Abstract
The ribosomal P-site hosts the peptidyl-tRNAs during translation elongation. Which P-site elements support these tRNA species to maintain codon-anticodon interactions has remained unclear. We investigated the effects of P-site features of methylations of G966, C967, and the conserved C-terminal tail sequence of Ser, Lys, and Arg (SKR) of the S9 ribosomal protein in maintenance of the translational reading frame of an mRNA. We generated Escherichia coli strains deleted for the SKR sequence in S9 ribosomal protein, RsmB (which methylates C967), and RsmD (which methylates G966) and used them to translate LacZ from its +1 and −1 out-of-frame constructs. We show that the S9 SKR tail prevents both the +1 and −1 frameshifts and plays a general role in holding the P-site tRNA/peptidyl-tRNA in place. In contrast, the G966 and C967 methylations did not make a direct contribution to the maintenance of the translational frame of an mRNA. However, deletion of rsmB in the S9Δ3 background caused significantly increased −1 frameshifting at 37°C. Interestingly, the effects of the deficiency of C967 methylation were annulled when the E. coli strain was grown at 30°C, supporting its context-dependent role.
INTRODUCTION
Ribosomes use elegant mechanisms to ensure that only the correct aminoacyl-tRNAs with anticodons cognate to the codon triplets displayed in the A-site are selected and accommodated to extend the polypeptide chain (1). However, for the accuracy of protein synthesis, maintenance of the codon-anticodon interactions during translocation and subsequently during the residency of the peptidyl-tRNA in the ribosomal P-site is also crucial. Any failures in maintenance of the codon-anticodon interactions, once established in the A-site, may lead to the display of an unintended triplet in the ribosomal A-site leading to mistranslation of the mRNA. Thus, the ribosomal P-site plays an important role not only at the step of initiation in selecting the initiator tRNA (fMet-tRNAfMet) to establish the translational reading frame but also at the step of elongation in hosting the peptidyl-tRNAs. Which P-site elements support these tRNA species to maintain codon-anticodon interactions is unclear.
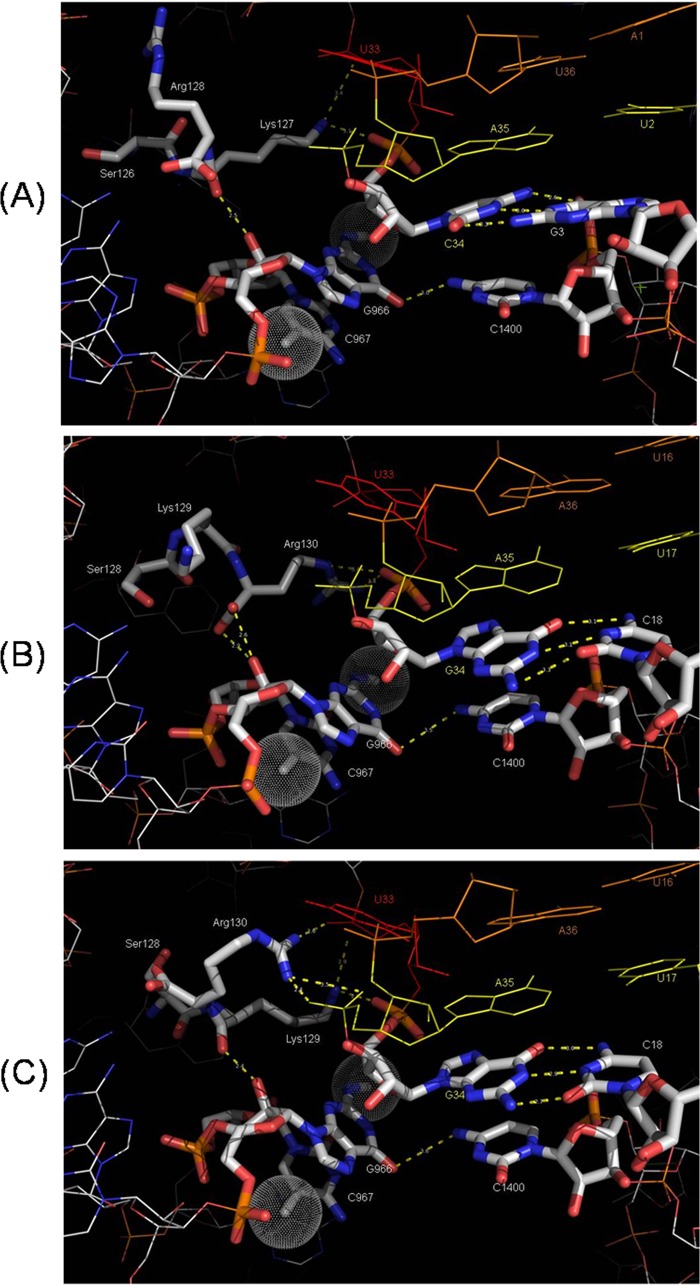
Cocrystal structures of the ribosomes have provided snapshots (Fig. 1) of interactions of the P-site elements with the tRNA and mRNA both in the classical and in the hybrid states of the ribosome (2, 3). These studies have shown that the 16S rRNA base G966 stacks against the ribose of the anticodon position 34 of the P-site tRNA and the tails of two of the ribosomal proteins, S9 and S13, contact the initiator or the P-site bound tRNA (2). The G966 and the adjacent base C967 are posttranscriptionally modified by site-specific methyltransferases, RsmD (4) and RsmB (5), respectively. RsmB is a conserved methyltransferase located adjacent to the fmt operon (containing def [N-formylaminoacyl deformylase] and fmt [methionyl-tRNA formyltransferase] genes) and is transcribed in the same direction. Intriguingly, the C-terminal tail sequence (Ser, Lys and Arg [SKR]) of the S9 protein, which contacts the P-site tRNA at positions 33 and 34 is also well conserved in bacteria (2). In vitro approaches have shown that the C-terminal SKR sequence of S9 is important in binding a set of elongator tRNAs at the P-site (6, 7). Methylated G966 