cells were suspended at an OD600 of 5.0 in 50 mM sodium phosphate buffer, pH 7.0, and the metabolites generated after 30 min of incubation at 37°C were extracted and analyzed by GC-MS.
GC-MS analyses were performed using a Hewlett Packard HP6980 series gas chromatograph interfaced with an HP5973 mass selective detector (Agilent Technologies). The mass selective detector was operated in electron impact (EI) mode and used a quadrupole mass analyzer. Under these conditions, the instrument resolution is 0.1 atomic mass units, which is sufficient to clearly distinguish between two compounds of atomic masses differing by a single atomic mass unit.
Assays to identify diphenylmethane and benzophenone metabolites produced from BphAEB356, BphAELB400, and BphAEp4 and to determine their steady-state kinetics.
Reconstituted His-tagged BPDO preparations were used in these experiments. His-tagged purified enzyme components were produced and purified by following protocols published previously (31). The enzyme assays were performed in a volume of 200 μl in 50 mM morpholineethanesulfonic (MES) buffer, pH 6.0, at 37°C as described previously (32). For metabolite analyses, the reaction medium was incubated for 10 min and the metabolites were extracted at pH 6.0 with ethyl acetate, and then they were treated with nBuB or TMS for GC-MS analyses as described above. The steady-state kinetics were determined by recording the oxygen consumption rates according to a protocol described previously, using a Clarke-type Hansatech model DW1 oxygraph (33). They were determined from three separately prepared purified preparations of the enzymes.
Purification and NMR analysis of 2,2′,3,3′-tetrahydroxybenzophenone.
2,2′,3,3′-Tetrahydroxybenzophenone was prepared using a coupled reaction composed of His-tagged purified preparations of B356 BPDO (BphAEFGB356) plus BphBB356. Each enzyme reaction mixture contained 50 nmol benzophenone, 0.6 nmol of each B356 BPDO component (BphAEB356, BphFB356, and BphGB356), 2 nmol BphBB356, 100 nmol NADH, and 100 nmol NAD in 200 μl (total volume) of 50 mM MES buffer (pH 6.0). The mixture was incubated for 15 min at 37°C and then extracted at pH 6.0 with ethyl acetate. The extract was concentrated 20-fold by evaporation under a stream of nitrogen, and this preparation was injected into an XDB-C8 column (4.6 by 150 mm). The column was eluted at 1 ml/min with a linear gradient starting from 80% high-performance liquid chromatography (HPLC)-grade water with 0.085% ortho-phosphoric acid and 20 to 50% acetonitrile at 12 min. The detector was set at a wavelength of 280 nm. The peak of the metabolite was collected, the solution was immediately adjusted to pH 6.0 with 0.1 M NaOH, and the metabolite was extracted with ethyl acetate. Its identity and purity were verified by GC-MS analysis of its TMS derivative before running the nuclear magnetic resonance (NMR) analysis. The NMR spectra were obtained at the Quebec/Eastern Canada High Field NMR Facility at McGill University (Montreal, Quebec, Canada) with a Bruker 500-mHz spectrometer. The analyses were carried out in deuterated acetone at room temperature.
Docking and structure analysis.
Dimer AB of BphAELB400 (RCBS Protein Data Bank [PDB] entry 2XRX) and of BphAEB356 (PDB entry 3GZX) were used as protein targets, and they were prepared as previously described (16). Ligands all were downloaded as sdf files from PubChem (http://pubchem.ncbi.nlm.nih.gov) and converted into pdb format in Discover Studio Visualizer 2.5. Both proteins and ligands were processed with AutoDockTools to obtain their proper pdbqt format. The searching space for the ligand was centered on mononuclear iron and contained 20 Å in each x, y, and z direction. Autodock 4 (34) with default parameters was used to perform the automatic docking.
RESULTS
Growth of P. pnomenusa B356 and B. xenovorans LB400 on diphenylmethane and benzophenone.
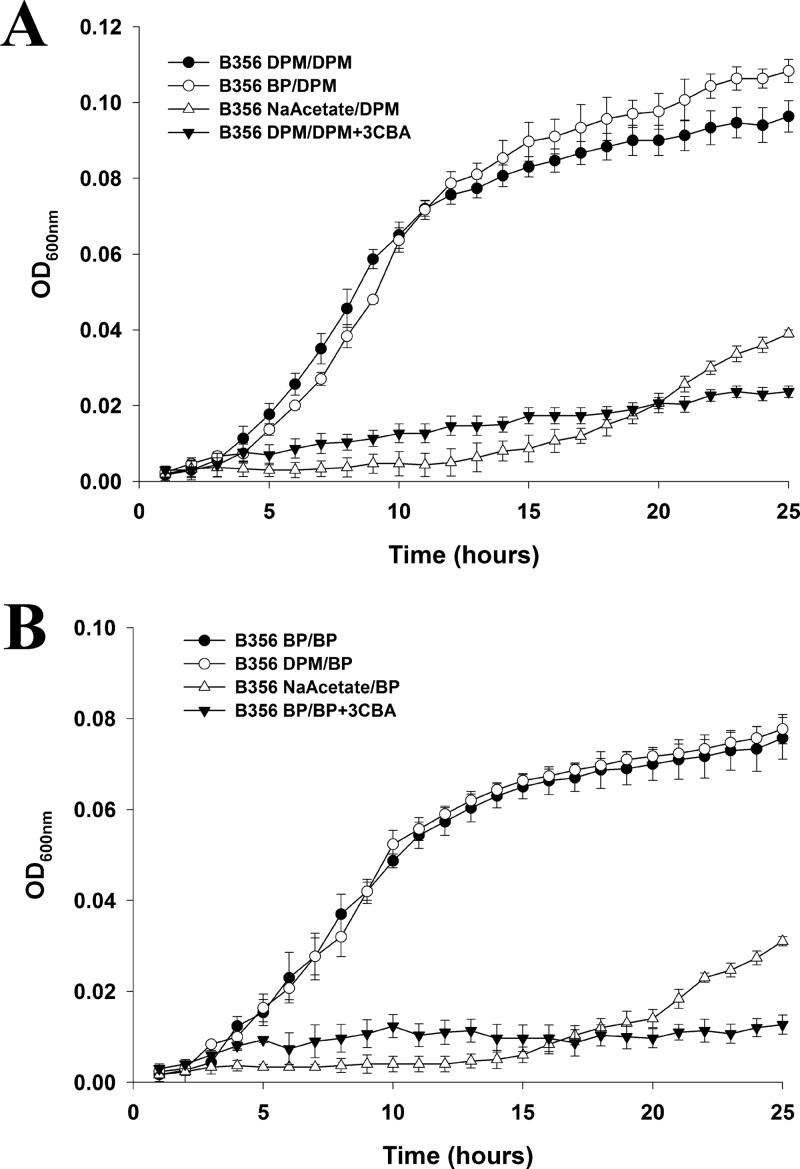
Neither P. pnomenusa B356 nor B. xenovorans LB400 grew when benzophenone was used as the sole growth substrate. Using a previously described 4-chlorobiphenyl conversion assay (14), we showed that benzophenone was unable to induce the biphenyl catabolic pathway of both strains. Similarly, diphenylmethane did not serve as the growth substrate for strain LB400 and did not induce its biphenyl catabolic pathway. However, remarkably, strain B356 grew very well in MM30 containing 2 mM diphenylmethane as the sole growth substrate. Under those conditions, CFU values of up to 4 × 109 cells/ml were obtained within 36 h at 28°C. In addition, using the 4-chlorobiphenyl conversion assay (14), we found that a suspension of log-phase cells grown on 2 mM diphenylmethane and adjusted to an OD600 of 1.0 produced 329 ± 20 μM 4-chlorobenzoate from 1.25 mM 4-chlorobiphenyl after 2 h of incubation. In comparison, a resting suspension of log-phase cells of strain B356 grown on 2 mM biphenyl and tested under the same conditions produced 330 ± 17 μM 4-chlorobenzoate. This suggested that the biphenyl catabolic pathway of B356 was induced during growth on diphenylmethane. Using the Bioscreen C system, we have compared the growth kinetics of strain B356 according to the substrate used, biphenyl or diphenylmethane. In addition, we have determined the effect of interchanging the growth substrate on growth kinetics (biphenyl or diphenylmethane) to prepare the inocula. Results shown in Fig. 2 clearly demonstrate that strain B356 grows as well on diphenylmethane as on biphenyl, sometimes even better. Furthermore, replacing diphenylmethane with biphenyl to prepare the inoculum for the Bioscreen C experiments did not affect significantly the growth kinetics on diphenylmethane; the lag phases were identical and the growth rates were similar (0.35 ± 0.01 h−1 and 0.34 ± 0.02 h−1 for diphenylmethane- or biphenyl-induced cells, respectively). Likewise, when biphenyl was used as the growth substrate, the lag phases were very similar whether the inocula were prepared by growing the cells on biphenyl or diphenylmethane, and the growth rates were identical (0.25 ± 0.02 h−1). Conversely, when cells were grown on sodium acetate to prepare the inoculum, the cells grew very poorly whether the Bioscreen C experiments were run using biphenyl or diphenylmethane as the substrate (Fig. 2), exhibiting long lag phases and low growth rates (0.14 h−1), and the CFU count did not exceed 5 × 107 cells/ml at the end of the log phase. We have no data explaining why B356 grew better on diphenylmethane than on biphenyl. This result shows that the combined upper and lower pathways metabolized diphenylmethane more efficiently than biphenyl. However, the facts that interchanging the substrates to prepare the inocula for the Bioscreen C experiments did not affect the growth kinetics and did not prolong the lag phase provide strong evidence that the two substrates were degraded by the same pathway, which they both induced.
