Abstract
Promoter recognition in bacteria is mediated primarily by the σ subunit of RNA polymerase (RNAP), which makes sequence-specific contacts with the promoter −10 and −35 elements in the context of the RNAP holoenzyme. However, the RNAP α subunit can also contribute to promoter recognition by making sequence-specific contacts with upstream (UP) elements that are associated with a subset of promoters, including the rRNA promoters. In Escherichia coli, these interactions between the RNAP α subunit (its C-terminal domain [CTD], in particular) and UP element DNA result in significant stimulation of rRNA transcription. Among the many cellular and bacteriophage-encoded regulators of transcription initiation that have been functionally dissected, most exert their effects via a direct interaction with either the σ or the α subunit. An unusual example is provided by a phage-encoded inhibitor of RNA synthesis in Staphylococcus aureus. This protein, phage G1 gp67, which binds tightly to σ in the context of the S. aureus RNAP holoenzyme, has recently been shown to exert selective effects on transcription by inhibiting the function of the α subunit CTD (αCTD). Here we report the development of a gp67-responsive E. coli-based transcription system. We examine transcription in vitro from promoters that do or do not carry the UP element associated with a well-characterized E. coli rRNA promoter. Our findings indicate that the αCTD can increase promoter activity significantly even in the absence of an UP element. We also find that gp67 can exert αCTD-dependent or αCTD-independent effects on transcription depending on the particular promoter, indicating that the mechanism of gp67 action is context dependent.
INTRODUCTION
Transcription in bacteria depends on a single multisubunit core enzyme with the subunit structure α2ββ′ω (1). This core enzyme is catalytically proficient but incapable of recognizing specific promoter sequences until it combines with a σ factor, forming the holoenzyme. All bacteria contain a primary σ factor responsible for the bulk of transcription during the exponential phase of growth, as well as one or more alternative σ factors that have more-specialized functions (2). Primary σ factors share four regions of conserved sequence (regions 1 to 4) (3, 4). Regions 2 and 4 contain the critical promoter recognition modules (5–7) as well as essential core interaction determinants, with region 2 binding the clamp helices of the β′ subunit and region 4 binding the flap domain of the β subunit (the β-flap) (8–10). Within the promoter, regions 2 and 4 contact the conserved −10 and −35 elements, respectively (6). Certain promoters contain additional elements that are recognized by σ, including the extended −10 motif (11, 12) and the discriminator (13, 14), which are located just upstream and downstream of the −10 element, respectively. In conjunction with some subset of these σ recognition elements (which necessarily includes the −10 element), certain promoters contain an A+T-rich element located upstream of the −35 element, known as the UP element, which is recognized by the C-terminal domains (CTDs) of the two α subunits (15, 16). Among those promoters that depend on an UP element for full activity are the rRNA promoters in both Escherichia coli (16) and Bacillus subtilis (17).
The study of bacteriophage-encoded regulators of transcription has provided invaluable insight into the function of the bacterial transcription apparatus. In particular, the identification of regulators that interact directly with the bacterial RNA polymerase (RNAP) and alter its function to support phage growth has provided a set of tools for probing RNAP function during transcription initiation, elongation, and termination (18, 19). Phage G1 gp67 was originally identified on the basis of its ability to inhibit cell growth and RNA synthesis when produced in Staphylococcus aureus (20). Consistent with its effect on transcription, gp67 was subsequently shown to associate with Staphylococcus aureus RNAP (21). In particular, gp67 interacts directly with region 4 of the primary σ factor in S. aureus, σA, and this interaction is stable in the context of the S. aureus holoenzyme (21, 22). Thus, gp67 exerts its effect on transcription as a component of the σA-containing S. aureus holoenzyme. Furthermore, experiments performed with S. aureus and in vitro with S. aureus RNAP indicate that gp67 functions selectively, inhibiting transcription from the rRNA promoters and certain others but evidently not influencing transcription from the majority of cellular promoters (22). Detailed analysis suggests that the explanation for this selectivity is that gp67 interferes with the function of the α subunit CTD (αCTD), preventing it from interacting productively with UP element DNA (22).
Although E. coli RNAP and E. coli promoters are much better characterized than their S. aureus counterparts, the use of wild-type E. coli RNAP for the analysis of gp67 function has not been feasible, because gp67 does not interact with E. coli σ70 (the primary σ factor in E. coli) (21, 22). Furthermore, although it was originally reported that a hybrid holoenzyme consisting of the E. coli core in complex with S. aureus σA is susceptible to the action of gp67 (21), subsequent work has indicated that gp67 is unable to interact with this hybrid holoenzyme (22). Having previously identified a limited number of amino acid substitutions in σ70 region 4 that enable an interaction with gp67 (22), we now show that gp67 can associate with the corresponding mutant E. coli RNAP holoenzyme. We use this E. coli-based transcription system to investigate the effect of gp67 on transcription from otherwise identical rRNA promoters that either do or do not contain an UP element, as well as from several other promoters. Our findings indicate that (i) the αCTD can increase promoter activity significantly even in the absence of an UP element and (ii) gp67 can exert αCTD-dependent and/or αCTD-independent effects on transcription as determined by the promoter context.
MATERIALS AND METHODS
Plasmids, strains, and growth conditions.
Complete lists of the bacterial strains and plasmids used in this study are provided in Tables S1 and S2 in the supplemental material. E. coli NEB 5-α F′Iq (New England BioLabs) was used as the recipient strain for all plasmid constructions. All E. coli strains were grown at 37°C in Luria-Bertani (LB) broth or on LB plates supplemented with the appropriate antibiotic(s) at the following concentrations: kanamycin, 50 μg/ml; chloramphenicol, 25 μg/ml; carbenicillin, 100 μg/ml; spectinomycin, 50 μg/ml. The σ70 depletion strain NUN449 (a derivative of strain CAG20153 [23]) was grown in liquid culture as a series of dilutions to prevent entry into stationary phase and was grown on plates for fewer than 16 h; both the liquid broth and the plates were supplemented with the inducer of the trp promoter, indole-3-acrylic acid (IAA), at 200 μM as necessary.
Functionality of E. coli σ70-quint and gp67-mediated growth inhibition.
To assess the functionality of E. coli σ70-quint, the σ70 depletion strain NUN449 was transformed with a plasmid encoding wild-type E. coli σ70 (σ70-WT), E. coli σ70-quint (a σ70 mutant bearing the corresponding S. aureus σA residues at five positions), or no protein under the control of a weak constitutive promoter and was grown in the presence of IAA. Unsaturated overnight cultures were serially diluted and were spotted onto plates with or without IAA. To assess the toxicity of gp67 in E. coli cells, the σ70 depletion strain was cotransformed with two compatible multicopy plasmids, one encoding either E. coli σ70-WT or E. coli σ70-quint under the control of a weak constitutive promoter and the other encoding either gp67-His6 or no protein under the control of an isopropyl-β-d-thiogalactopyranoside (IPTG)-inducible promoter. Cultures that were in mid-exponential phase (optical density at 600 nm [OD600], 0.3 to 0.6) after overnight growth were then serially diluted in fresh LB broth and were spotted onto plates supplemented with chloramphenicol, carbenicillin, and spectinomycin either without IPTG or with 20 μM IPTG in the absence of IAA. Plates were incubated at 37°C for fewer than 16 h.