is also reported to be important for the stability of a subset of elongator tRNAs at the P-site (7). Recently, we have shown that the m2G966, m5C967 and the C-terminal tail (SKR) of S9 make distinct contributions in maintaining fidelity of initiation in Escherichia coli (8). It was also reported that the loss of methylations at G966 and C967 leads to defective in vitro binding stability of initiator tRNA (∼2-fold) (9). In Salmonella enterica serovar Typhimurium, mutations in S9 protein have been shown to induce +1 frameshifting (10, 11). However, in these studies on S. Typhimurium, −1 frameshifting events were not investigated. The two methylated nucleosides G966 and C967 are reported to contact S9 via backbone interactions (12). C967 is also important in correct positioning of G966 to interact with P-site tRNA. The methylated nucleosides and the S9 tail form a network of interactions important in scrutiny of codon-anticodon pair during initiation. However, the relevance of these elements in the residency of the peptidyl-tRNAs, and thus in the maintenance of the translational reading frame, if any, is not known.
Fig 1.
Models showing the interaction of G966, C967, and the S9 C-terminal tail residues (Ser, Lys, and Arg [SKR]) with the anticodon region of the tRNA. (A) Structure corresponding to the interactions with tRNAfMet in P/P state in T. thermophilus ribosome (anticodon bases C34, A35, and U36 are paired with codon bases A1, U2, and G3 in mRNA). (B and C) Structures corresponding to the interactions with tRNAPhe in the P/P and P/E states, respectively, of E. coli ribosomes (anticodon bases G34, A35, and A36 are paired with codon bases U16, U17, and C18 in mRNA). The S9 C-terminal residues, SKR, are numbered 126, 127, and 128 in Thermus thermophilus (A) and 128, 129, and 130 in E. coli (B and C). Residues Lys and/or Arg contacting the anticodon loop of the resident tRNAs are in proximity to G966 and C967. Dotted spheres on G966 or C967 indicate methyl groups. The models were generated with PyMOL v1.5.0.3 using PDB accession numbers 2J00, 3R8O, and 3R8N (2, 3). The image in panel A was adapted from an earlier work (8), with permission of the publisher.
A C-terminal S9 tail deletion strain of E. coli (S9Δ3), wherein the three C-terminal amino acids (SKR) of the S9 protein are deleted, is known to be cold sensitive (6). Likewise, strains lacking KsgA (a methyltransferase conserved across the three domains of life and which methylates A1518 and A1519 in the 3′ minor domain of 16S rRNA) also imparts cold sensitivity to E. coli (13). The absence of methylation of A1518 and A1519 has also been reported to have subtle effects on the readthrough of nonsense codons and in frameshift errors (14). Thus, it was of interest to understand the role of methylations of G966 and C967 present in the 3′ major domain of 16S rRNA and the S9 tail on the residency of the peptidyl-tRNAs in the ribosomal P-site in E. coli.
MATERIALS AND METHODS
Strains, plasmids and DNA oligomers.
The E. coli strains and plasmids used in this study are listed in Tables 1 and 2. Bacteria were cultured in Luria-Bertani (LB) broth or LB agar (LB broth containing 1.8% agar; Difco) at 37°C or as indicated with constant shaking at 200 rpm. Ampicillin (100 μg ml−1), kanamycin (25 μg ml−1), tetracycline (7.5 μg ml−1), or IPTG (isopropyl-β-d-thiogalactopyranoside) were used as needed.
