Abstract
Borrelia species of relapsing fever (RF) and Lyme disease (LD) lineages have linear chromosomes and both linear and circular plasmids. Unique to RF species, and little characterized to date, are large linear plasmids of ∼160 kb, or ∼10% of the genome. By a combination of Sanger and next-generation methods, we determined the sequences of large linear plasmids of two New World species: Borrelia hermsii, to completion of its 174-kb length, and B. turicatae, partially to 114 kb of its 150 kb. These sequences were then compared to corresponding sequences of the Old World species B. duttonii and B. recurrentis and to plasmid sequences of LD Borrelia species. The large plasmids were largely colinear, except for their left ends, about 27 kb of which was inverted in New World species. Approximately 60% of the B. hermsii lp174 plasmid sequence was repetitive for 6 types of sequence, and half of its open reading frames encoded hypothetical proteins not discernibly similar to proteins in the database. The central ∼25 kb of all 4 linear plasmids was syntenic for orthologous genes for plasmid maintenance or partitioning in Borrelia species. Of all the sequenced linear and circular plasmids in Borrelia species, the large plasmid's putative partition/replication genes were most similar to those of the 54-kb linear plasmids of LD species. Further evidence for shared ancestry was the observation that two of the hypothetical proteins were predicted to be structurally similar to the LD species' CspA proteins, which are encoded on the 54-kb plasmids.
INTRODUCTION
Plasmids are rich sources of variation for the adaptive evolution of bacteria (1). These extrachromosomal replicons are usually circular in structure, but linear plasmids have been noted in some bacterial taxa, including the genus Borrelia, which has linear chromosomes as well (2–4). Other bacterial genera with linear plasmids include Streptomyces and Rhodococcus species (5, 6). Some of the larger plasmids of these genera have been well characterized and shown to be occupied largely by genes involved in antibiotic production and metabolic processes (7, 8). Comparatively less is known about the linear plasmids of Borrelia species.
Borreliae are host-associated, arthropod-transmitted spirochetes. The genus includes the agents of Lyme disease (LD), such as B. burgdorferi, B. afzelii, and B. garinii, and of relapsing fever (RF), such as B. hermsii and B. turicatae of the New World and B. duttonii and B. crocidurae of the Old World. Although Borrelia species also have circular plasmids, most of their extrachromosomal content is in the form of linear replicons that range in length from 5 to ∼200 kb (2–4, 9). The individual plasmids exist in approximately the same copy number as the chromosome in the cells and, as a set with a single chromosome, constitute one of the genomes in these polyploid organisms (3, 10, 11).
The telomeres of the Borrelia linear plasmids are hairpins that form dimer junctions during replication, which are recognized and resolved by a telomere resolvase, ResT (12, 13). Plasmid compatibility of the segmented Borrelia genome involves orthologous sets of proteins, one of which, designated Pfam32, is homologous to ParA, a plasmid partitioning protein found in other bacteria (14–16). Other Borrelia plasmid-encoded proteins of these sets, such as Pfam49, appear to be unique to this genus, are widely distributed on both linear and circular replicons, and presumably are also involved in replication and compatibility. One isolate of B. hermsii had a large plasmid that existed as a circle (17), but more typically these replicons are stably maintained in their full-length linear forms.
While LD and RF species share many characteristics, such as a dependence on arthropods for transmission and most of their nutritional requirements, they differ in biology in other respects. LD Borrelia species in their vertebrate hosts cause a transient, low-grade bacteremia followed by a persistent infection in the skin and various organs (18). They are transmitted between a variety of different host species by hard ticks, and transovarial transmission is nonexistent or rare (reviewed in reference 19). In contrast, RF Borrelia species cause infections of comparatively shorter duration, but cell densities in the blood reach 107 per milliliter or higher. With the exception of B. recurrentis, which uses the human body louse as a vector, RF Borrelia species are transmitted by soft tick vectors, and transovarial transmission is common (19).
Compared to LD Borrelia species, for which genome sequences are available for 20 or more strains among 5 species (20–23), less is known about the genomes of RF Borrelia species. Reportedly complete chromosome and plasmid sequences are available for only three Old World species: B. duttonii, B. recurrentis, and B. crocidurae (24, 25). The report on the genomes for B. duttonii and B. recurrentis focused on the chromosome and some of the smaller plasmids, with limited annotation of the features of the large linear plasmids of those species (24). To date, only a few genes have been mapped to the large RF species linear plasmid, leaving much of the plasmid undefined (26–30).