Protein expression and purification.
E. coli σ70-WT and E. coli σ70-quint proteins bearing an N-terminal His6 tag were purified from inclusion bodies in BL21(DE3) cells transformed with plasmid pLHN12-His6-Eco σ70-WT or pLHN12-His6-Eco σ70-quint as described previously (24). Wild-type E. coli core RNAP for in vitro transcription experiments was purchased from Epicentre. Plasmid-encoded gp67-His6 was purified from BL21(DE3) cells as described previously (22), and E. coli core RNAP lacking the αCTD (EαΔCTD) was prepared as described previously (25).
In vitro transcription assays.
Multiround in vitro transcription reactions using supercoiled templates bearing the promoters shown in Fig. S1 in the supplemental material were performed essentially as described previously (15) with some modifications. Purified E. coli σ70-WT or E. coli σ70-quint (40 nM) was preincubated either in the absence of gp67 or with 50 or 100 nM gp67 for 10 min on ice in 1× transcription buffer (30 mM KCl, 40 mM Tris-acetate [pH 7.9], 10 mM MgCl2, 1 mM dithiothreitol [DTT], and bovine serum albumin [BSA] at 100 μg/ml). Wild-type or αΔCTD E. coli core RNAP (4 nM) was then added, and the mixture was incubated for 10 min at 24°C to form the holoenzyme. The holoenzyme was added to a reaction mixture containing 1× transcription buffer, 50 ng (0.6 nM) supercoiled plasmid DNA, 500 μM ATP, 200 μM CTP, 200 μM GTP, 10 μM UTP, and 10 μCi/μl [α-32P]UTP (Perkin-Elmer) to initiate transcription (final reaction volume, 20 μl), and the reaction was allowed to proceed at 24°C for 15 min. Reactions were stopped by the addition of 20 μl of loading buffer (95% [vol/vol] formamide, 20 mM EDTA, 0.05% [wt/vol] bromophenol blue, and 0.05% [wt/vol] xylene cyanol), and the reaction mixture was first heated for 1 min at 90°C and then electrophoresed on a 6% urea-polyacrylamide gel. Supercoiled plasmid template DNA was purified as described previously (26). Bands were visualized by phosphorimaging, and the data were quantified using ImageQuant software. The data reported for all experiments are the averages for at least three independent experiments with standard deviations, normalized at each promoter to the band intensity in the absence of gp67.
One- and two-hybrid assays.
For the bacterial two-hybrid assay (27), reporter strain FW102 OL2–62 (28) was cotransformed with two compatible multicopy plasmids, one encoding either the α subunit alone or the indicated fusion protein (see Fig. 1 and 4) consisting of α and region 4 of σ (α–σ4) and another encoding a λ CI–S. aureus β-flap fusion protein. These plasmids direct the synthesis of the fusion proteins (or α) under the control of IPTG-inducible promoters. Individual transformants were selected on plates, and three independent isolates of each were grown overnight in LB broth supplemented with kanamycin, carbenicillin, and chloramphenicol in the absence of IPTG. Overnight cultures were diluted 1:100 and were grown in LB medium supplemented with kanamycin, carbenicillin, chloramphenicol, and 20 μM IPTG in microtiter plates to mid-exponential phase (OD600, 0.3 to 0.6). For the bacterial one-hybrid assays to detect −35 element binding (28, 29), reporter strain FW102 placCons−35C (BN317) (29), BN318, or BN299 (30) was transformed with a single multicopy plasmid encoding either α alone or the indicated α–σ4 fusion protein under the control of an IPTG-inducible promoter. Individual transformants were selected on plates, and three independent isolates of each were grown and assayed as described for the two-hybrid assays with the corresponding antibiotics and IPTG concentrations. To assess the effect of unfused gp67 on −35 element binding by σ4, strain BN317 was cotransformed with a second compatible multicopy plasmid encoding either unfused C-terminally His6-tagged gp67 or no protein under the control of an IPTG-inducible promoter. β-Galactosidase assays were performed as described elsewhere using microtiter plates and a microtiter plate reader (31), and Miller units were calculated as described previously (28, 31). The results presented below are the averages for the three independent isolates with standard deviations.
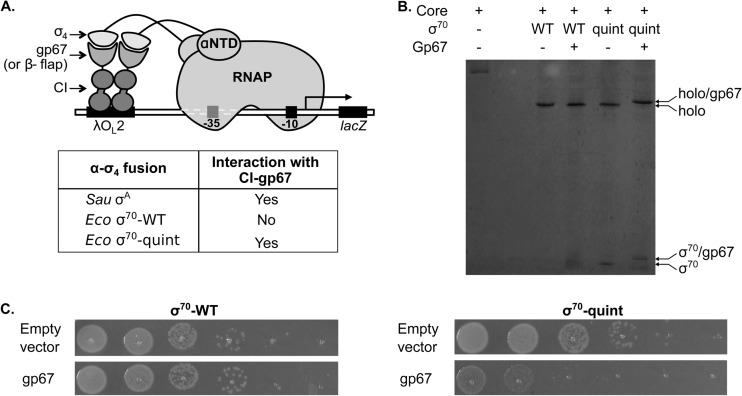
Fig 1.
gp67 binds to a mutant E. coli RNAP holoenzyme. (A) Bacterial two-hybrid assay used to dissect the gp67–σ4 interaction. The cartoon shows how the interaction between σ4 fused to the RNAP α N-terminal domain (αNTD) and gp67 fused to the bacteriophage λ CI protein activates the transcription of a lacZ reporter gene from test promoter placOL2–62, which bears λ operator OL2 upstream of the lac core promoter. Whereas this assay detects an interaction between gp67 and region 4 of S. aureus σA, no interaction is detected between gp67 and region 4 of E. coli σ70. Genetic analysis led to the identification of a limited number of specificity-determining residues, enabling the construction of a σ704 mutant bearing five S. aureus residues that interacts efficiently with gp67 (22). The two-hybrid assay also detects an interaction between the σ4 moiety of the α–σ4 fusion protein and the β-flap moiety of a λ CI–β-flap fusion protein (9); this interaction serves as a control for the structural integrity of σ4 mutants. Sau, S. aureus; Eco, E. coli. (B) Native electrophoretic mobility shift analysis showing the interaction between gp67 and the mutant E. coli RNAP holoenzyme. Wild-type E. coli σ70 (WT) or the E. coli σ70 quintuple mutant (quint) (each at 2 μM) was incubated with gp67 (2 μM) for 10 min on ice before the addition of E. coli RNAP core (1 μM). Complex formation was analyzed by native gel electrophoresis on a 4-to-12% PhastGel. Only the mutant reconstituted holoenzyme contains gp67. (C) gp67 production is toxic in E. coli cells containing the σ70 quintuple mutant but not in cells containing wild-type σ70. The E. coli σ70 depletion strain NUN449 was cotransformed with compatible plasmids, one encoding either wild-type σ70 or the σ70 quintuple mutant under the control of a weak constitutive promoter and the other encoding C-terminally His6-tagged gp67 (or no protein) under the control of the IPTG-inducible lacUV5 promoter. In the absence of indole-3-acrylic acid, the chromosomal rpoD gene in strain NUN449 is repressed, and functional plasmid-encoded σ70 must be provided to permit cell growth. Strain survival was assayed by spotting serial dilutions of cell cultures onto LB agar plates supplemented with 20 μM IPTG.