Table 1.
Strains used in this study
| Strain | Relevant features | Source or reference |
|---|---|---|
| BW25113 (or BW) | Δ(araD-araB)567 ΔlacZ4787(::rrnB-3) λ− rph-1 Δ(rhaD-rhaB)568 hsdR514 | Nara Institute, Japan |
| BW ΔrsmB::kan (or BW ΔrsmB) | BW derivative with rsmB deletion | This study |
| BW ΔrsmD::kan (or BW ΔrsmD) | BW derivative with rsmD deletion | This study |
| BW ΔrsmBΔrsmD::kan (or BW ΔrsmBΔrsmD) | BW derivative with deletion of rsmB and rsmD | This study |
| BW S9Δ3::kan (or BW S9Δ3) | BW derivative with a deletion of C terminus of S9 tail | This study |
Table 2.
Plasmids and DNA oligomers used in this study
| Plasmid or DNA oligomer | Relevant features or sequence (5′–3′) | Reference |
|---|---|---|
| Plasmids | ||
| pCATam1 | Renamed from pRSVCATam1.2.5; pBR322 derivative harboring a CAT reporter gene with UAG as an initiation codon | 24 |
| pCATam1metYCUA | Renamed from pRSVCATam1.2.5trnfMU35A36; pBR322 derivative harboring a CAT reporter gene with UAG as an initiation codon and expressing tRNAfMet with a CUA anticodon | 24 |
| pCATam1metY | Renamed from pRSVCATam1.2.5trnfM; a pBR322 derivative harboring a CAT reporter gene with UAG as an initiation codon and expressing tRNAfMet | 24 |
| pSG25-lacZ | Construct with wild-type lacZ | 16 |
| pSG-lac7(+1) | Production of functional LacZ from this construct requires +1 frameshifting in the frameshift window | 16 |
| pSG12DP(−1) | Production of functional LacZ from this construct requires −1 frameshifting in the frameshift window | 16 |
| pSGlac10(−1) | Production of functional LacZ from this construct requires −1 frameshift in the frameshift window; the frameshift occurs at a codon different from pSG12DP | 15 |
| pCP20 | Ampr and Cmr plasmid that shows tempurature-sensitive replication and thermal induction of flippage recombinase (Flp) synthesisa | 25 |
| DNA oligomers | ||
| rsmDfp | TTCATGGCACAGCGTTAACG | 8 |
| rsmDrp | AACGAAGATATTCAGCGGGC | 8 |
| rsmBfp | CTCTCGTCGGGAATGGTTTG | 8 |
| rsmBrp | CTGCTCTGGTGCGATCAGAT | 8 |
| S9del3fp | GTCGGTCTGCGTAAAGCACGTCGTCGTCCGCAGTTCTAGTAAGCTTGTGTAGGCTGGAGCTGCTTCG | 8 |
| S9del3rp | TTTTCGAAAATTGTTTTCTGCCGGAGCAGAAGCCAACATATGAATATCCTCCTTA | 8 |
Cmr, chloramphenicol resistance; Ampr, ampicillin resistance.
Generation of strains and the lacZ reporter constructs.
The required strains (Table 1) were generated by P1 phage-mediated transductions (8). The lacZ constructs (Table 2) used for the −1 and +1 frameshift assays were kindly provided by A. Dahlberg (15, 16).
β-Galactosidase assay.
The standard Miller's protocol was used (17). The strains (four to five replicates each) were cultured overnight in LB broth at 37°C. A 1% inoculum from these was used to subculture and, at an optical density at 600 nm (OD600) of ∼0.4, the cultures were induced with 1 mM IPTG for 1 h. Cells were pelleted from 1.0 ml of culture, and the pellet was resuspended in 1 ml of Z buffer (60 mM Na2HPO4, 40 mM NaH2PO4, 10 mM KCl, 0.1 mM MgSO4, 50 mM β-mercaptoethanol; pH 7.4). The assays were performed using ortho-nitrophenyl-β-galactoside (ONPG; 4 mg ml−1) as a substrate for β-galactosidase at 30°C. The reaction was stopped by adding 500 μl of 1 M Na2CO3, and the product intensity was estimated at OD420. The β-galactosidase activity in Miller units was calculated using the formula: Miller units = 1,000 [OD420 − (1.75·OD550)/OD600·time (t)·volume (V)], where OD420 is the ONPG absorbance and OD550 is the scatter absorbance from cell debris. OD600 is the cell density, t is time of reaction in minutes, and V is the volume of culture used in milliliters.
RESULTS
Experimental design.