Fig 2.
Growth curves of strain B356 on diphenylmethane when the inocula were prepared by growing the cells on either diphenylmethane (DPM/DPM), biphenyl (BP/DPM), or sodium acetate (NaAcetate/DPM) (A) or on biphenyl when the inocula were prepared by growing the cells on either biphenyl (BP/BP), diphenylmethane (DPM/BP), or sodium acetate (NaAcetate/BP) (B). A growth curve also was obtained for cells pregrown on diphenylmethane and then grown on diphenylmethane plus 3-chlorobenzoate (DPM/DPM+3CBA) or cells pregrown on biphenyl and then grown on biphenyl plus 3-chlorobenzoate (BP+3CBA). The growth curves were obtained using a Bioscreen C system as described in Materials and Methods.
As further evidence that growth on diphenylmethane proceeded through the biphenyl catabolic pathway, we examined the effect of adding 3-chlorocatechol and 3-chlorobenzoate to the growth medium. In the Bioscreen C experiments, 0.2 mM 3-chlorocatechol completely inhibited growth of strain B356 on both biphenyl and diphenylmethane (data not shown). In a previous work, we showed that B356 BphC is very sensitive to 3-chlorocatechol (35), and its presence in the growth medium strongly inhibits biphenyl metabolism (36).
It is also noteworthy that in the Bioscreen C experiments, when 3-chlorobenzoate (2 mM) was added as a cosubstrate, cell growth was strongly inhibited (Fig. 2). The same inhibition was observed whether the cells were pregrown on diphenylmethane or biphenyl or whether the Bioscreen C experiments were run using biphenyl or diphenylmethane as the substrate. This inhibition was not observed when the Bioscreen C experiments were run using sodium acetate as the growth substrate (not shown). The significant inhibitory effect of 3-chlorobenzoate on the metabolism of biphenyl by strain B356 has been reported previously (36). It was attributed to the ability of the lower biphenyl catabolic pathway to convert 3-chlorobenzoate into 3-chlorocatechol, which strongly inhibits the upper biphenyl catabolic pathway (36). Therefore, data obtained in the Bioscreen C experiments suggest that a pathway able to metabolize 3-chlorobenzoate into 3-chlorocatechol is induced during growth on diphenylmethane, and 3-chlorocatechol strongly impairs the metabolism of the upper pathway. The fact that both the diphenylmethane and biphenyl degradation pathways responded similarly to the presence of 3-chlorocatechol and 3-chlorobenzoate provides additional evidence that both substrates are metabolized by the same pathway.
Because phenylacetate would be the expected end product if diphenylmethane was metabolized by the enzymes of the upper biphenyl pathway, we have determined whether it could serve as the growth substrate for strains B356 and LB400. In both cases, cells grew very well, reaching CFU values exceeding 109 cells/ml within 18 h at 28°C.
Metabolism of diphenylmethane and benzophenone by P. pnomenusa B356.
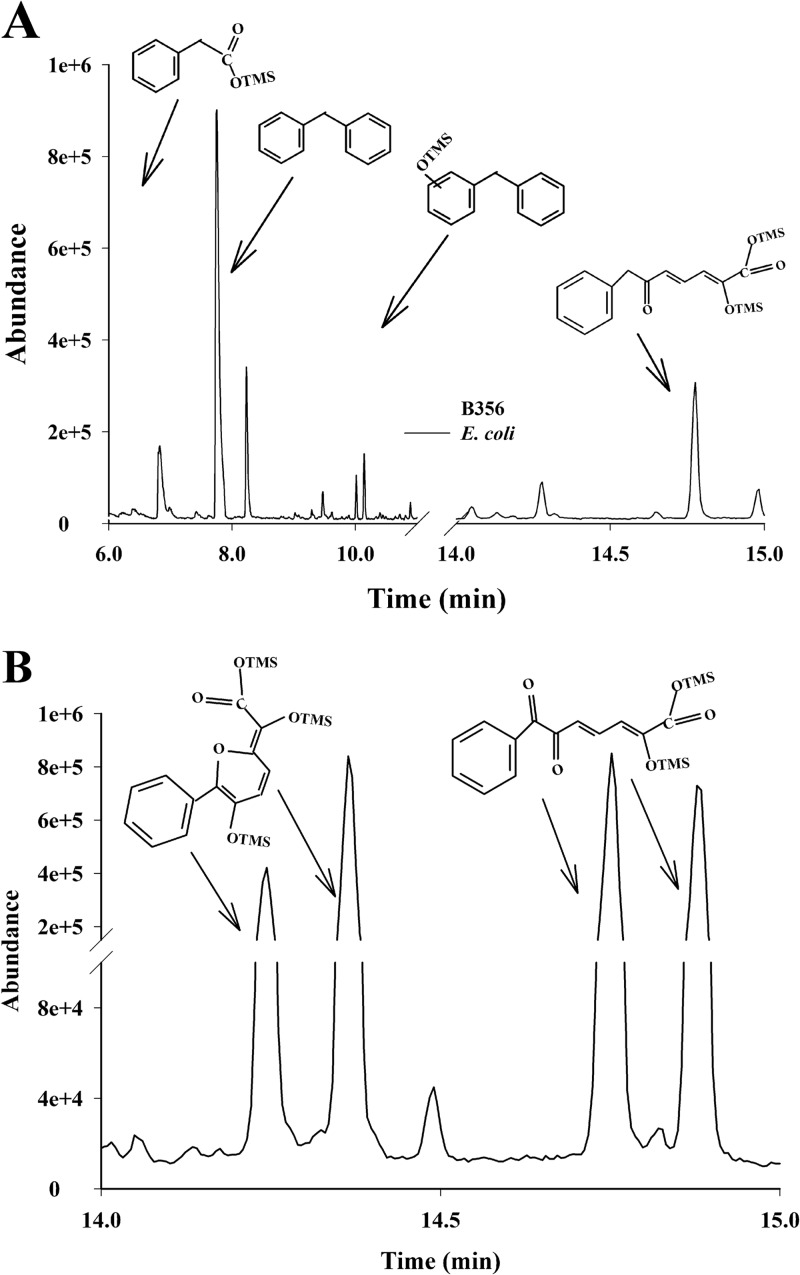
When diphenylmethane-grown cells of strain B356 were inoculated in MM30 containing 2 mM diphenylmethane, the substrate (approximately 40 μmol) was almost completely metabolized after 22 h. The mass spectral features of the major metabolite in the acidic ethyl acetate extract were identical to those of an authentic standard of phenylacetic acid (Fig. 3A; spectra are not shown). Based on the area under the GC-MS peak, approximately 15 nmol was present in the 22-h-old cultures. Among the minor metabolites, one peak eluting at 14.76 min (Fig. 3A) exhibited mass spectral features that correspond to those of a meta-fission metabolite resulting from the catalytic cleavage of a catechol derivative of diphenylmethane. This metabolite was tentatively identified as 2-hydroxy-6-oxo-7-phenylhepta-2,4-dienoic acid (7-phenyl HODA) {m/z 376 (M+), m/z 361 (M+ − CH3), m/z 333 (M+ − CH3 − CO), and m/z 259 [M+ − COO(CH3)3Si]} (see Fig. S1 in the supplemental material for the spectrum). This is the metabolite that would be expected if the hydroxylation reaction had occurred on the ortho-meta carbons of one benzene ring of diphenylmethane to generate 3-benzylcyclohexa-3,5-diene-1,2-diol. The identity of the acidic metabolite as 7-phenyl HODA is consistent with the fact that a metabolite exhibiting the same mass spectral features was produced from diphenylmethane by resting cells of E. coli pQE31[B356-bphAE]+pYH31[LB400-bphFGBC] (Fig. 3A). The E. coli resting cell assay also shows that BphBLB400 and BphCLB400 can further metabolize 3-benzylcyclohexa-3,5-diene-1,2-diol generated by BphAEB356.