RESULTS
Construction of a gp67-responsive E. coli RNAP variant.
We previously used a bacterial two-hybrid system to genetically dissect the protein-protein interaction between gp67 and S. aureus σA region 4 (σA4) (Fig. 1A) (22). With this system, we identified a minimal set of amino acid differences between σA4 and σ70 region 4 (σ704) that accounts for the inability of gp67 to interact with σ704 (22). We used this information to construct a σ70 variant that was efficiently bound by gp67; this variant (σ70-quint) bears the corresponding S. aureus σA residues at five positions (A553D, A556E, K557N, Q579V, D581G) (22). We reconstituted the mutant E. coli RNAP holoenzyme (Eσ70-quint) in vitro and compared its activity to that of the reconstituted wild-type RNAP holoenzyme (Eσ70-WT). Transcription assays performed with several different promoters revealed that Eσ70-quint was fully functional (see below). We used native gel shift analysis to determine whether or not gp67 could associate with Eσ70-quint to form a stable ternary complex. The results of this analysis indicated that gp67 associated with Eσ70-quint but not with Eσ70-WT (Fig. 1B).
gp67 toxicity in a modified strain of E. coli.
Based on the results of the two-hybrid and native gel shift analyses, we predicted that gp67 would be toxic when produced in E. coli cells containing σ70-quint but not in cells containing wild-type σ70. To test this prediction, we first assessed the ability of σ70-quint to complement the effects of σ70 depletion. To do this, we introduced a plasmid encoding either wild-type σ70 or σ70-quint (under the control of a constitutive promoter) into a σ70 depletion strain in which the chromosomal rpoD gene is under the control of the trp promoter (23). Under repressing conditions (in tryptophan-replete medium), rpoD expression is greatly reduced and cell growth is severely compromised. We found that cell growth was restored in the presence of either plasmid, indicating that σ70-quint was fully capable of supporting cell growth (see Fig. S2 in the supplemental material). We then introduced a compatible plasmid directing the inducible synthesis of gp67 into depletion strain cells containing either plasmid-encoded wild-type σ70 or plasmid-encoded σ70-quint. We found that gp67 was toxic when produced in depletion strain cells grown in tryptophan-replete medium in the presence of plasmid-encoded σ70-quint, but we detected no toxicity when the cells were grown under the same conditions in the presence of plasmid-encoded wild-type σ70 (Fig. 1C).
Effects of gp67 on UP element-dependent and UP element-independent transcription.
Next, we tested whether or not gp67 exerted promoter-specific effects on transcription in the context of Eσ70-quint. We began by assaying transcription in vitro with supercoiled DNA templates bearing the well-characterized rRNA promoter rrnB P1, either lacking (rrnB −41) or containing (rrnB −61) its UP element (15). These plasmids carry a transcription terminator downstream of the test promoter, enabling the identification of a specific transcript by denaturing gel electrophoresis; in addition, they encode the 108-nucleotide (nt) RNA-I transcript, which derives from the plasmid origin of replication and serves as a control −10/−35 promoter that does not bear an UP element. We performed the transcription assays with RNAP reconstituted with either wild-type σ70 or σ70-quint (Eσ70-WT or Eσ70-quint). As expected, gp67 did not inhibit transcription from any of the promoters when tested with Eσ70-WT (see Fig. S3A and B in the supplemental material). In contrast, gp67 inhibited transcription from all of the promoters when tested with Eσ70-quint (Fig. 2A and B; see also Fig. S4A and B in the supplemental material). The effect of gp67 on transcription from the promoters lacking UP elements (rrnB −41 and RNA-I) was unexpected, all the more so because the magnitude of the inhibition was somewhat greater with rrnB −41 (as much as 7.6-fold) than with rrnB −61 (as much as 4.9-fold).
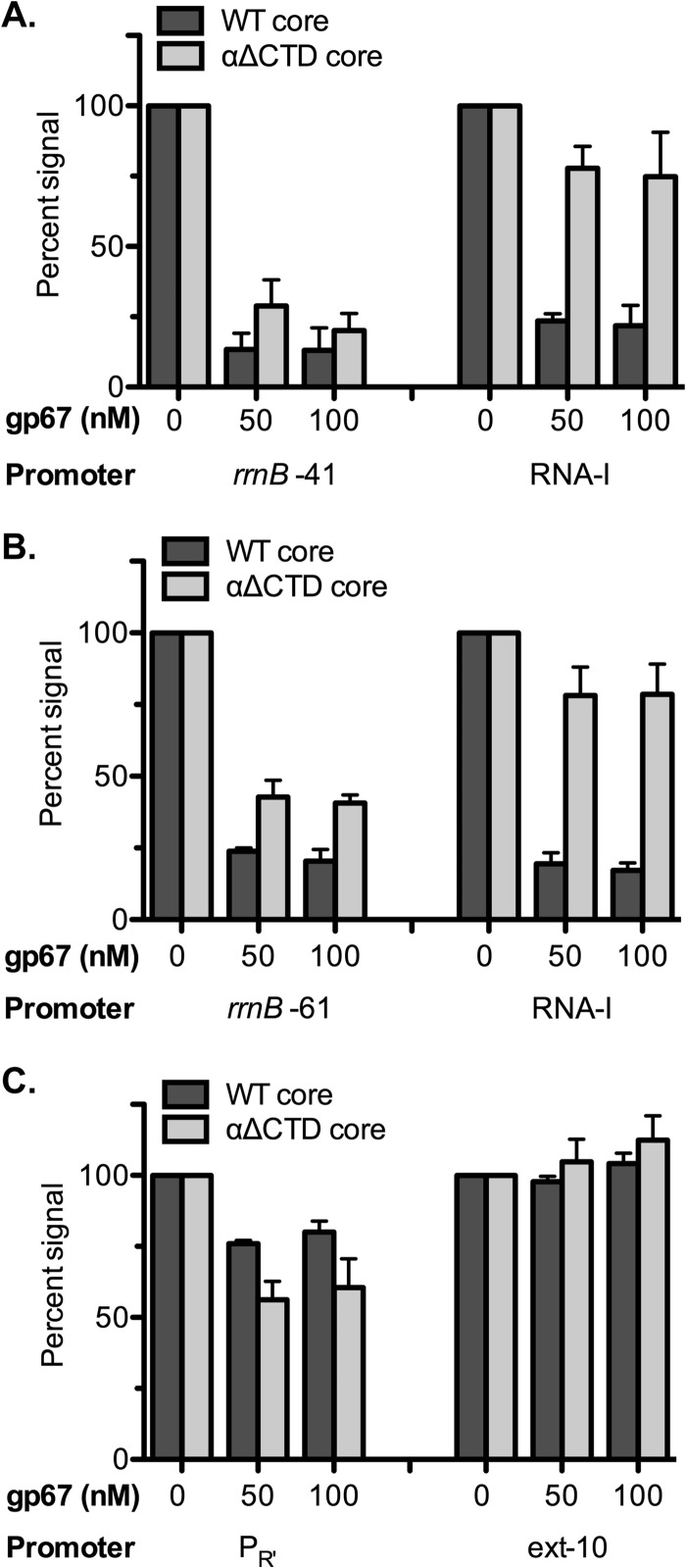
Fig 2.