Recently, we investigated the role of the ribosomal P-site elements in pairing of several combinations of the wild type or the mutant anticodons in initiator tRNAs and the initiation codons in the reporter mRNA constructs (8). We showed that, while the RsmB deficiency (lack of C967 methylation) in general did not impact initiation from most codon-anticodon pairs, the RsmD deficiency (lack of G966 methylation) did. Also, it was shown that the S9 C-terminal SKR sequence played an important role in the fidelity of the initiator tRNA selection in the P-site. As shown in Fig. 1, as well as based on our molecular dynamics studies (8), whereas the finer details of the interactions with the C-terminal SKR tail of the S9 differ, the contacts of the tail and the methylated G966 and C967 in the P-site are, in general, preserved irrespective of its (P-site) residency by the initiator tRNA (Fig. 1A) or the elongator tRNA in a classical or the hybrid state (Fig. 1B and C). Thus, we surmised that the S9 tail and the G966 and C967 methylations might play a role in the maintenance of interactions of the anticodon of the P-site bound peptidyl-tRNA with the cognate codon of the mRNA during the step of elongation. To investigate such a role, we exploited the phenomenon of frameshifting using lacZ constructs (Fig. 2 and Table 2) whose utility had already been validated for such studies (15, 16, 18). As shown in Fig. 2, we used four reporter constructs: a wild type lacZ, a +1 lacZ construct where LacZ activity is produced only upon +1 frameshifting and two constructs where LacZ activity is produced only upon −1 frameshifting. LacZ activities from the +1, and the −1 frameshift constructs provide a readout of any deficiencies in the maintenance of the interaction of the peptidyl-tRNA anticodon with its cognate codon in the mRNA. Moreover, the wild-type lacZ construct serves as a control for the translation efficiency in each strain.
Fig 2.

Schematics of the LacZ constructs used to investigate frameshift errors. The frameshift window in each construct, within which the ribosome must shift to resume the translational frame of LacZ, is indicated in boldface (15, 16).
Generation of the S9 C-terminal tail deletion (S9Δ3) and rsmB rsmD deletion (S9Δ3ΔrsmBΔrsmD) strains and their characterization.
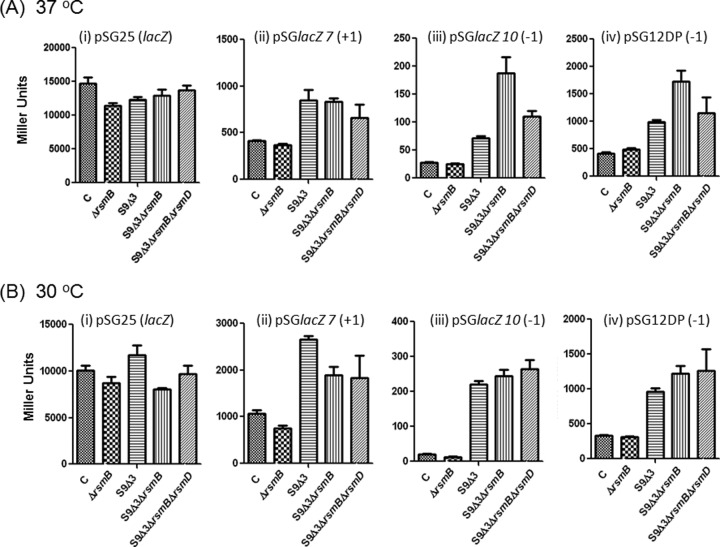
To carry out the frameshift assays, we used E. coli BW (deleted of its chromosomal lacZ) and its derivatives. The strains were generated by moving the S9Δ3 allele (S9 lacking the C-terminal tripeptide sequence [SKR]) and the deletion alleles of rsmB and/or rsmD by P1 phage-mediated transductions (8). Double- and triple-deletion strains generated by combining S9Δ3 with rsmB and rsmD deletion(s) were also used. Consistent with an earlier report (6), we observed that the S9Δ3 derivative of E. coli BW was cold sensitive (Fig. 3, right panel, growth at 30°C), and the introduction of the ΔrsmB allele led to enhanced cold sensitivity of the double-deletion (S9Δ3ΔrsmB) strain. A double deletion of S9Δ3 and rsmD was also made, but this did not alter the cold sensitivity of the S9Δ3 strain. Finally, a triple-deletion strain (S9Δ3ΔrsmBΔrsmD) was not significantly different from the double-deletion strain S9Δ3ΔrsmB. The growth defect was efficiently rescued when the strains were grown at 37°C (Fig. 3, left panel).
Fig 3.