Fig 3.
Total ion chromatograms of acidic metabolites detected in 22-h-old cultures of strain B356 grown on diphenylmethane (black line) or in resting cell suspensions of E. coli carrying plasmids pQE31[B356-bphAE] and pYH31[LB400-bphFGBC] incubated for 30 min with diphenylmethane (gray line) (A) or acidic metabolites produced by a suspension of biphenyl-induced cells of strain B356 incubated for 60 min with benzophenone (B). The protocols are described in Materials and Methods. Shown are the peaks of the TMS-treated metabolites that were identified from their mass spectral features. The unlabeled peaks were also detected in controls unexposed to the substrate. OTMS, trimethylsilyl-oxygen complex.
The acidic extracts of cells of strain B356 grown on diphenylmethane also contained a metabolite whose TMS derivative exhibited a molecular mass at m/z 256 corresponding to a mono-hydroxy-diphenylmethane (Fig. 3A; spectrum not shown). This metabolite was presumably generated from the corresponding dihydrodiol metabolite during the extraction procedure at pH 4.
In the culture extracts prepared at neutral pH, 3-benzylcyclohexa-3,5-diene-1,2-diol and 2,3-dihydroxydiphenylmethane were detected only in trace amounts (not shown). The fact that only small amounts of acidic and neutral metabolites were detected in the culture medium of 22-h-old cultures shows diphenylmethane metabolism is very efficient, and this is consistent with the rapid growth on this substrate.
Strain B356 was unable to grow on benzophenone. However, biphenyl- or diphenylmethane-grown resting cell suspensions metabolized benzophenone and produced a yellow meta-fission metabolite which was detected within minutes after substrate addition. Two metabolites (Fig. 3B) with similar spectral features were detected by GC-MS analysis of their TMS derivatives {m/z 390 (M+), m/z 375 (M+ − CH3), m/z 347 (M+ − CH3 − CO), m/z 273 [M+ − COO(CH3)3Si], m/z 258 [M+ − COO(CH3)3Si − CH3], m/z 184 [M+ − COO(CH3)3Si − O(CH3)3Si]} (see Fig. S2 in the supplemental material), and their concentrations increased steadily between 5 and 30 min of incubation (not shown). These metabolites were tentatively identified as isomers of 2-hydroxy-6,7-dioxo-7-phenylheptanoic acid (7-phenyl DODA) that should be expected from the meta-fission of 2,3-dihydroxybenzophenone. Although the formation of isomers of the meta-fission products resulting from the BphC reaction has been reported in several other investigations (33, 37, 38), their mechanism of formation has not yet been elucidated. However, as proposed previously (38) for the meta-fission products of chlorobiphenyls, these isomers may be generated during the oxidative cleavage of catechol, where the formation of C-7 keto next to the phenyl ring and C-1 carboxylic functions may promote the isomerization of the double bonds at C-2 and C-4.
No 2,3-dioxo-3-phenylpropanoic acid, which presumably is the metabolite produced from 7-phenyl DODA by the phenyl HODA hydrolase (BphD), was detected, even after 1 h of incubation in the presence of the substrate. Therefore, we were unable to obtain evidence that benzophenone metabolism goes beyond the meta-fission reaction. On the other hand, two metabolites (Fig. 3B) that exhibited spectral features that could correspond to pyranol isomers {m/z 462 (M+), m/z 447 (M+ − CO), m/z 419 (M+ − CH3 − CO), m/z 345 [M+ − COO(CH3)3Si]} (Fig. 3B; also see Fig. S3 in the supplemental material for the spectrum) were detected. These pyranol isomers may have been generated from a cyclization reaction through intramolecular rearrangements. At this time, there is no documented evidence of spontaneous tautomerization or cyclization of phenyl HODAs. The fact that the same metabolites were produced by an IPTG-induced resting cell suspension of a 1:1 mixture of recombinant E. coli pQE31[B356-bphAE]+pYH31[LB400-bphFGB] plus E. coli pQE31[B356-bphC] (not shown) suggests they were produced spontaneously from 7-phenyl DODA in the cell suspensions or during the TMS reaction. However, we cannot exclude other mechanisms of formation involving unspecific enzymatic reactions occurring in both strain B356 and E. coli. Therefore, their production remains unexplained at this time.
Metabolism of diphenylmethane by purified BphAEB356, BphAELB400, and BphAEp4.
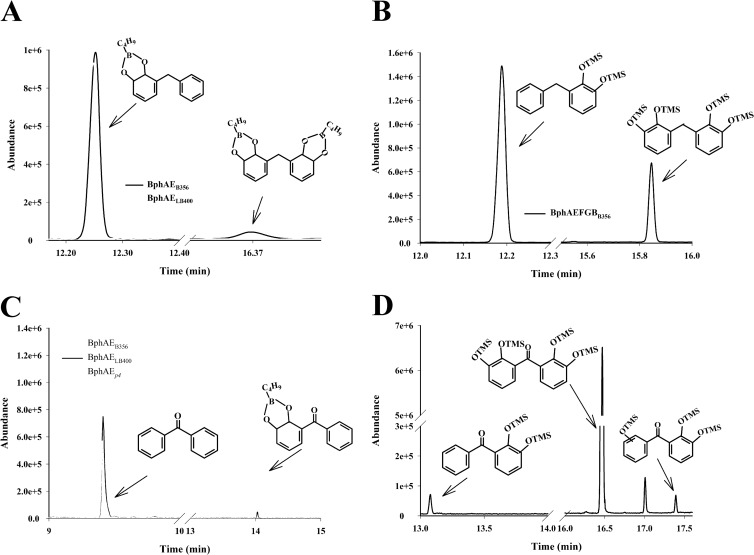
To confirm that the biphenyl catabolic enzymes of strain B356 metabolize diphenylmethane efficiently, we have examined the catalytic properties of BphAEB356 toward diphenylmethane, and we have compared them to those of BphAELB400 and BphAEp4. GC-MS analysis of the nBuB-treated diphenylmethane metabolites revealed two metabolites (Fig. 4A). Since nBuB reacts only with vicinal hydroxyl groups, the mass spectral features (see Fig. S4 in the supplemental material) of the major metabolite (m/z 268 [M+], m/z 211 [M+ − nBu], m/z 184 [M+ − nBuBO], m/z 177 [M+ − C7H7], m/z 168 [M+ − nBuBO2], m/z 156 [M+ − nBuBO − CO]) must correspond to a dihydrodiol. Docking experiments (see below) suggested the oxygenation occurred on the ortho-meta carbons. Therefore, on the basis of its mass spectral features, the major metabolite was tentatively identified as 3-benzylcyclohexa-3,5-diene-1,2-diol.