UP element-independent effects of gp67 on transcription in vitro from promoter rrnB P1. Multiround transcription assays were performed with supercoiled plasmid templates containing the indicated promoter (rrnB −41, rrnB −61, or λ PR′). The template containing the λ PR′ promoter also contains the extended −10 promoter (ext−10). The templates also contain the RNA-I promoter within the plasmid origin of replication. The transcription assays were performed with E. coli RNAP (4 nM) reconstituted with the σ70 quintuple mutant (40 nM). For each promoter, the amount of transcription detected in the presence of gp67 (at either 50 nM or 100 nM) was normalized to that observed in the absence of gp67, which was set to 100%. Representative gels are shown in Fig. S4. The data shown in the graphs represent the results of at least three independent experiments that were quantified by phosphorimaging and averaged (error bars indicate standard deviations). Assays were performed with the RNAP holoenzyme containing either wild-type core (Eσ70-quint) (dark shaded bars) or αΔCTD core (EαΔCTDσ70-quint) (light shaded bars). (A and B) Effects of gp67 on transcription from the rrnB P1 core and full promoters. The inhibitory effects of gp67 on transcription from the rrnB −41 promoter (A) and the rrnB −61 promoter (B) were only modestly dependent on the αCTD (compare the WT core with the αΔCTD core). In contrast, the inhibitory effect of gp67 on transcription from the RNA-I promoter was almost completely dependent on the αCTD. (C) Effect of gp67 on transcription from the λ PR′ promoter. gp67 exerted only a weak inhibitory effect on transcription from the λ PR′ promoter and no inhibitory effect on transcription from the associated extended −10 promoter.
In order to assess the extent to which the observed inhibitory effects of gp67 depended on the αCTD, we made use of a mutant RNAP core enzyme lacking the αCTD (EαΔCTD). Previous studies have established that the EαΔCTD RNAP holoenzyme is transcriptionally active and capable of promoter-specific transcription initiation (15, 32). We used the EαΔCTD core RNAP to reconstitute a holoenzyme containing either wild-type σ70 or σ70-quint (EαΔCTD σ70-WT or EαΔCTD σ70-quint). As expected, gp67 did not inhibit transcription from any of the promoters when tested with EαΔCTD σ70-WT (see Fig. S3A and B in the supplemental material). Furthermore, in the case of the RNA-I promoter, gp67 exerted only a slight inhibitory effect when tested with EαΔCTD σ70-quint (≤25%) (Fig. 2A and B; see also Fig. S4A and B in the supplemental material), suggesting that transcription from this promoter depends, in part, on sequence-nonspecific interactions of the αCTD with upstream DNA that are disrupted in the presence of gp67. In contrast, the use of EαΔCTD σ70-quint revealed that the inhibitory effects of gp67 on transcription from the rRNA promoters (rrnB −61 and rrnB −41) were only partially dependent on the αCTD and that the extents of αCTD dependence were similar for the two promoters (Fig. 2A and B; see also Fig. S4A and B in the supplemental material). We conclude that gp67 can inhibit transcription in a manner that does not necessarily depend on the αCTD and that the rrnB P1 core promoter is susceptible to this activity of gp67. Our results also reveal UP element-independent effects of the αCTD, which are particularly evident at the RNA-I promoter. Although UP element-independent effects of the αCTD on transcription from the RNA-I promoter have not been reported previously, previous kinetic experiments have uncovered a substantial effect of sequence-nonspecific interactions between the αCTD and upstream DNA on the rate of RNAP association at two well-characterized promoters that lack UP elements (33; see also reference 34).
We tested the effect of gp67 on transcription from another −10/−35 promoter by constructing a plasmid vector bearing the bacteriophage λ late promoter PR′ (which lacks an UP element) in place of the rrnB promoter. This vector also contained a consensus extended −10 promoter positioned within the PR′ transcribed region (35). Extended −10 promoters are defined by the presence of a TG dinucleotide 1 bp upstream of the −10 hexamer and typically lack a −35 element (12). With this plasmid vector, transcription initiation at PR′ produces a transcript of 202 nt, whereas initiation at the extended −10 promoter produces a transcript of 170 nt. We found that gp67 exerted no inhibitory effect on transcription from the extended −10 promoter (which lacks both a −35 element and an UP element) when tested with either EαΔCTD σ70-WT or EαΔCTD σ70-quint (Fig. 2C; see also Fig. S4C in the supplemental material). The resistance of the extended −10 promoter to the action of gp67 confirmed that gp67 does not function by sequestering σ70-quint and preventing its association with the core enzyme in the manner of a classic anti-σ factor (36). Transcription from PR′ was also relatively resistant to the action of gp67; when tested with Eσ70-quint, gp67 reduced transcription by only ∼22%, and when tested with EαΔCTD σ70-quint, gp67 had a somewhat larger effect, reducing transcription by ∼42% (Fig. 2C; see also Fig. S4C in the supplemental material). These results indicate that different promoters are differentially sensitive to gp67 and, importantly, that gp67 does not interfere with holoenzyme formation.