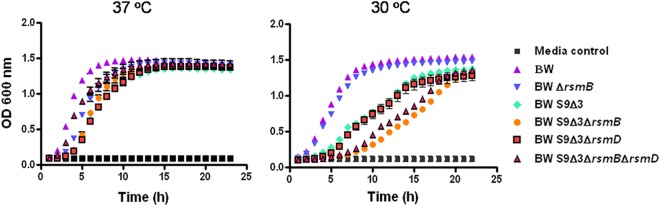
Growth curve analysis of E. coli BW and its derivatives—S9Δ3ΔrsmB, S9Δ3ΔrsmD, and S9Δ3ΔrsmBΔrsmD—at 30°C (right) and 37°C (left). Overnight cultures grown in LB broth were diluted a thousand-fold in LB broth, and their growths were monitored in a Bioscreen C growth reader.
S9 tail plays a role in maintenance of the reading frame.
To assess the effects of S9 tail on the maintenance of the translational reading frame, we assayed frameshift errors in the presence or absence of the S9 tail. LacZ reporter constructs that allow the assessment of frameshifting (either +1 or −1) were introduced into E. coli BW, BW ΔrsmD, BW S9Δ3, and BW S9Δ3ΔrsmD strains, and the LacZ activities of the transformants were assayed. As shown in Fig. 4, deletion of the S9 tail (in S9Δ3) led to increased frameshift errors relative to the wild-type parent (E. coli BW control [C]) in either +1 or the −1 frameshift constructs (Fig. 4ii to iv). As a control, LacZ activity from the wild-type lacZ construct (requiring no frameshifting) was nearly the same in all the strains (Fig. 4i). It may be mentioned that the extent of frameshifting in the constructs varies because of the differences in the frameshift windows within which the ribosome must shift the reading frame (Fig. 2). Thus, these observations suggest that the S9 tail plays a general role in the maintenance of the reading frame during elongation of the polypeptide.
Fig 4.
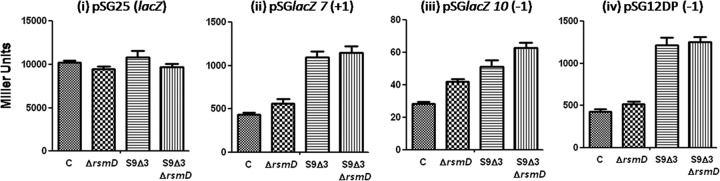
LacZ assays in various strains harboring various constructs as indicated. Cultures were grown to log phase at 37°C, induced with IPTG, and assayed for LacZ (see Materials and Methods).
Absence of G966 methylation alone or together with the S9 tail deletion does not significantly impact frameshifting.
Assays to assess the impact of methylation of G966 on frameshifting showed that deletion of rsmD had only a subtle effect on frameshifting (Fig. 4ii to iv, compare the control strain [C] to the ΔrsmD strain). However, in one of the −1 frameshift constructs (Fig. 4iiii and Table 3), we did see an ∼1.4-fold increase in frameshifting. This small difference between the two −1 frameshift constructs could be due to their slightly different frameshift windows (Fig. 2). Further, a double deletion of S9Δ3 and rsmD did not show a significant change in frameshifting compared to that seen in the S9Δ3 strain (Fig. 4ii to iv, compare S9Δ3ΔrsmD to S9Δ3).
Table 3.
Fold changes in β-galactosidase activity at 37°C
| Frameshift construct | Mean fold change in β-galactosidase activity ± SD at 37°Ca |
||||||
|---|---|---|---|---|---|---|---|
| C | ΔrsmB | ΔrsmD | S9Δ3 | S9Δ3ΔrsmD | S9Δ3ΔrsmB | S9Δ3ΔrsmBΔrsmD | |
| pSG25 (WT) | 1 | 0.73 ± 0.06 | 0.92 ± 0.023 | 1 ± 0.09 | 1 ± 0.03 | 0.84 ± 0.07 | 0.89 ± 0.05 |
| pSG-lac7(+1) | 1 | 1.04 ± 0.1 | 1.29 ± 0.14 | 2.7 ± 0.26 | 2.6 ± 0.26 | 2.9 ± 0.13 | 2.35 ± 0.08 |
| pSG12DP(−1) | 1 | 1.13 ± 0.06 | 1 ± 0.05 | 2.71 ± 0.29 | 2.9 ± 0.14 | 4.13 ± 0.29 | 3.73 ± 0.23 |
| pSGlac10(−1) | 1 | 0.9 ± 0.03 | 1.43 ± 0.1 | 1.8 ± 0.13 | 2.2 ± 0.2 | 6.7 ± 1.8 | 4 ± 0.47 |
Fold differences in β-galactosidase activities were calculated from pooled data (from independent experiments) for each of the plasmid constructs in the various strains, and values shown are relative to that for E. coli BW (column C, taken as 1).
The absence of C967 methylation alone does not impact frameshifting but enhances the impact of the S9 tail deletion in −1 constructs.