Fig 4.
Total ion chromatograms of metabolites produced from diphenylmethane by purified preparations of BphAEB356 (black line) and BphAELB400 (gray line) (A) or by a coupled reaction composed of purified BphAEB356 plus BphBB356 (B). (C) Total ion chromatograms of metabolites produced from benzophenone by purified preparations of BphAEB356 (light gray line), BphAELB400 (black line), and BphAEp4 (dark gray line). (D) Total ion chromatograms of metabolites produced from benzophenone by a coupled reaction composed of purified BphAEB356 plus BphBB356. The procedures to purify the enzymes, to set up the reactions, and to extract the metabolites for GC-MS analysis are described in Materials and Methods. Shown are the peaks of nBuB- or TMS-treated metabolites that were identified from their mass spectral features. The unlabeled peaks were also present in controls unexposed to the substrate.
The minor metabolite contains two pairs of vicinal hydroxyl groups (Fig. 4A). This metabolite presumably was generated from the catalytic ortho-meta hydroxylation of the nonhydroxylated ring of 3-benzylcyclohexa-3,5-diene-1,2-diol. It was tentatively identified as 3-[(5,6-dihydroxycyclohexa-1,3-dien-1-yl)methyl]cyclohexa-3,5-diene-1,2-diol, showing ions at m/z 368 (M+), 284 (M+ − nBuBO), 200 (M+ − nBuBO − nBuBO), 184 (M+ − nBuBO − nBuBO2), 177 (M+ − nBuBO2 − C7H7), 168 (M+ − nBuBO2 − nBuBO2), and 156 (M+ − nBuBO2 − nBuBO − CO) (see Fig. S5 in the supplemental material). The fragmentation ion at m/z 177 provides evidence that both rings were oxidized.
The production of 3-benzylcyclohexa-3,5-diene-1,2-diol and 3-[(5,6-dihydroxycyclohexa-1,3-dien-1-yl)methyl]cyclohexa-3,5-diene-1,2-diol as sole products of the B356-BPDO reaction was confirmed by the fact that a coupled reaction composed of purified preparations of BphAEB356 plus BphBB356 generated two metabolites. On the basis of the mass spectra of their TMS derivatives, the major one was identified as 2,3-dihydroxydiphenylmethane (3-benzylbenzene-1,2-diol) and the minor one as 2,2′,3,3′-tetrahydroxydiphenylmethane {3-[(2,3-dihydroxyphenyl)methyl]benzene-1,2-diol} (Fig. 4B). The diagnostically important ions for the TMS-derived 2,3-dihydroxydiphenylmethane (see Fig. S6 in the supplemental material) comprise the molecular ion at m/z 344 and fragmentation ions at m/z 329 (M+ − CH3), 271 [M+ − (CH3)3Si], 255 [M+ − O(CH3)3Si], 225 [M+ − O(CH3)3Si − (CH3)2]. For the TMS-derived 2,2′,3,3′-tetrahydroxydiphenylmethane (see Fig. S7), they are m/z 520 (M+), 505 (M+ − CH3), 447 [M+ − (CH3)3Si], 431 [M+ − O(CH3)3Si], 342 [M+ − O(CH3)3Si − O(CH3)3Si], 312 [M+ − O(CH3)3Si − O(CH3)3Si − (CH3)2], 253 [M+ − O(CH3)3Si − O(CH3)3Si − O(CH3)3Si]. The vicinity of the two hydroxyl groups in these metabolites was confirmed by the mass spectra of the nBuB derivatives (not shown).
When BphAELB400 was incubated for 10 min with the substrate, it produced approximately the same amount of 3-benzylcyclohexa-3,5-diene-1,2-diol from diphenylmethane as BphAEB356, but no 3-[(5,6-dihydroxycyclohexa-1,3-dien-1-yl)methyl]cyclohexa-3,5-diene-1,2-diol was produced (Fig. 4A). The metabolic pattern of BphAEp4 toward diphenylmethane was very similar to that of the parental enzyme BphAELB400 (not shown). Therefore, BphAEB356 was the only one of the three enzymes to further oxidize the dihydroxylated metabolite.
The steady-state kinetic parameters of purified preparations of BphAEB356, BphAEp4, and BphAELB400 toward diphenylmethane were calculated from the initial oxygen consumption, and they were consistent with the GC-MS analysis. Diphenylmethane was as good a substrate as biphenyl for all three enzymes. Thus, the kcat and kcat/Km values for BphAEB356 (Table 2) were in the range reported for biphenyl (4.3 s−1 and 63 × 103 M−1 s−1, respectively) (16) when this enzyme was used under the same reaction conditions. Similarly, the kcat and kcat/Km values for BphAELB400 and for BphAEp4 (Table 2) were very close to those obtained when biphenyl was the substrate and the reactions were run under identical conditions (the reported values were 0.9 s−1 and 41 × 103 M−1 s−1 for BphAELB400, respectively, and 1.0 s−1 and 31 × 103 M−1 s−1 for BphAEp4, respectively) (31).
Table 2.
Steady-state kinetic parametersa of BphAEB356, BphAELB400, and BphAEp4 toward diphenylmethane and benzophenone
| Substrate and enzyme | Km (mM) | kcat (s−1) | kcat/Km (103 · M−1 · s−1) |
|---|---|---|---|
| Diphenylmethane | |||
| BphAEB356 | 63.0 (5.6) | 8.9 (0.3) | 141.3 (7.5) |
| BphAELB400 | 15.4 (1.1) | 1.0 (0.1) | 64.9 (2.7) |
| BphAEp4 | 17.9 (2.2) | 1.6 (0.1) | 89.4 (5.5) |
| Benzophenone | |||
| BphAEB356 | 65.3 (8.5) | 1.2 (0.2) | 18.4 (0.6) |
| BphAELB400 | NDb | ND | ND |
| BphAEp4 | 9.1 (2.1) | 0.1 (0.0) | 11.0 (1.6) |
The steady-states kinetics were determined from the oxygen consumption rates as described in Materials and Methods. The values are results ± standard deviations from three independently produced enzyme preparations.
ND, not determined; metabolism was too slow to determine values accurately.
Benzophenone metabolism by purified BphAEB356, BphAELB400, and BphAEp4.
Unlike diphenylmethane, benzophenone is metabolized differently by BphAEB356 and BphAELB400. A purified preparation of BphAEB356 completely oxidized 50 nmol benzophenone in 10 min to generate a single metabolite (Fig. 4C), tentatively identified as 3-benzoylcyclohexa-3,5-diene-1,2-diol from the GC-MS spectrum of its nBuB derivative (see Fig. S8 in the supplemental material) (ions at m/z 282 [M+] and at 225 [M+ − nBu], 198 [M+ − nBuBO], 182 [M+ − nBuBO2], and 170 [M+ − nBuBO − CO]). Under the same conditions, BphAELB400 and BphAEp4 oxidized only a fraction of the added 50 nmol benzophenone, but both enzymes produced the same metabolite as BphAEB356 (Fig. 4C). This is consistent with the steady-state kinetics shown in Table 2, where the turnover rate of the reaction of BphAEB356 toward benzophenone was 12 times higher than that for BphAEp4, and the turnover rate of the reaction of BphAELB400 was too low to obtain reliable values.