Our goal was to provide for a fuller understanding of these large linear plasmids. To that end, we determined the complete sequence of the 174-kb linear plasmid of B. hermsii and most of the sequence of the 150-kb plasmid of B. turicatae, another New World species. A combination of Sanger sequencing and next-generation sequencing of genomic DNA and gap-spanning PCR products was performed for this work. These sequences were compared with the those of similarly sized linear plasmids of B. duttonii, B. recurrentis, and B. crocidurae. We found that the large plasmids of New World and Old World RF species were syntenic over most of their lengths and predominantly conserved in the central regions of their linear sequence. Comparisons with a wide variety of Borrelia plasmids revealed large portions of these large plasmids to be orthologous to genes of the ubiquitous lp54-type plasmids of LD Borrelia species.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
B. hermsii strain HS1 serotypes 7 and 33 (31) and strain DAH (32) from eastern Washington, B. turicatae strain 91E135 (also known as “Oz1”) from western Texas (33, 34), and B. parkeri strain HR1 from northern California were used. Isolates HS1 and DAH are the same strain on the basis of restriction fragment length polymorphism analysis and multilocus sequence typing (35, 36). Their ∼950-kb-long chromosomes are >99.99% identical in sequence (A. G. Barbour, unpublished data). B. turicatae and B. parkeri are closely related on the basis of sequences of several genes (36). Low-passage isolates were cultured in BSK II medium with 6% or 12% rabbit serum and grown at 34°C, unless otherwise noted (37). Total DNA of B. duttonii strain Ly was kindly provided by Sven Bergström, Umeå University, Sweden. Spirochetes were harvested by centrifugation at 9,500 × g for 20 min and washed twice with phosphate-buffered saline (PBS)–5 mM MgCl2 at pH 7.4 (PBS-Mg).
Mouse infections.
Female 5- to 6-week-old CB17 severe combined immunodeficiency (SCID) mice (Charles River Laboratories, Wilmington, MA) were inoculated intraperitoneally with 10 to 50 cells of serotype 7 of B. hermsii strain HS1. The infection was monitored by phase-contrast microscopy of a wet mount of tail vein blood. The mice were euthanized when there were ∼108 spirochetes per ml of blood, and blood was collected by cardiac puncture with syringes coated with 3% sodium citrate solution. Animal work was approved by the Institutional Animal Care and Use Committee (IACUC) at the University of California, Irvine.
Pulsed-field gel electrophoresis.
Agarose plugs of genomic DNA were prepared as described previously with modifications (17, 31). In brief, low-passage-number cells were harvested by centrifugation for 15 min at 8,000 × g at room temperature and washed with 150 mM NaCl–50 mM Tris–1 mM EDTA (TN buffer), with final resuspension in TN buffer. An equal volume of molten 2% low-melting-point SeaPlaque GTG agarose (Lonza, Rockland, ME) in TN buffer was added, giving a final concentration of 109 cells ml−1; 80 μl of the mixture was poured into each casting well (Bio-Rad, Hercules, CA). Agarose plugs were submerged in lysis solution containing 1 mg ml−1 proteinase K (Roche, Mannheim, Germany) in 50 mM Tris–50 mM EDTA–1% sodium dodecyl sulfate (SDS). Lysis was performed at 50°C for 24 h. Treated agarose blocks were washed twice in 10 mM Tris–1 mM EDTA (TE) buffer, and one-third of the plug was loaded into wells of a 0.8% agarose gel. Pulsed-field gel electrophoresis was performed in circulating 0.5× TBE buffer (45 mM Tris-borate and 1 mM EDTA) at 14°C for 21 h on a CHEF Mapper XA apparatus with parameters for separating 50- to 1,000-kb DNA molecules per the manufacturer's instructions: 6 V cm−1, included angle of 120°, and initial switch time of 50 s ramped to a final switch time of 90 s.
Restriction digestion and Southern blot analysis.
For restriction enzyme digestion, total genomic DNA was isolated using a DNeasy tissue kit per the manufacturer's instructions; approximately 1 μg of DNA was used in each restriction digestion reaction. Digested fragments were separated on a 0.7% TBE gel, and Southern blot analysis was carried out as described previously (27). Prehybridization and hybridization solutions contained 6× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate), 5× Denhardt's solution, 0.5% SDS, 100 μg ml−1 salmon sperm DNA, and 50% formamide. A probe for the 5′ end of the plasmid and corresponding to the fhbA gene (see Table S1 in the supplemental material) was made using PCR amplicons as template DNA, purified using the Zymo DNA Clean and Concentrator kit (Zymo Research, Irvine, CA), and subsequently biotinylated by random labeling using the Phototope labeling kit (New England BioLabs, Ipswich, MA) per the manufacturer's instructions. Detection of the hybridized probe was performed with the Phototope detection kit (New England BioLabs). A 60-mer oligonucleotide probe for the 3′ end of the plasmid corresponding to part of bha167 was biotinylated at the 5′ end (Sigma-Aldrich, St. Louis, MO) (see Table S1 in the supplemental material).
PCR procedures.