Like the effect of rsmD deletion, an independent deletion of rsmB (C967 methyltransferase) did not have a significant effect on frameshifting at either 37°C (Fig. 5A) or 30°C (Fig. 5Bii to iv, compare the control strain [C] to the ΔrsmB strain). As seen in Fig. 4, in this experiment (Fig. 5) also, deletion of the S9 tail increased +1 and −1 frameshift errors, whereas there were no significant changes in production of LacZ from the wild-type lacZ at either 37 or 30°C (Fig. 5A and B, panels i, compare the control strain [C] to the other strains). Also, at least in the case of the +1 frameshift construct, similar to the deletion of rsmD in the S9Δ3 background, deletion of rsmB in the S9Δ3 background did not significantly impact frameshifting (Fig. 5A and B, panels ii, compare S9Δ3 to S9Δ3ΔrsmB) at either 37 or 30°C. Interestingly, in the case of both −1 frameshift constructs, deletion of rsmB in the S9Δ3 background resulted in a significantly increased frameshifting at 37°C but not at 30°C (Fig. 5A and B, panels iii and iv). However, such an impact of rsmB deletion in the S9Δ3ΔrsmD background, in both the −1 frameshift constructs, was diminished at 37°C (compare S9Δ3ΔrsmB to S9Δ3ΔrsmBΔrsmD) but not at 30°C (see Discussion).
Fig 5.
LacZ assays in various strains harboring various constructs as indicated. Cultures were grown to log phase at 37°C (A) or 30°C, induced with IPTG, and assayed for LacZ (see Materials and Methods).
DISCUSSION
Unlike the ribosomal A-site, the P-site hosts both the initiator and the elongator tRNAs. Recently, we showed that P-site features such as the S9 tail and methylated G966 and C967, which form a network of interactions within themselves and with the codon-anticodon pair, play a role at the step of initiation in selecting a correct reading frame (8). In the present study, we investigated the role of the same P-site features of the ribosome in maintenance of the reading frame during translation of an mRNA. We exploited one +1 and two −1 out-of-frame constructs of LacZ in these assays, such that any deficiencies in the fidelity of maintenance of a reading frame would result in the production of LacZ from the out-of-frame constructs. As summarized in Tables 3 and 4, the effects of S9 C-terminal tripeptide (SKR) deletion on increased frameshifting are consistent in both the +1 and −1 frame constructs at either 30 or 37°C. However, the effects of the methylations at G966 and C967 positions are context dependent. These observations are, by and large, reminiscent of what we observed for the role of the same P-site elements in fidelity to the initiator tRNA selection (8).
Table 4.
Fold changes in β-galactosidase activity at 30°C
| Frameshift construct | Mean fold change in β-galactosidase activity ± SD at 30°Ca |
||||
|---|---|---|---|---|---|
| C | ΔrsmB | S9Δ3 | S9Δ3ΔrsmB | S9Δ3ΔrsmBΔrsmD | |
| pSG25 (WT) | 1 | 0.9 ± 0.05 | 1.13 ± 0.09 | 0.8 ± 0.05 | 1 ± 0.09 |
| pSG-lac7(+1) | 1 | 0.7 ± 0.047 | 2.7 ± 0.12 | 1.9 ± 0.17 | 2.26 ± 0.28 |
| pSG12DP(−1) | 1 | 0.92 ± 0.03 | 2.9 ± 0.15 | 3.7 ± 0.38 | 4.25 ± 0.78 |
| pSGlac10(−1) | 1 | 0.7 ± 0.12 | 11.45 ± 1.5 | 12.19 ± 0.93 | 13 ± 1.6 |
Fold differences in β-galactosidase activities were calculated from pooled data (from independent experiments) for each of the plasmid constructs in the various strains, and values shown are relative to that for E. coli BW (column C, taken as 1).
The deletion of the C-terminal SKR sequence of the S9 led to increased +1 and −1 frameshift errors at 37°C, as well as at 30°C (Fig. 4 and 5). Mechanisms of +1 and −1 frameshifting, at least based on the impact of nucleoside modifications in tRNAs, are thought to be different (19). Hence, our observations using the S9 C-terminal tail deletion suggest that the mechanisms of +1 and −1 frameshifting need not be different in all cases. In the present case, these are both due to common deficiencies of the intrinsic features (the P-site elements) of the ribosome. In the case of S9, its C-terminal tail, as deduced from the tRNAPhe-bound crystal structure (3), contacts the elongator tRNA in both the P/P and the P/E states (Fig. 1), suggesting that the C-terminal floppy tail of the S9 might offer a common mechanism to hold the tRNA in place, preventing its anticodon from slippage on the mRNA in the P-site. Such an inference is analogous to the proposal that the maintenance of the codon-anticodon interaction by the specific contacts the tip of domain IV of EFG makes with the tRNA and mRNA to prevent frameshifting during translocation of the tRNA from the A-site to the P-site (20). It would be interesting to determine whether there is a cross talk between the S9 C-terminal tail contacts and the contacts EFG make with the tRNA and mRNA, or the other ribosomal features proposed to or known to prevent frameshifting. For example, the structural analyses of ribosomes complexed with mRNA and tRNA have detailed several other mechanisms such as the formation of a kink in the mRNA between the A-site and the P-site codons or the contacts of the S13 ribosomal protein with the P-site tRNA. However, it should also be said that the functional analyses of the cross talk between these ribosomal elements in the maintenance of the reading frame of an mRNA have not been carried out.