When BphBB356 was used to oxidize 3-benzoylcyclohexa-3,5-diene-1,2-diol, 2,3-dihydroxybenzophenone was produced (not shown) and was identified from the GC-MS spectral features of its TMS derivative {ions at m/z 358 (M+) and m/z 343 (M+ − CH3), 270 [M+ − (CH3)4Si], and 212 [M+ − (CH3)4Si − (CH3)2Si]} (see Fig. S9 in the supplemental material).
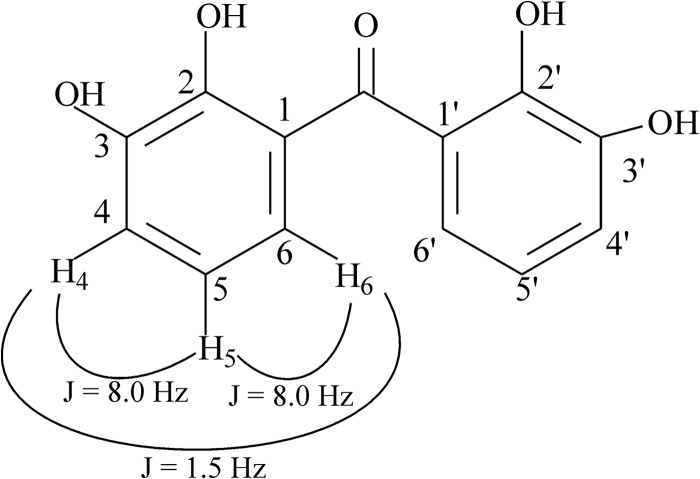
Remarkably, when benzophenone at a concentration of 50 nmol was the substrate for the coupled reaction composed of BphAEB356 plus BphBB356, it was almost completely metabolized to 2,2′,3,3′-tetrahydroxybenzophenone (Fig. 4D), which was identified from its GC-MS and NMR spectra. The other two metabolites, 2,3-dihydroxybenzophenone and 2,2′,3- or 2,3,3′-trihydroxybenzophenone, represented less than 1% of total metabolites produced by this reaction. The spectral features of the TMS-treated major metabolite (see Fig. S10 in the supplemental material) was comprised of a molecular ion at m/z 534 plus fragmentation ions at m/z 519 (M+ − CH3), 446 [M+ − (CH3)4Si], and 358 [M+ − (CH3)4Si − (CH3)4Si]. The position of the hydroxyl groups was confirmed by NMR analysis. The spectrum showed only three signals for six protons between 6.80 and 7.10 ppm. This indicates that there are two identical aromatic rings, and each one contains 3 protons. The chemical shifts for the protons labeled H4, H5, and H6 in Fig. 5 were recorded at 6.824 ppm (triplet) (H5), at 7.096 (doublet of doublets), and 6.969 (doublet of doublets) (H4 and H6). The coupling constant between the three protons (Fig. 5) and the shape of the peaks (1 triplet and 2 doublet of doublets) revealed that the 3 protons of each aromatic ring were vicinal to each other. With this in mind, the only possibility for the structure of the metabolite was 2,2′,3,3′-tetrahydroxybenzophenone.
Fig 5.
Structural features of the hydroxylated metabolite obtained from benzophenone by the coupled reaction of BphAEB356 plus BphBB356, which was identified as 2,2′-3,3′-tetrahydroxybenzophenone by NMR analysis.
Docking experiments with diphenylmethane and benzophenone.
We docked diphenylmethane and benzophenone in the substrate-bound form of BphAEB356 and BphAELB400 after removing biphenyl. For both substrates, the conformation of the top-ranked docked molecules in BphAEB356 exhibited an orientation that would enable oxygenation of the ortho-meta carbons of the phenyl ring. In both cases, the ortho-meta carbons closely aligned with carbons 2 and 3 of the oxidized ring of biphenyl in the complexed form (Fig. 6A and B). This suggests BphAEB356 produces 3-benzylcyclohexa-3,5-diene-1,2-diol from diphenylmethane and 3-benzoylcyclohexa-3,5-diene-1,2-diol from benzophenone, and it is consistent with the NMR spectral data that confirm that 2,2′,3,3′-tetrahydroxybenzophenone was produced from benzophenone by the combined reaction of BphAEB356 plus BphBB356.
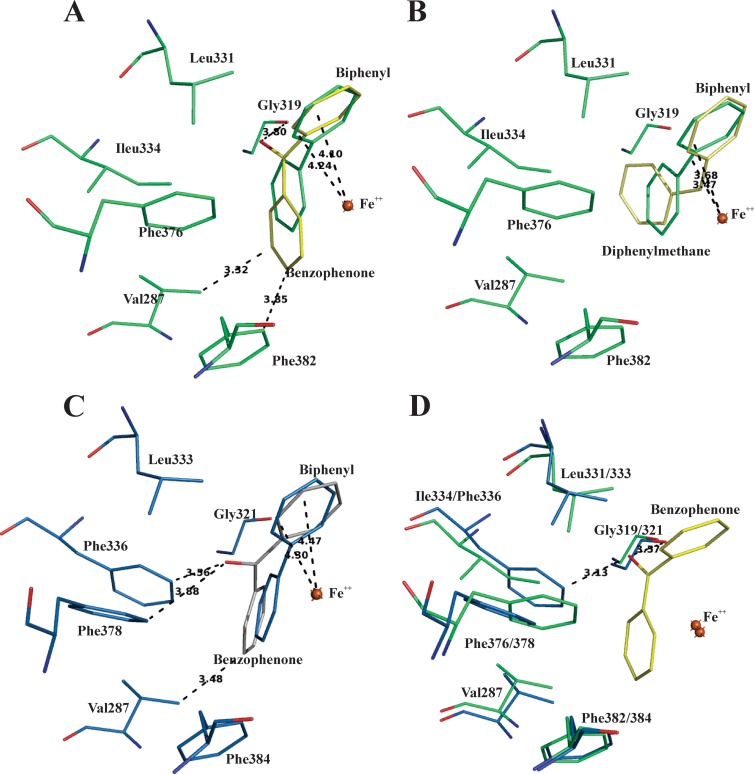
Fig 6.
(A) Superposition of catalytic center residues of benzophenone-docked (yellow) and biphenyl-bound (green) forms of BphAEB356. (B) Superposition of catalytic center residues of diphenylmethane-docked (yellow) and biphenyl-bound (green) forms of BphAEB356. (C) Superposition of catalytic center residues of benzophenone-docked (white) and biphenyl-bound (blue) forms of BphAELB400. (D) Superposition of the biphenyl-bound form of BphAELB400 (blue) after removal of biphenyl substrate and the benzophenone-docked (yellow) BphAEB356 (green). The oxygen atoms are in red.