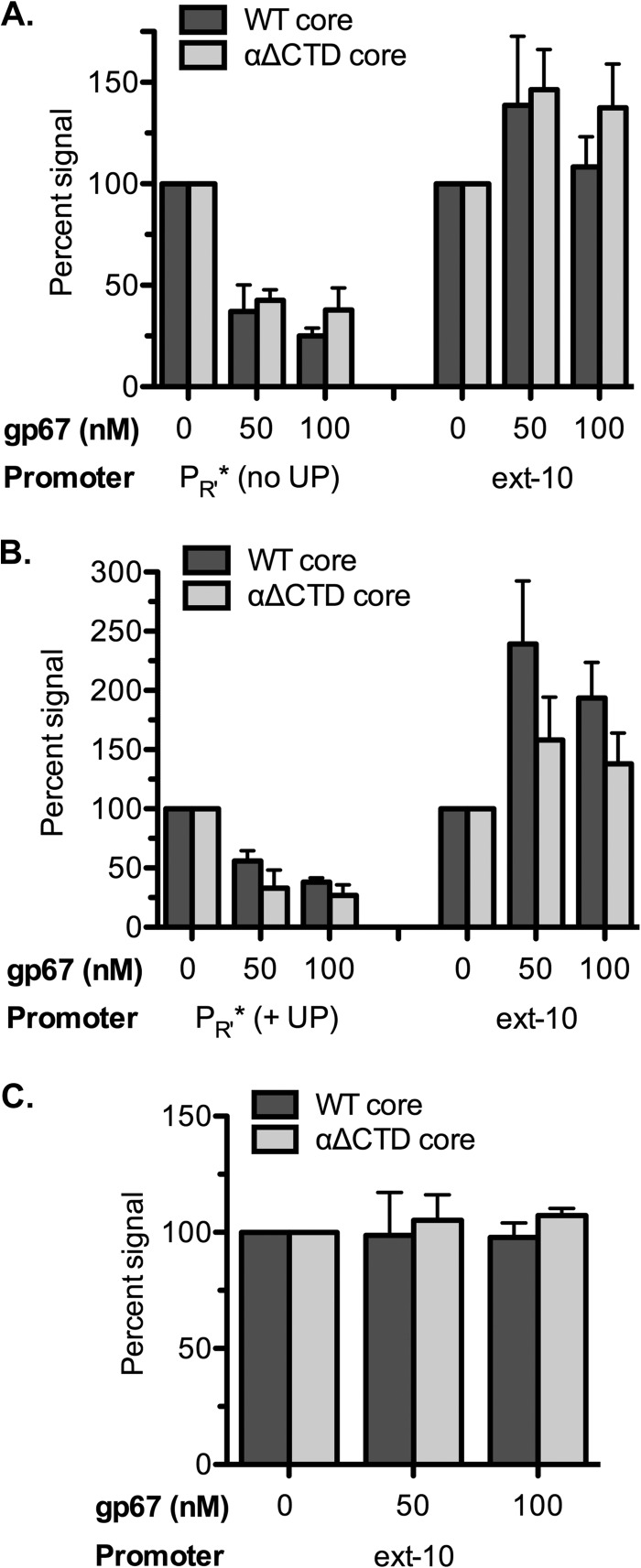
We wondered whether or not the addition of an UP element to PR′ would confer gp67 sensitivity on this promoter. To test this possibility, we fused the rrnB P1 UP element to the PR′ core promoter, but we found that the UP element did not increase transcription from this promoter. However, when we created a PR′ variant (designated PR′*) with a weakened −35 element (TTGACT to TTGATA), we were able to detect a modest (∼3-fold) stimulatory effect of the appended UP element (see Fig. S5A in the supplemental material). We then tested the effect of gp67 on transcription from this weakened PR′ variant with either the native PR′ upstream sequence or the UP element; like the wild-type PR′ template, these modified templates bore the consensus extended −10 promoter in the PR′ transcribed region. We found that gp67 inhibited transcription from these PR′ derivatives as much as 3.7-fold and that this inhibitory effect was essentially independent of the αCTD (Fig. 3A and B; see also Fig. S5A and B in the supplemental material); in fact, in the case of the UP element-containing derivative, the magnitude of the inhibitory effect was slightly larger when transcription was assayed with EαΔCTD σ70-quint (Fig. 3B). In these assays, we found that transcription from the extended −10 promoter increased in the presence of gp67. We suspected that this apparent stimulatory effect of gp67 was due to promoter competition, because the extended −10 promoter is only 30 bp downstream of PR′ (see Fig. S1 in the supplemental material). To eliminate the potential for competition, we inactivated PR′, leaving only the extended −10 promoter intact (see Fig. S1). As expected, gp67 had no effect on transcription from the extended −10 promoter when the template was modified in this manner (Fig. 3C; see also Fig. S5A and B in the supplemental material). These experiments reinforce our conclusion that gp67 can exert inhibitory effects on transcription that are independent of the αCTD. Furthermore, because we found that weakening the −35 element can enhance the sensitivity of a promoter to gp67, we suggest that gp67 can compromise the ability of σ704 to engage in optimal interactions with the −35 element.
Fig 3.
UP element-independent effects of gp67 on transcription in vitro from a weakened λ PR′ promoter. Multiround transcription assays were performed with supercoiled plasmid templates, and the data were quantified, as described in the legend to Fig. 2. (A and B) Effects of gp67 on transcription from a weakened λ PR′ promoter (λ PR′*) without or with an appended UP element. The inhibitory effects of gp67 on transcription from the λ PR′* promoters were essentially independent of the αCTD. (C) Effect of gp67 on transcription from the extended −10 promoter. gp67 had no stimulatory effect on transcription from the extended −10 promoter when the competing λ PR′ promoter was inactivated (see the text).
Effects of gp67 on −35 element binding.
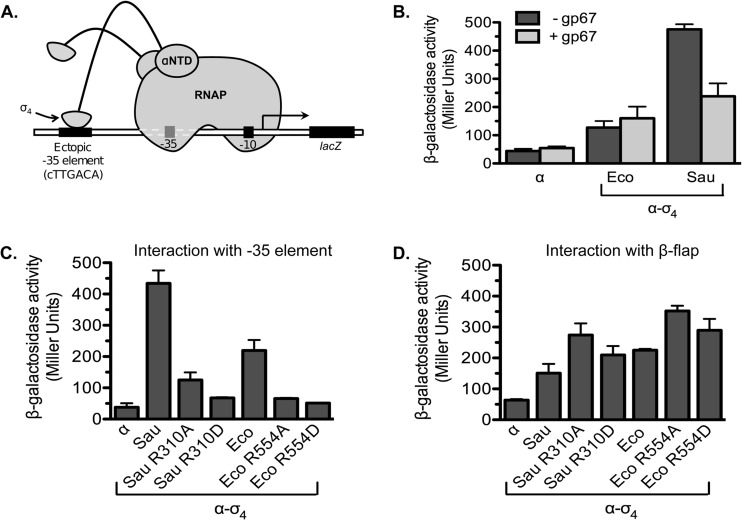
To examine the potential of gp67 to modulate the binding of σ4 to the −35 element, we employed a bacterial one-hybrid assay that enables the detection of an interaction between σ704 and the −35 element (29). In this system, interaction between σ4 fused to the α subunit of RNAP and a consensus −35 element positioned upstream of the lac core promoter activates the transcription of a lacZ reporter gene (Fig. 4A; see also Fig. S6 in the supplemental material). We therefore tested the effect of producing unfused gp67 in the one-hybrid reporter strain containing either α–S. aureus σA4 or α–E. coli σ704. Whereas gp67 reduced transcription by a factor of ∼2 in cells containing α–S. aureus σA4, gp67 had no inhibitory effect on reporter gene expression in cells containing α–E. coli σ704, a finding consistent with the inability of gp67 to interact with wild-type σ704 (Fig. 4B). We conclude that when bound to σ4, gp67 can compromise −35 element binding, at least in the context of the one-hybrid system, which detects the interaction between σ4 and the −35 element in the absence of other potentially stabilizing interactions.