When we deleted the RsmD or RsmB methyltransferases, which specifically methylate G966 and C967, respectively, we observed that while the modification of G966 has a subtle effect on maintenance of the reading frame, that of C967 did not have a detectable impact, at least on its own (Fig. 4 and 5). Since G966 and C967 are a part of the network of interactions with the S9 tail, to investigate whether these modifications had a greater impact in conjunction with the S9 tail, we combined the RsmB/RsmD deletions with the deletion of S9 C-terminal tail. In these strains also, we did not detect a significant impact of G966 modification (Fig. 4). Interestingly, deletion of RsmB, together with the deletion of S9 tail (S9Δ3), resulted in a synergistic impact on frameshifting of the −1 constructs but not of the +1 construct (Fig. 5A). Moreover, even though the polarities of frameshifting are different, such an observation of the impact of this 16S rRNA residue modification is, in essence, similar to that observed earlier for the role of the modified nucleoside of tRNA in frameshifting (19). Nonetheless, the interpretation of the effect of this ribosomal modification is complicated by the fact that its synergistic impact was lost when the assays were performed at 30°C (Fig. 5B), a temperature that significantly slows down the growth of the strains. It is reasonable to propose that the slow-growth phenotype of the strain may itself be a consequence of altered ribosome function. Thus, it seems prudent to suggest that the roles of at least the G966 and C967 methylations are context dependent, and their significance may only be in the form of fine-tuning the codon-anticodon interactions and in maintenance of the fidelity of the translational frame. Such an interpretation of the role of methylations of G966 and C967 residues is consistent with their roles proposed in other studies (8). Interestingly, such a role for these methylations is quite distinct from the role of the two adjacent dimethylations of A1518 and A1519, which affect frameshifting and readthrough in vivo (14).
In conclusion, of the ribosomal features analyzed, although the S9 protein has a general role in holding the P-site tRNA/peptidyl-tRNA in place, the function of G966 and C967 methylations is context dependent. A better understanding of the cross talk between these and the other intrinsic features of the ribosome would help to elucidate the maintenance of translational reading frame, as well as the events that take place during the efficient programmed frameshifting in translational regulation of mRNAs in various organisms (21–23).
ACKNOWLEDGMENTS
We thank our laboratory coworkers for their suggestions and A. E. Dahlberg of the Brown University, Providence, RI, for the generous gift of the lacZ constructs.
This study was supported by grants from the Department of Science and Technology (DST) and the Department of Biotechnology, New Delhi, India. U.V. is a J. C. Bose fellow of the DST. S.A. was a senior research fellow of the Council of Scientific and Industrial Research, New Delhi, India.