As shown in Fig. 6A and B, the conformation of benzophenone inside the catalytic pocket of BphAEB356 differs from that of diphenylmethane. Therefore, the carbonyl group strongly influences the interaction between the substrate and the amino acid residues that line the catalytic pocket. However, the ortho-meta carbons of the docked benzophenone aligned very well with the reactive carbons of biphenyl in the biphenyl-bound form of BphAEB356 (Fig. 6A). In the case of BphAELB400, the top-ranked conformations for benzophenone were at a distance from the catalytic iron that may have allowed catalytic hydroxylation (Fig. 6C). However, the reactive ring did not align as well as for BphAEB356 with the reactive ring of biphenyl, and this may explain why BphAELB400 catalyzed the oxygenation of benzophenone poorly. On the other hand, we cannot exclude the possibility that other types of interactions between protein structures and the substrate, which could not be reproduced in the docking experiments, prevent its productive binding to the BphAELB400 catalytic pocket. Since the conformation of benzophenone in the docked BphAEB356 structure appears to be more favorable for the catalytic reaction, we have superposed the benzophenone docked structure of BphAEB356 on the structure of the biphenyl-bound BphAELB400 after biphenyl removal (Fig. 6D). As shown in the figure, the benzophenone carbonyl oxygen was very close to both Gly321 and Phe336 of BphAELB400. Therefore, in this conformation of the substrate, the proximity of these two residues relative to the carbonyl oxygen may hinder proper binding. This is consistent with previous observations (8, 16) where Gly321 and Phe336, two residues lining the catalytic pocket, appeared to play a significant role in binding the substrate's analogs. It is also consistent with the observation that BphAEp4, the doubly substituted Thr335Ala/Phe336Met mutant of BphAELB400, metabolized benzophenone more efficiently than its parent (Table 2). In this case, the Thr335Ala mutation releases constraints imposed by Thr335 on Gly321, allowing movement of its carbonyl group during substrate binding to create more space to accommodate larger substrates.
On the other hand, in the case of 2,3-dihydroxybenzophenone, none of the 20 top-ranked conformations of the docked substrate were at a distance from BphAEB356 catalytic iron that could have allowed a catalytic reaction. Therefore, perhaps structural features involving an induced-fit mechanism that the docking experiment could not reproduce are required to allow productive binding of this substrate to the catalytic pocket of BphAEB356.
DISCUSSION
Unlike B. xenovorans LB400, P. pnomenusa B356 grew remarkably well on diphenylmethane, and many observations made in this work are supportive of the idea that it was metabolized by the enzymes of the biphenyl catabolic pathway to produce phenylacetate, which was further metabolized by a lower pathway. Cells of strain B356 produced the same metabolites from diphenylmethane during their growth on this substrate as those isolated from enzymes of the biphenyl catabolic pathway (Fig. 7). Furthermore, the kinetic parameters of BphAEB356 toward diphenylmethane were in the same range as those for biphenyl, suggesting this enzyme metabolizes diphenylmethane during growth on this substrate. It is noteworthy that when purified preparations of B356 BPDO or B356 BPDO plus BphBB356 were used to metabolize diphenylmethane, significant amounts of tetrahydroxylated metabolites were detected in the reaction medium. However, the tetrahydroxylated metabolites were present in very small amounts in cultures of B356 growing on diphenylmethane (not shown). This shows that during growth of strain B356 on diphenylmethane, 3-benzylcyclohexa-3,5-diene-1,2-diol and subsequent metabolites were oxidized very efficiently by the downstream enzymes of the catabolic pathway, thus preventing further BPDO oxidation of the dihydrodiol and the catechol metabolites.
Fig 7.
Profile of metabolites generated during the oxidation of diphenylmethane (upper) or benzophenone (lower) by diphenylmethane- or biphenyl-induced cells of strain B356 or by isolated enzymes of its biphenyl catabolic pathway.
Judging by its steady-state kinetics toward diphenylmethane, BphAELB400 metabolized this substrate as well as biphenyl. In addition, we found that BphCLB400 catalyzed the ring cleavage of 2,3-dihydroxydiphenylmethane, and the strain grew well on phenylacetate. However, unlike strain B356, cells of strain LB400 grown cometabolically on diphenylmethane plus sodium acetate were unable to metabolize 4-chlorobiphenyl, showing that diphenylmethane did not induce the biphenyl catabolic pathway of strain LB400. Therefore, the inability of diphenylmethane to induce the biphenyl catabolic pathway in strain LB400 most likely is the reason why this strain is unable to grow on this substrate.
Growth kinetics data provided strong evidence that both diphenylmethane and biphenyl induced the same catabolic enzymes during growth of strain B356. Uninduced cells grew poorly on diphenylmethane or biphenyl, which was the case when the inocula were prepared by growing the cells on sodium acetate. However, interchanging the growth substrates to prepare the inocula did not affect the growth kinetics on diphenylmethane or on biphenyl, which strongly suggests that both substrates induced the same catabolic pathway.
Remarkably, although further work will be needed to confirm the conclusion, data suggest that a benzoate pathway is induced when cells are grown on diphenylmethane. This was supported by the fact that 3-chlorobenzoate strongly inhibited growth on diphenylmethane. Based on a previous report (36), in order to inhibit growth on biphenyl, 3-chlorobenzoate must be converted to 3-chlorocatechol, which is a very potent inhibitor of BphC. In this study, we have not elucidated the enzymatic steps involved in phenylacetate catabolism during growth of strain B356 on diphenylmethane. However, since phenylacetate is a key metabolite in the metabolism of many aromatic chemicals, including phenylalanine, most bacteria have the ability to metabolize it through a pathway that proceeds via the formation of a coenzyme A (CoA)-thioester derivative (39). Strain LB400 can metabolize 3- and 4-hydroxyphenylacetate through the homogentisate pathway (40), but the ability of this strain to metabolize phenylacetate through this pathway has not been demonstrated. Focht and Alexander (22, 41) have described a Hydrogenomonas isolate that metabolizes diphenylmethane through a pathway that presumably involves the metacleavage of a catechol metabolite, which is further metabolized to generate phenylacetate. Since cells grown on diphenylmethane readily metabolized homogentisate, they postulated that phenylacetate was metabolized via a homogentisic pathway in that Hydrogenomonas strain. However, neither the phenylacetyl-coenzyme A nor the homogentisate pathway involves production of a 2,3-catechol derivative, as is the case for the metabolism of benzoic acid in strain B356. Therefore, the growth inhibition caused by 3-chlorobenzoate suggests that diphenylmethane or one of its metabolites induces a pathway involving a benzoate 2,3-dioxygenase, namely, the biphenyl-associated benzoate pathway. Indeed, strain B356 carries two benzoate pathways, only one of which, the one induced when cells are grown on biphenyl, is able to produce 3-chlorocatechol from 3-chlorobenzoate (36).
Few investigations have assessed the ability of the bacterial biphenyl catabolic enzymes to metabolize diphenylmethane or one of its derivatives, such as DDT or 1,1-dichloro-2,2-(4-chlorophenyl)ethane (DDD) (16, 19, 42–44), but no studies have examined the ability of biphenyl-degrading bacteria to grow on diphenylmethane. Therefore, strain B356 might not be the only one with the ability to use the biphenyl catabolic pathway to grow on this substrate.
Neither strain B356 nor LB400 could use benzophenone as a growth substrate; however, BphAEB356 metabolized this diphenylmethane analog significantly more efficiently than BphAELB400. Structural analysis of the docked substrate showed that residues Phe336 and Gly321 were partly responsible for preventing productive binding of this substrate with BphAELB400. Structural analyses of substrate-bound crystals and docking experiments have shown that the combined effect of these two substitutions increases the space required in the catalytic pocket to accommodate bulkier substrates (8, 16). The replacement of Thr335 of BphAELB400 by Gly333 in BphAEB356 produces a similar effect (16). However, other structural features that our docking experiments could not identify also may have an influence on the ability of the enzyme to metabolize this substrate, since, on the basis of their steady-state kinetics, BphAEB356 metabolized benzophenone more efficiently than BphAEp4, which is a doubly substituted Thr335Ala/Phe336Met mutant of BphAELB400.