Fig 4.
Effects of gp67 and amino acid substitutions on the binding of σ4 to the −35 element. (A) A one-hybrid assay detects the interaction between σ4 and a −35 element. The cartoon shows how the interaction between the σ4 moiety of the α–σ4 fusion protein and a consensus −35 element positioned upstream of the lac core promoter activates the transcription of a lacZ reporter gene. (B) Effects of unfused gp67 on the interaction between σ4 and the −35 element as assessed using a bacterial one-hybrid assay. Reporter strain FW102 placCons−35 was transformed with two compatible plasmids, one encoding the indicated α–σ4 fusion protein (either α–S. aureus σA4 or α–E. coli σ704) or unfused α and the other encoding C-terminally His6-tagged gp67 (light shaded bars) or no protein (dark shaded bars). Cells were grown in the presence of 5 μM IPTG and were assayed for β-galactosidase. The graph shows the averages for three biological replicates assayed in the same experiment (error bars indicate standard deviations). (C) One-hybrid interactions of α–σ4 fusion proteins bearing amino acid substitutions at S. aureus σA residue R310 or the corresponding E. coli σ70 residue, R554. One-hybrid reporter strain FW102 placCons−35 was transformed with a control plasmid encoding unfused α (α) or an experimental plasmid encoding either the α–S. aureus σA4 (Sau) or the α–E. coli σ704 (Eco) fusion protein bearing the indicated amino acid substitution. Cells were grown in the presence of 20 μM IPTG and were assayed for β-galactosidase. The graph shows the averages for three biological replicates assayed in the same experiment (with standard deviations). (D) Two-hybrid interactions of α–σ4 fusion proteins bearing amino acid substitutions at S. aureus σA residue R310 and the corresponding E. coli σ70 residue, R554. Two-hybrid reporter strain FW102 OL2–62 was transformed with two compatible plasmids. The first encoded unfused α or one of the two α–σ4 fusion proteins (either α–S. aureus σA4 or α–E. coli σ704) bearing the indicated amino acid substitution, and the second encoded the λ CI–β-flap fusion protein (see Fig. 1A).
The previously determined crystal structure of gp67 in complex with S. aureus σA4 suggests that the binding of gp67 to σA4 is compatible with the binding of σA4 to the −35 element (22). Among the 10 residues of σA4 that are predicted to make direct contact with −35 element DNA (5, 37), only one (R310) is part of the interface with gp67. This exceptional residue makes contact with a DNA phosphate just upstream of the −35 element, based on high-resolution structures of Thermus aquaticus σA4 in complex with −35 element DNA (5, 37). However, in the gp67–S. aureus σA4 complex, R310 lies within a deep pocket of gp67, where it would be inaccessible to the DNA (22). We suggest, therefore, that the inhibitory effect of gp67 on −35 element binding as detected in the one-hybrid assay might be due to the disruption of R310 (R554 in E. coli σ70) contact with the DNA phosphate backbone. In agreement with this possibility, we found that replacement of this arginine by either alanine or aspartic acid in the context of either σA4 or σ704 reduced −35 element binding in the one-hybrid assay (Fig. 4C). The same substitutions did not compromise the interaction between either σA4 or σ704 and the β-flap, as assayed in the two-hybrid system (Fig. 1A and Fig. 4D), indicating that their effects on −35 element binding are not explained by nonspecific effects on fusion protein stability.
DISCUSSION
Here we describe an E. coli-based transcription system that is responsive to the transcription inhibitor gp67, encoded by S. aureus phage G1. Previous work conducted with S. aureus and with a native S. aureus-based transcription system revealed that gp67 selectively inhibits transcription from rRNA promoters and from a minority of other promoters that are proposed to depend on UP element-like sequences for full activity (22). Furthermore, gp67 was found to disrupt the contacts of RNAP with upstream DNA (22). The E. coli-based system enabled us to take advantage of specific well-characterized reagents developed in order to study UP element-dependent transcription, including the αΔCTD-RNAP core enzyme and a model rRNA promoter (rrnB P1) with or without its associated UP element. Our findings revealed a strong inhibitory effect of gp67 on rrnB P1 transcription, but unexpectedly, this effect was not dependent on the UP element and was largely independent of the αCTD. Contrasting results were obtained with another previously studied −10/−35 promoter (RNA-I), which lacks an UP element. In this case, gp67 exerted a strong inhibitory effect on transcription, which was, however, almost completely dependent on the αCTD. We note that both the rrnB P1 and RNA-I promoters have suboptimal −35 elements (see Fig. S1 in the supplemental material), indicating that this characteristic alone is not sufficient to explain the αCTD-independent effect of gp67 on rrnB P1 transcription. Taken together, our findings with these and several other promoters indicate that gp67 can exert αCTD-independent effects on transcription, which we suggest are mediated through σ4. At the same time, our findings with the RNA-I promoter illustrate the use of gp67 as a tool that can reveal sequence-nonspecific effects of the αCTD on promoter activity.
Sequence-specific and sequence-nonspecific interactions of the αCTD with upstream promoter DNA.
The UP element was originally identified in the context of the E. coli rrnB P1 promoter, where it was shown to function as a binding site for the αCTD, increasing transcription substantially both in vitro and in vivo (15). Although the αCTD is essential for cell viability, the demonstration that a transcriptionally active αΔCTD-RNAP that retains the ability to recognize core promoters could be reconstituted in vitro provided a critical tool for demonstrating the αCTD dependence of UP element utilization. Thus, unlike wild-type RNAP, the αΔCTD-RNAP cannot distinguish between otherwise identical promoters bearing or lacking an UP element (15). However, evaluation of the potential contribution of sequence-nonspecific interactions of the αCTD to promoter activity is less straightforward than evaluation of the potential contribution of sequence-specific interactions of the αCTD to promoter activity because of the need to control for possible activity differences between the wild-type and mutant enzyme preparations when one is performing transcription assays with wild-type RNAP and the αΔCTD-RNAP. Kinetic studies provide one way to circumvent this difficulty, and the results of such an analysis performed with two well-characterized promoters that lack UP elements revealed a substantial effect of sequence-nonspecific interactions of the αCTD with upstream promoter DNA on the rate of RNAP association, raising the possibility that the αCTD contributes to transcription initiation at all promoters (33). Our finding that gp67 inhibits transcription from the RNA-I promoter in a manner that depends on the αCTD provides support for the idea that sequence-nonspecific interactions of the αCTD with upstream promoter DNA can contribute to promoter function.
Comparison between the effects of gp67 in the context of S. aureus RNAP and in the context of E. coli RNAP.