Footnotes
Published ahead of print 31 May 2013
REFERENCES
- 1. Ogle JM, Brodersen DE, Clemons WM, Jr, Tarry MJ, Carter AP, Ramakrishnan V. 2001. Recognition of cognate transfer RNA by the 30S ribosomal subunit. Science 292:897–902 [DOI] [PubMed] [Google Scholar]
- 2. Selmer M, Dunham CM, Murphy FV, Weixlbaumer A, Petry S, Kelley AC, Weir JR, Ramakrishnan V. 2006. Structure of the 70S ribosome complexed with mRNA and tRNA. Science 313:1935–1942 [DOI] [PubMed] [Google Scholar]
- 3. Dunkle JA, Wang L, Feldman MB, Pulk A, Chen VB, Kapral GJ, Noeske J, Richardson JS, Blanchard SC, Cate JH. 2011. Structures of the bacterial ribosome in classical and hybrid states of tRNA binding. Science 332:981–984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Lesnyak DV, Osipiuk J, Skarina T, Sergiev PV, Bogdanov AA, Edwards A, Savchenko A, Joachimiak A, Dontsova OA. 2007. Methyltransferase that modifies guanine 966 of the 16 S rRNA: functional identification and tertiary structure. J. Biol. Chem. 282:5880–5887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Gu XR, Gustafsson C, Ku J, Yu M, Santi DV. 1999. Identification of the 16S rRNA m5C967 methyltransferase from Escherichia coli. Biochemistry 38:4053–4057 [DOI] [PubMed] [Google Scholar]
- 6. Hoang L, Fredrick K, Noller HF. 2004. Creating ribosomes with an all-RNA 30S subunit P site. Proc. Natl. Acad. Sci. U. S. A. 101:12439–12443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Shoji S, Abdi NM, Bundschuh R, Fredrick K. 2009. Contribution of ribosomal residues to P-site tRNA binding. Nucleic Acids Res. 37:4033–4042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Arora S, Bhamidimarri SP, Bhattacharyya M, Govindan A, Weber MHW, Vishveswara S, Varshney U. 2013. Distinctive contributions of the ribosomal P-site elements m2G966, m5C967, and the C-terminal tail of the S9 protein in the fidelity of initiation of translation in Escherichia coli. Nucleic Acids Res. 25:2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Burakovsky DE, Prokhorova IV, Sergiev PV, Milon P, Sergeeva OV, Bogdanov AA, Rodnina MV, Dontsova OA. 2012. Impact of methylations of m2G966/m5C967 in 16S rRNA on bacterial fitness and translation initiation. Nucleic Acids Res. 40:7885–7895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Nasvall SJ, Nilsson K, Bjork GR. 2009. The ribosomal grip of the peptidyl-tRNA is critical for reading frame maintenance. J. Mol. Biol. 385:350–367 [DOI] [PubMed] [Google Scholar]
- 11. Jager G, Nilsson K, Bjork GR. The phenotype of many independently isolated +1 frameshift suppressor mutants supports a pivotal role of the P-site in reading frame maintenance. PLoS One 8:e60246. 10.1371/journal.pone.0060246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Saraiya AA, Lamichhane TN, Chow CS, SantaLucia J, Jr, Cunningham PR. 2008. Identification and role of functionally important motifs in the 970 loop of Escherichia coli 16S rRNA. J. Mol. Biol. 376:645–657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Connolly K, Rife JP, Culver G. 2008. Mechanistic insight into the ribosome biogenesis functions of the ancient protein KsgA. Mol. Microbiol. 70:1062–1075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. van Buul CP, Visser W, van Knippenberg PH. 1984. Increased translational fidelity caused by the antibiotic kasugamycin and ribosomal ambiguity in mutants harbouring the ksgA gene. FEBS Lett. 177:119–124 [DOI] [PubMed] [Google Scholar]
- 15. O'Connor M, Goringer HU, Dahlberg AE. 1992. A ribosomal ambiguity mutation in the 530 loop of Escherichia coli 16S rRNA. Nucleic Acids Res. 20:4221–4227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. O'Connor M, Thomas CL, Zimmermann RA, Dahlberg AE. 1997. Decoding fidelity at the ribosomal A and P sites: influence of mutations in three different regions of the decoding domain in 16S rRNA. Nucleic Acids Res. 25:1185–1193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Miller JH. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY [Google Scholar]
- 18. Connolly K, Culver G. 2013. Overexpression of RbfA in the absence of the KsgA checkpoint results in impaired translation initiation. Mol. Microbiol. 87:968–981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Urbonavicius J, Stahl G, Durand JM, Ben Salem SN, Qian Q, Farabaugh PJ, Bjork GR. 2003. Transfer RNA modifications that alter +1 frameshifting in general fail to affect −1 frameshifting. RNA 9:760–768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Gao YG, Selmer M, Dunham CM, Weixlbaumer A, Kelley AC, Ramakrishnan V. 2009. The structure of the ribosome with elongation factor G trapped in the posttranslocational state. Science 326:694–699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Ivanov IP, Atkins JF. 2007. Ribosomal frameshifting in decoding antizyme mRNAs from yeast and protists to humans: close to 300 cases reveal remarkable diversity despite underlying conservation. Nucleic Acids Res. 35:1842–1858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Baranov PV, Gesteland RF, Atkins JF. 2004. P-site tRNA is a crucial initiator of ribosomal frameshifting. RNA 10:221–230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Leger M, Dulude D, Steinberg SV, Brakier-Gingras L. 2007. The three transfer RNAs occupying the A, P and E sites on the ribosome are involved in viral programmed −1 ribosomal frameshift. Nucleic Acids Res. 35:5581–5592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Varshney U, RajBhandary UL. 1990. Initiation of protein synthesis from a termination codon. Proc. Natl. Acad. Sci. U. S. A. 87:1586–1590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Cherepanov PP, Wackernagel W. 1995. Gene disruption in Escherichia coli: TcR and KmR cassettes with the option of Flp-catalyzed excision of the antibiotic-resistance determinant. Gene 158:9–14 [DOI] [PubMed] [Google Scholar]