Generally, the investigations related to the biphenyl-degrading bacteria during the last 5 decades were initiated with the objective of designing a biological process to degrade PCBs and other chlorinated aromatics, such as chlorodibenzofurans (45, 46). These bacteria were obtained by enrichments on biphenyl, and traditionally it was believed that the four-enzymatic-step biphenyl pathway evolved in bacteria primarily to transform biphenyl into benzoic acid. However, data presented in the current and previous works (14) bring us to question this belief. Biphenyl is a naturally occurring chemical, but it is not universally distributed in nature. It is noteworthy that bacteria carrying the biphenyl catabolic pathway enzymes were obtained from pristine soils not exposed to biphenyl or chlorobiphenyls (10).
On the basis of their primary amino acid sequences, BphAEB356 and BphAELB400 belong to distinct phylogenetic clusters of BphAE (9–11). Recent data suggested that each has acquired a distinct PCB-degrading pattern (9, 15), as well as distinct abilities to metabolize simple flavonoids (14), DDT (16), or DDD (43). In this work, we found that BphAEB356 and BphAELB400 metabolized diphenylmethane similarly but benzophenone and 2,3-dihydroxybenzophenone very differently. Furthermore, the fact that the biphenyl catabolic pathway of strain B356 is inducible by diphenylmethane allows the strain to use it as a growth substrate. As a result, under natural conditions, strain B356 has the potential to metabolize or cometabolize many chemicals that strain LB400 metabolizes poorly.
Although the enzymes of the peripheral pathways, such as those of the biphenyl catabolic pathway, have evolved to metabolize a broad range of substrates, on the basis of the observations made in this work and a previous one (14), we postulate that the biphenyl catabolic pathway enzymes have evolved divergently in bacteria, in such a way that each phylogenetic branch has specialized to play distinct ecophysiological functions with regard to chemicals naturally found in nature.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by the Natural Sciences and Engineering Research Council of Canada (NSERC) (grant RGPIN/39579-2012).
We thank Sameer Al-Abdul-Wahid, QANUC NMR Facility (McGill University, Montreal, Quebec, Canada), for his help in NMR analysis. We also thank Eric Déziel, INRS-Institut Armand-Frappier, for the use of his Bioscreen C system.
Footnotes
Published ahead of print 7 June 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JB.00161-13.
REFERENCES
- 1. Sylvestre M. 2013. Prospects for using combined engineered bacterial enzymes and plant systems to rhizoremediate polychlorinated biphenyls. Environ. Microbiol. 15:907–915 [DOI] [PubMed] [Google Scholar]
- 2. Boyd DR, Bugg TDH. 2006. Arene cis-dihydrodiol formation: from biology to application. Org. Biomol. Chem. 4:181–192 [DOI] [PubMed] [Google Scholar]
- 3. Chun HK, Ohnishi Y, Shindo K, Misawa N, Furukawa K, Horinouchi S. 2003. Biotransformation of flavone and flavanone by Streptomyces lividans cells carrying shuffled biphenyl dioxygenase genes. J. Mol. Catal. B Enzym. 21:113–121 [Google Scholar]
- 4. Kagami O, Shindo K, Kyojima A, Takeda K, Ikenaga H, Furukawa K, Misawa N. 2008. Protein engineering on biphenyl dioxygenase for conferring activity to convert 7-hydroxyflavone and 5,7-dihydroxyflavone (chrysin). J. Biosci. Bioeng. 106:121–127 [DOI] [PubMed] [Google Scholar]
- 5. Misawa N, Nakamura R, Kagiyama Y, Ikenaga H, Furukawa K, Shindo K. 2005. Synthesis of vicinal diols from various arenes with a heterocyclic, amino or carboxyl group by using recombinant Escherichia coli cells expressing evolved biphenyl dioxygenase and dihydrodiol dehydrogenase genes. Tetrahedron 61:195–204 [Google Scholar]
- 6. Seeger M, Gonzalez M, Camara B, Munoz L, Ponce E, Mejias L, Mascayano C, Vasquez Y, Sepulveda-Boza S. 2003. Biotransformation of natural and synthetic isoflavonoids by two recombinant microbial enzymes. Appl. Environ. Microbiol. 69:5045–5050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Seo J, Kang SI, Ryu JY, Lee YJ, Park KD, Kim M, Won D, Park HY, Ahn JH, Chong Y, Kanaly RA, Han J, Hur HG. 2010. Location of flavone B-ring controls regioselectivity and stereoselectivity of naphthalene dioxygenase from Pseudomonas sp. strain NCIB 9816-4. Appl. Microbiol. Biotechnol. 86:1451–1462 [DOI] [PubMed] [Google Scholar]
- 8. Kumar P, Mohammadi M, Viger JF, Barriault D, Gomez-Gil L, Eltis LD, Bolin JT, Sylvestre M. 2011. Structural insight into the expanded PCB-degrading abilities of a biphenyl dioxygenase obtained by directed evolution. J. Mol. Biol. 405:531–547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Standfuß-Gabisch C, Al-Halbouni D, Hofer B. 2012. Characterization of biphenyl dioxygenase sequences and activities encoded by the metagenomes of highly polychlorobiphenyl-contaminated soils. Appl. Environ. Microbiol. 78:2706–2715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Vézina J, Barriault D, Sylvestre M. 2008. Diversity of the C-terminal portion of the biphenyl dioxygenase large subunit. J. Mol. Microbiol. Biotechnol. 15:139–151 [DOI] [PubMed] [Google Scholar]
- 11. Witzig R, Junca H, Hecht HJ, Pieper DH. 2006. Assessment of toluene/biphenyl dioxygenase gene diversity in benzene-polluted soils: links between benzene biodegradation and genes similar to those encoding isopropylbenzene dioxygenases. Appl. Environ. Microbiol. 72:3504–3514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Colbert CL, Agar NY, Kumar P, Chakko MN, Sinha SC, Powlowski JB, Eltis LD, Bolin JT. 2013. Structural characterization of Pandoraea pnomenusa B-356 biphenyl dioxygenase reveals features of potent polychlorinated biphenyl-degrading enzymes. PLoS One 8:e52550. 10.1371/journal.pone.0052550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Furusawa Y, Nagarajan V, Tanokura M, Masai E, Fukuda M, Senda T. 2004. Crystal structure of the terminal oxygenase component of biphenyl dioxygenase derived from Rhodococcus sp. strain RHA1. J. Mol. Biol. 342:1041–1052 [DOI] [PubMed] [Google Scholar]
- 14. Pham TTM, Tu Y, Sylvestre M. 2012. Remarkable ability of Pandoraea pnomenusa B356 biphenyl dioxygenase to metabolize simple flavonoids. Appl. Environ. Microbiol. 78:3560–3570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Gomez-Gil L, Kumar P, Barriault D, Bolin JT, Sylvestre M, Eltis LD. 2007. Characterization of biphenyl dioxygenase of Pandoraea pnomenusa B-356 as a potent polychlorinated biphenyl-degrading enzyme. J. Bacteriol. 189:5705–5715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. L'Abbée JB, Tu YB, Barriault D, Sylvestre M. 2011. Insight into the metabolism of 1,1,1-trichloro-2,2-bis(4-chlorophenyl)ethane (DDT) by biphenyl dioxygenases. Arch. Biochem. Biophys. 516:35–44 [DOI] [PubMed] [Google Scholar]
- 17. Anonymous 2012. Some chemicals present in industrial and consumer products, food and drinking-water, p 285–305 IARC monographs on the evaluation of carcinogenic risks to humans, vol 101 World Health Organization Press, Geneva, Switzerland: [PMC free article] [PubMed] [Google Scholar]