Despite binding to σ4 in the context of the RNAP holoenzyme, gp67 did not appear to inhibit −35 element recognition when tested in a native S. aureus-based transcription system (22). Instead, the evidence suggests that gp67 exerts its effect on S. aureus transcription indirectly, by disrupting interactions of the αCTD with upstream promoter DNA (22). In contrast, however, when tested with our modified E. coli RNAP, gp67 inhibited transcription from the rrnB P1 promoter (with or without its UP element) in a manner only partially dependent on the αCTD. In particular, the use of the αΔCTD-RNAP enabled us to show that gp67 inhibited transcription from the rrnB P1 promoter even in the absence of the αCTD. Similarly, gp67 exerted αCTD-independent inhibitory effects on transcription from a weakened version of the λ late promoter PR′ with or without an UP element appended. Importantly, however, gp67 did not inhibit −35 element-independent transcription from a consensus extended −10 promoter in the E. coli system, indicating that its effects on transcription from −10/−35 promoters are specific and are not due to σ70 sequestration.
We suggest that in our modified E. coli-based system, gp67 exerts effects on −35 element binding as well as effects on upstream DNA interactions with the αCTD. We consider several possible explanations for the apparently more restrictive action of gp67 on transcription in the S. aureus system. As noted above, the crystal structure of the gp67/S. aureus σA4 complex reveals that only 1 of 10 conserved σA residues that make direct contact with −35 element DNA lies at the gp67–σA4 interface (22). This residue, S. aureus σA R310 (corresponding to E. coli σ70 R554), is seen to contact a DNA phosphate just upstream of the −35 element (at position −36) in two high-resolution crystal structures of σ4 in complex with −35 element DNA (5, 37). Furthermore, we have shown that this contact contributes to the stability of −35 element binding as assayed in our one-hybrid system using either an α–S. aureus σA4 or an α–E. coli σ704 fusion protein. Thus, one possible explanation for the difference between the actions of gp67 in the E. coli and S. aureus systems is that this conserved arginine residue contributes more significantly to promoter binding in the context of the specific E. coli promoters we examined than in the context of most S. aureus promoters.
A second possible explanation for the more restrictive action of gp67 on transcription in the S. aureus system involves potential interactions of gp67 with promoter DNA. In particular, we suggest that sequence-independent interactions of gp67 with the DNA upstream of the −35 element may, depending on the context, compensate for the loss of the contact between S. aureus σA R310 (E. coli σ70 R554) and the phosphate backbone. In support of this possibility, a model of the gp67/σA4 complex bound to promoter DNA in the context of the RNAP open complex reveals that the gp67 N-terminal domain presents a very basic molecular surface to the DNA just upstream of the −35 element (22). If gp67 indeed has the potential to make stabilizing interactions with the DNA, then that activity may be optimized in the context of the S. aureus RNAP holoenzyme and/or may be highly dependent on promoter context.
Finally, we note that structure-based modeling of the gp67/RNAP holoenzyme/open promoter complex revealed a minor clash with DNA at the downstream end of the −35 element (22), suggesting a third possible explanation for the difference between the actions of gp67 in the E. coli and S. aureus systems. Although only small rearrangements in either the protein or the DNA would be required to relieve this clash, which involves the helical tower of the gp67 C-terminal domain (22), it is possible that these accommodations are more readily made in the context of the S. aureus RNAP holoenzyme.
Phage-encoded transcription regulators as probes of RNAP function.
Because bacteriophages typically depend on the host multisubunit RNAP for the transcription of at least a subset of their genes, they often encode regulators that enable them to subvert the cellular transcription program and compete effectively for components of the transcription machinery. In particular, phage-encoded inhibitors of cellular transcription can bias transcription away from highly transcribed cellular genes to facilitate the transcription of the phage genome. Such inhibitors, which can be thought of as promoter selectivity factors, can potentially be used as tools for promoter classification. In S. aureus, a global transcriptome analysis identified a subset of cellular promoters that are particularly sensitive to the inhibitory action of gp67 (22). Performing an analogous analysis in a suitably modified strain of E. coli would reveal whether or not the effects of gp67 are more widespread in E. coli and would aid in the identification of promoter characteristics that specify gp67 susceptibility in both the presence and the absence of the αCTD.
Supplementary Material
ACKNOWLEDGMENTS
We thank Wilma Ross for invaluable advice and for providing the rrnB −41 and rrnB −61 vectors. We thank Simon Dove for helpful comments on the manuscript.
This work was supported by NIH R01 GM044025 (to A.H.) and NIH R01 GM053759 (to S.A.D.).
Footnotes
Published ahead of print 7 June 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JB.00499-13.
REFERENCES
- 1. Darst SA. 2001. Bacterial RNA polymerase. Curr. Opin. Struct. Biol. 11: 155– 162 [DOI] [PubMed] [Google Scholar]
- 2. Gruber TM, Gross CA. 2003. Multiple sigma subunits and the partitioning of bacterial transcription space. Annu. Rev. Microbiol. 57: 441– 466 [DOI] [PubMed] [Google Scholar]
- 3. Lonetto M, Gribskov M, Gross CA. 1992. The σ70 family: sequence conservation and evolutionary relationships. J. Bacteriol. 174: 3843– 3849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Paget MS, Helmann JD. 2003. The σ70 family of sigma factors. Genome Biol. 4: 203. 10.1186/gb-2003-4-1-203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Campbell EA, Muzzin O, Chlenov M, Sun JL, Olson CA, Weinman O, Trester-Zedlitz ML, Darst SA. 2002. Structure of the bacterial RNA polymerase promoter specificity σ subunit. Mol. Cell 9:527– 539 [DOI] [PubMed] [Google Scholar]
- 6. Murakami KS, Darst SA. 2003. Bacterial RNA polymerases: the wholo story. Curr. Opin. Struct. Biol. 13: 31– 39 [DOI] [PubMed] [Google Scholar]
- 7. Feklistov A, Darst SA. 2011. Structural basis for promoter −10 element recognition by the bacterial RNA polymerase σ subunit. Cell 147: 1257– 1269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Arthur TM, Burgess RR. 1998. Localization of a σ70 binding site on the N terminus of the Escherichia coli RNA polymerase β′ subunit. J. Biol. Chem. 273: 31381– 31387 [DOI] [PubMed] [Google Scholar]