Genomic DNA, which was isolated using a DNeasy tissue kit (Qiagen, Germantown, MD), was used as the template for amplification using Phusion DNA polymerase and HF buffer (Finnzymes, Woburn, MA). For all PCRs, the initial denaturation step at 98°C for 3 min was followed by 35 to 40 cycles of denaturation at 98°C for 10 s, annealing for 30 s, and extension at 72°C for 2 min with a final 7-min extension at 72°C. Primer sets and annealing temperatures are listed in Table S1 in the supplemental material. For reverse transcription and quantitative PCR (qPCR), either (i) total RNA from cultured B. hermsii cells was stabilized in RNAprotect Bacteria reagent and then extracted using the Qiagen RNeasy Mini Kit with automation by the QiaCube (Qiagen) or (ii) RNA from cells in mouse blood was isolated using the Mouse RiboPure-Blood RNA isolation kit (Ambion, Austin, TX). RNA was DNase treated with the DNA-free kit (Ambion) for 30 min at 37°C according to the manufacturer's instructions. The RNA was then reverse transcribed to cDNA using random hexamers as primers and a TaqMan Gold reverse transcriptase PCR (RT-PCR) kit (Ambion) with and without reverse transcriptase at 42°C for 30 min followed by incubation at 95°C. After cDNA synthesis, real-time PCR amplification was performed using Eurogentec's qPCR Master Mix Plus (AnaSpec, Fremont, CA) and a Rotor-Gene 3000 thermal cycler (Qiagen). The real-time PCR mixture contained cDNA, 0.3 μM each primer, 0.25 μM dually labeled probe, and 0.5 volume qPCR Master Mix Plus. Samples and standards were run in duplicate. Reaction conditions were as follows: 50°C for 2 min, 95°C for 10 min, 40 cycles at 95°C for 15 s, and 60°C for 1 min. Primer and probe sets are listed in Table S1 in the supplemental material. Amplification curves were analyzed using Rotor Gene software version 6.0 (Qiagen). Standard curves using 10-fold dilutions of circular plasmid DNA with known concentrations were generated to determine the relative amounts of cDNA. Statistical analysis, including a generalized linear model (GLM) procedure with default settings, was carried out with STATA version 10 (STATA Corp., College Station, TX).
Genome sequencing of B. hermsii strain DAH and B. turicatae strain 91E135.
Sanger sequencing of shotgun libraries of 2- to 3-kb fragments of genomic DNA was carried out as described previously (31, 38). For the present study, pyrosequencing using 454 FLX technology (Roche, Branford, CT) also was performed following the manufacturer's recommended protocols. Fragment- and mate-paired libraries, each at 30× coverage, were obtained for both genomes. De novo assemblies were generated using Newbler (Roche).
The complete large-plasmid sequence for B. hermsii strain HS1 was obtained by next-generation Illumina sequencing with a final coverage of ≥10× (described in more detail below). As studies have previously shown, B. hermsii strains HS1 and DAH, both originating from the same location in eastern Washington, are very close genetically and are thought perhaps to be isolates of the same strain. Initially, the B. hermsii DAH genome was sequenced by a combination of Sanger and 454-based methods, resulting in sequences with ≥30× coverage, and assembled de novo at Rocky Mountain Laboratories (Hamilton, MT). The assembly of the large plasmid for B. hermsii HS1 was achieved by mapping against the preliminary assembly of the DAH large linear plasmid. The latter sequence contained gaps for which primers were designed for use in PCR.
Genome sequencing of B. hermsii strain HS1.
Genomic DNA was isolated using a DNeasy tissue kit (Qiagen). DNA libraries were sheared to an average size of between 150 and 200 bp and repaired to generate blunt ends, and adapters were ligated to ends. Fragments with ligated adapter sequences were then amplified and validated with an Agilent Bioanalyzer (Santa Clara, CA) and subsequently quantified by qPCR using a KAPA library quantification kit (Kapa Biosystems, Woburn, MA). Based on qPCR concentrations, libraries were normalized to 10 nM in a 10-μl volume and from there were cluster generated for data processing with an Illumina HiSeq 2000 NGS platform. The depth of coverage was ≥10×. Sequences were then mapped to a reference assembly of the B. hermsii DAH large plasmid (described below) using CLC Genomics Workbench version 5.1 (CLC bio, Aarhus, Denmark). After the final assembly of the B. hermsii HS1 large linear plasmid sequence was done, remaining gaps were manually closed by PCR followed by primer-directed Sanger sequencing of the products at Genewiz (La Jolla, CA). Sequence was confirmed by resequencing of the genome as follows. DNA extracted from culture harvest was first treated with RNase I (Fermentas, Burlington, Ontario), purified as described above, and then enzymatically sheared with an Ion Express Plus fragment library kit (Life Technologies, Grand Island, NY). Products were size selected by gel purification with the E-Gel system (Invitrogen). Templates were prepared by emulsion PCR on a Ion Torrent OneTouch apparatus (Life Technologies), and these were sequenced on an Ion Torrent Personal Genome Machine with Ion 314 chips (Life Technologies). Single reads of ∼100 bases were assembled into de novo contigs or mapped to reference sequences using CLC Genomics Workbench version 5.1.