- 18. Hemshekhar M, Sunitha K, Santhosh MS, Devaraja S, Kemparaju K, Vishwanath BS, Niranjana SR, Girish KS. 2011. An overview on genus Garcinia: phytochemical and therapeutical aspects. Phytochem. Rev. 10:325–351 [Google Scholar]
- 19. Misawa N, Shindo K, Takahashi H, Suenaga H, Iguchi K, Okazaki H, Harayama S, Furukawa K. 2002. Hydroxylation of various molecules including heterocyclic aromatics using recombinant Escherichia coli cells expressing modified biphenyl dioxygenase genes. Tetrahedron 58:9605–9612 [Google Scholar]
- 20. Focht DD, Alexander M. 1971. Aerobic cometabolism of DDT analogues by Hydrogenomonas sp. J. Agric. Food Chem. 19:20–22 [DOI] [PubMed] [Google Scholar]
- 21. Liu YS, Ying GG, Shareef A, Kookana RS. 2012. Biodegradation of the ultraviolet filter benzophenone-3 under different redox conditions. Environ. Toxicol. Chem. 31:289–295 [DOI] [PubMed] [Google Scholar]
- 22. Focht DD, Alexander M. 1970. DDT metabolites and analogs: ring fission by Hydrogenomonas. Science 170:91–92 [DOI] [PubMed] [Google Scholar]
- 23. Lin JJ, Smith M, Jessee J, Bloom F. 1992. DH11s: an E. coli strain for preparation of single-stranded DNA from phagemid vectors. Biotechniques 12:718–721 [PubMed] [Google Scholar]
- 24. Miroux B, Walker JE. 1996. Over-production of proteins in Escherichia coli: mutant hosts that allow synthesis of some membrane proteins and globular proteins at high levels. J. Mol. Biol. 260:289–298 [DOI] [PubMed] [Google Scholar]
- 25. Erickson BD, Mondello FJ. 1992. Nucleotide sequencing and transcriptional mapping of the genes encoding biphenyl dioxygenase, a multicomponent polychlorinated-biphenyl-degrading enzyme in Pseudomonas strain LB400. J. Bacteriol. 174:2903–2912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Barriault D, Pelletier C, Hurtubise Y, Sylvestre M. 1997. Substrate selectivity pattern of Comamonas testosteroni strain B-356 towards dichlorobiphenyls. Int. Biodeterior. Biodegradation 39:311–316 [Google Scholar]
- 27. Hurtubise Y, Barriault D, Powlowski J, Sylvestre M. 1995. Purification and characterization of the Comamonas testosteroni B-356 biphenyl dioxygenase components. J. Bacteriol. 177:6610–6618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Sambrook J, Fritsch EF, Maniatis T. 1989. Molecular cloning: a laboratory manual. Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 29. Sylvestre M. 1980. Isolation method for bacterial isolates capable of growth on p-chlorobiphenyl. Appl. Environ. Microbiol. 39:1223–1224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Toussaint JP, Pham TTM, Barriault D, Sylvestre M. 2012. Plant exudates promote PCB degradation by a rhodococcal rhizobacteria. Appl. Microbiol. Biotechnol. 95:1589–1603 [DOI] [PubMed] [Google Scholar]
- 31. Mohammadi M, Viger JF, Kumar P, Barriault D, Bolin JT, Sylvestre M. 2011. Retuning Rieske-type oxygenases to expand substrate range. J. Biol. Chem. 286:27612–27621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Hurtubise Y, Barriault D, Sylvestre M. 1996. Characterization of active recombinant his-tagged oxygenase component of Comamonas testosteroni B-356 biphenyl dioxygenase. J. Biol. Chem. 271:8152–8156 [DOI] [PubMed] [Google Scholar]
- 33. Mohammadi M, Sylvestre M. 2005. Resolving the profile of metabolites generated during oxidation of dibenzofuran and chlorodibenzofurans by the biphenyl catabolic pathway enzymes. Chem. Biol. 12:835–846 [DOI] [PubMed] [Google Scholar]
- 34. Morris GM, Huey R, Lindstrom W, Sanner MF, Belew RK, Goodsell DS, Olson AJ. 2009. AutoDock4 and AutoDockTools4: automated docking with selective receptor flexibility. J. Comput. Chem. 30:2785–2791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Hein P, Powlowski J, Barriault D, Hurtubise Y, Ahmad D, Sylvestre M. 1998. Biphenyl-associated meta-cleavage dioxygenases from Comamonas testosteroni B-356. Can. J. Microbiol. 44:42–49 [PubMed] [Google Scholar]
- 36. Sondossi M, Sylvestre M, Ahmad D. 1992. Effects of chlorobenzoate transformation on the Pseudomonas testosteroni biphenyl and chlorobiphenyl degradation pathway. Appl. Environ. Microbiol. 58:485–495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Barriault D, Durand J, Maaroufi H, Eltis LD, Sylvestre M. 1998. Degradation of polychlorinated biphenyl metabolites by naphthalene-catabolizing enzymes. Appl. Environ. Microbiol. 64:4637–4642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Massé R, Messier F, Ayotte C, Lévesque Sylvestre M-FM. 1989. A Comprehensive gas chromatographic/mass spectrometric analysis of 4-chlorobiphenyl bacterial degradation products. Biomed. Environ. Mass Spectrom. 18:27–47 [Google Scholar]
- 39. Teufel R, Gantert C, Voss M, Eisenreich W, Haehnel W, Fuchs G. 2011. Studies on the mechanism of ring hydrolysis in phenylacetate degradation: a metabolic branching point. J. Biol. Chem. 286:11021–11034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Romero-Silva MJ, Mendez V, Agullo L, Seeger M. 2013. Genomic and functional analyses of the gentisate and protocatechuate ring-cleavage pathways and related 3-hydroxybenzoate and 4-hydroxybenzoate peripheral pathways in Burkholderia xenovorans LB400. PLoS One 8:e56038. 10.1371/journal.pone.0056038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Focht DD, Alexander M. 1970. Bacterial degradation of diphenylmethane, a DDT model substrate. Appl. Microbiol. 20:608–611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Hay AG, Focht DD. 1998. Cometabolism of 1,1-dichloro-2,2-bis(4-chlorophenyl)ethylene by Pseudomonas acidovorans M3GY grown on biphenyl. Appl. Environ. Microbiol. 64:2141–2146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Hay AG, Focht DD. 2000. Transformation of 1,1-dichloro-2,2-(4-chlorophenyl)ethane (DDD) by Ralstonia eutropha strain A5. FEMS Microbiol. Ecol. 31:249–253 [DOI] [PubMed] [Google Scholar]
- 44. Nadeau LJ, Menn FM, Breen A, Sayler GS. 1994. Aerobic degradation of 1,1,1-trichloro-2,2-bis(4-chlorophenyl)ethane (DDT) by Alcaligenes eutrophus A5. Appl. Environ. Microbiol. 60:51–55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Kumar P, Mohammadi M, Dhindwal S, Pham TT, Bolin JT, Sylvestre M. 2012. Structural insights into the metabolism of 2-chlorodibenzofuran by an evolved biphenyl dioxygenase. Biochem. Biophys. Res. Commun. 421:757–762 [DOI] [PubMed] [Google Scholar]
- 46. Pieper DH, Seeger M. 2008. Bacterial metabolism of polychlorinated biphenyls. J. Mol. Microbiol. Biotechnol. 15:121–138 [DOI] [PubMed] [Google Scholar]
- 47. Barriault D, Sylvestre M. 2004. Evolution of the biphenyl dioxygenase BphA from Burkholderia xenovorans LB400 by random mutagenesis of multiple sites in region III. J. Biol. Chem. 279:47480–47488 [DOI] [PubMed] [Google Scholar]
- 48. Barriault D, Plante MM, Sylvestre M. 2002. Family shuffling of a targeted bphA region to engineer biphenyl dioxygenase. J. Bacteriol. 184:3794–3800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Dhindwal S, Patil DN, Mohammadi M, Sylvestre M, Tomar S, Kumar P. 2011. Biochemical studies and ligand bound structures of biphenyl dehydrogenase from Pandoraea pnomenusa strain B-356 reveal a basis for broad specificity of the enzyme. J. Biol. Chem. 286:37011–37022 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.