- 9. Kuznedelov K, Minakhin L, Niedziela-Majka A, Dove SL, Rogulja D, Nickels BE, Hochschild A, Heyduk T, Severinov K. 2002. A role for interaction of the RNA polymerase flap domain with the σ subunit in promoter recognition. Science 295: 855– 857 [DOI] [PubMed] [Google Scholar]
- 10. Murakami KS, Masuda S, Darst SA. 2002. Structural basis of transcription initiation: RNA polymerase holoenzyme at 4 Å resolution. Science 296: 1280– 1284 [DOI] [PubMed] [Google Scholar]
- 11. Keilty S, Rosenberg M. 1987. Constitutive function of a positively regulated promoter reveals new sequences essential for activity. J. Biol. Chem. 262: 6389– 6395 [PubMed] [Google Scholar]
- 12. Bown JA, Barne KA, Minchin SD, Busby SJW. 1997. Extended −10 promoters, p 41–52 In Eckstein F, Lilley DMJ. (ed), Nucleic acids and molecular biology, vol 11. Mechanisms of transcription. Springer, Berlin, Germany [Google Scholar]
- 13. Feklistov A, Barinova N, Sevostyanova A, Heyduk E, Bass I, Vvedenskaya I, Kuznedelov K, Merkienė E, Stavrovskaya E, Klimašauskas S, Nikiforov V, Heyduk T, Severinov K, Kulbachinskiy A. 2006. A basal promoter element recognized by free RNA polymerase σ subunit determines promoter recognition by RNA polymerase holoenzyme. Mol. Cell 23: 97– 107 [DOI] [PubMed] [Google Scholar]
- 14. Haugen SP, Berkmen MB, Ross W, Gaal T, Ward C, Gourse RL. 2006. rRNA promoter regulation by nonoptimal binding of σ region 1.2: an additional recognition element for RNA polymerase. Cell 125: 1069– 1082 [DOI] [PubMed] [Google Scholar]
- 15. Ross W, Gosink KK, Salomon J, Igarashi K, Zou C, Ishihama A, Severinov K, Gourse RL. 1993. A third recognition element in bacterial promoters: DNA binding by the α subunit of RNA polymerase. Science 262: 1407– 1413 [DOI] [PubMed] [Google Scholar]
- 16. Gourse RL, Ross W, Gaal T. 2000. UPs and downs in bacterial transcription initiation: the role of the alpha subunit of RNA polymerase in promoter recognition. Mol. Microbiol. 37: 687– 695 [DOI] [PubMed] [Google Scholar]
- 17. Krásný L, Gourse RL. 2004. An alternative strategy for bacterial ribosome synthesis: Bacillus subtilis rRNA transcription regulation. EMBO J. 23: 4473– 4483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Ptashne M. 1992. A genetic switch: phage λ and higher organisms, 2nd ed. Blackwell Science, Malden, MA, and Cell Press, Cambridge, MA [Google Scholar]
- 19. Roberts JW, Shankar S, Filter JJ. 2008. RNA polymerase elongation factors. Annu. Rev. Microbiol. 62: 211– 233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Liu J, Dehbi M, Moeck G, Arhin F, Bauda P, Bergeron D, Callejo M, Ferretti V, Ha N, Kwan T, McCarty J, Srikumar R, Williams D, Wu JJ, Gros P, Pelletier J, DuBow M. 2004. Antimicrobial drug discovery through bacteriophage genomics. Nat. Biotechnol. 22: 185– 191 [DOI] [PubMed] [Google Scholar]
- 21. Dehbi M, Moeck G, Arhin FF, Bauda P, Bergeron D, Kwan T, Liu J, McCarty J, Dubow M, Pelletier J. 2009. Inhibition of transcription in Staphylococcus aureus by a primary sigma factor-binding polypeptide from phage G1. J. Bacteriol. 191: 3763– 3771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Osmundson J, Montero-Diez C, Westblade LF, Hochschild A, Darst SA. 2012. Promoter-specific transcription inhibition in Staphylococcus aureus by a phage protein. Cell 151: 1005– 1016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Lonetto MA, Rhodius V, Lamberg K, Kiley P, Busby S, Gross C. 1998. Identification of a contact site for different transcription activators in region 4 of the Escherichia coli RNA polymerase σ70 subunit. J. Mol. Biol. 284: 1353– 1365 [DOI] [PubMed] [Google Scholar]
- 24. Panaghie G, Aiyar SE, Bobb KL, Hayward RS, de Haseth PL. 2000. Aromatic amino acids in region 2.3 of Escherichia coli sigma 70 participate collectively in the formation of an RNA polymerase-promoter open complex. J. Mol. Biol. 299: 1217– 1230 [DOI] [PubMed] [Google Scholar]
- 25. Twist KA, Husnain SI, Franke JD, Jain D, Campbell EA, Nickels BE, Thomas MS, Darst SA, Westblade LF. 2011. A novel method for the production of in vivo-assembled, recombinant Escherichia coli RNA polymerase lacking the α C-terminal domain. Protein Sci. 20: 986– 995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Ross W, Thompson JF, Newlands JT, Gourse RL. 1990. E. coli Fis protein activates ribosomal RNA transcription in vitro and in vivo. EMBO J. 9: 3733– 3742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Dove SL, Hochschild A. 2004. A bacterial two-hybrid system based on transcription activation. Methods Mol. Biol. 261: 231– 246 [DOI] [PubMed] [Google Scholar]
- 28. Nickels BE. 2009. Genetic assays to define and characterize protein-protein interactions involved in gene regulation. Methods 47: 53– 62 [DOI] [PubMed] [Google Scholar]
- 29. Nickels BE, Dove SL, Murakami KS, Darst SA, Hochschild A. 2002. Protein-protein and protein-DNA interactions of σ70 region 4 involved in transcription activation by λcI. J. Mol. Biol. 324: 17– 34 [DOI] [PubMed] [Google Scholar]
- 30. Nickels BE, Roberts CW, Sun H, Roberts JW, Hochschild A. 2002. The σ70 subunit of RNA polymerase is contacted by the λQ antiterminator during early elongation. Mol. Cell 10: 611– 622 [DOI] [PubMed] [Google Scholar]
- 31. Thibodeau SA, Fang R, Joung JK. 2004. High-throughput β-galactosidase assay for bacterial cell-based reporter systems. Biotechniques 36: 410– 415 [DOI] [PubMed] [Google Scholar]
- 32. Igarashi K, Ishihama A. 1991. Bipartite functional map of the E. coli RNA polymerase α subunit: involvement of the C-terminal region in transcription activation by cAMP-CRP. Cell 65: 1015– 1022 [DOI] [PubMed] [Google Scholar]
- 33. Ross W, Gourse RL. 2005. Sequence-independent upstream DNA-αCTD interactions strongly stimulate Escherichia coli RNA polymerase-lacUV5 promoter association. Proc. Natl. Acad. Sci. U. S. A. 102: 291– 296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Davis CA, Capp MW, Record MT, Jr, Saecker RM. 2005. The effects of upstream DNA on open complex formation by Escherichia coli RNA polymerase. Proc. Natl. Acad. Sci. U. S. A. 102: 285– 290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Deighan P, Pukhrambam C, Nickels BE, Hochschild A. 2011. Initial transcribed region sequences influence the composition and functional properties of the bacterial elongation complex. Genes Dev. 25: 77– 88 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Campbell EA, Westblade LF, Darst SA. 2008. Regulation of bacterial RNA polymerase σ factor activity: a structural perspective. Curr. Opin. Microbiol. 11: 121– 127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Jain D, Nickels BE, Sun L, Hochschild A, Darst SA. 2004. Structure of a ternary transcription activation complex. Mol. Cell 13: 45– 53 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.