Annotation.
Open reading frames (ORFs) of ≥150 bp were identified with GLIMMER v. 3.02 and AMIGene and then manually curated for consensus start sites (39, 40). All putative ORFs were searched against the National Center for Biotechnology Information (NCBI) nonredundant protein database using BLASTp (see Tables S2 and S3 in the supplemental material) (41). The BLAST search criteria for designating an ORF as homologous to a deduced protein of an LD Borrelia species or another organism were the following: E values of <10−5, pairwise amino acid identities of >20%, and >50% coverage of the smaller protein (42, 43). ORFs were analyzed using a collection of protein families represented by hidden Markov models (HMM) constructed from family member seed alignments (protein family A) or from unannotated and automatically generated nonredundant clusters (protein family B) with Pfam version 24.0 using HMMER3 (http://pfam.sanger.ac.uk/) (44). Putative lipoproteins with signal peptidase II cleavage sites were identified using LipoP 1.0 (http://www.cbs.dtu.dk) (45). ORF names were assigned based on designations given to LD Borrelia plasmids, with the first two letters representing the genus and species abbreviation, a third letter representing the plasmid compatibility group, and a gene number, starting from the arbitrarily designated left end of a linear plasmid.
DNA sequence analysis.
Distance phylograms were constructed using the maximum-likelihood protocol of PhyML, as implemented by the SeaView suite version 4.2.12 (46). Global alignments to identify rearrangements and colinearity within the large plasmids were performed using Progressive MAUVE 2.1.0 aligner software according to the manufacturer's instructions with a seed weight of 17 and local colinear block (LCB) weight of 500 (47). Total GC skews of large plasmids or portions of plasmids were calculated by determining (G-C)/(G+C) with settings described in the text http://www.genomicsplace.com/gc_skew/gc_skew.html. DNA matrix dot plots were constructed using the multiple-alignment program MAFFT version 6 (http://mafft.cbrc.jp), which uses a fast Fourier transform (48). Default settings were a scoring matrix of 200 PAM/κ = 2, a gap opening penalty of 1.53, and a plot threshold score of 39 (E = 8.4e−11). For a deduced amino acid sequence, virtual protein folding and then comparison of the predicted structure with the protein structure database were carried out on the UC Santa Cruz Bioinformatics server (http://compbio.soe.ucsc.edu) as described by Karplus (49). Predicted tertiary structures were visualized with KiNG software (http://kinemage.biochem.duke.edu).
Nucleotide sequence accession numbers.
The complete sequence for B. hermsii HS1 lp174 plasmid and the partial sequence for B. turicatae 91E135 lp150 plasmid were assigned GenBank accession numbers HM008709 and HM008710, respectively. Sequences for B. hermsii HS1 bha064 and bha065 and B. turicatae 91E135 bta037 and bta038 have accession numbers GQ141864 to GQ141867, respectively. Sequences used in the phylogram for Pfam32 genes were the following: B. afzelii ACA-1 lp54 (CP001247), cp32-3 (CP001237), cp26 (CP001250), and lp28-7 (CP001242); B. afzelii PKo chromosome (CP002933); B. burgdorferi B31 lp54 (AE000790), cp32-3 (AE001576), cp26 (AE000792), lp28-1 (AE000794), and chromosome (AE000783); B. duttonii Ly lp165 (CP000979), lp26 (CP000982), lp35 (CP000985), cp26 (CP000980), and chromosome (CP000976); B. garinii PBr lp54 (CP001308), cp32-10 (CP001306), cp26 (CP001305), and lp28-1 (CP001310); B. hermsii HS1 lp174 (HM008709), cp32-like (AF209440), lp53 (JN232111), lp28-2 (DQ172919), and chromosome (CP000048); B. lonestari LS-1 lp28-like (EF507519); and B. turicatae 91E135 lp150 (HM008710) and chromosome (CP000049).
RESULTS
Sizes of the large linear plasmids.
We estimated the sizes of the large linear plasmids of B. hermsii, B. parkeri, and B. turicatae by pulsed-field gel electrophoresis under conditions optimized for separation of linear duplex DNA in the range of 50 to 1,000 kb. Size standards were bacteriophage lambda concatemers, Saccharomyces cerevisiae chromosomes, and the sized linear chromosome and plasmids of the B. burgdorferi type strain B31 (16, 43). Figure 1 shows a gel of the intact linear replicons of B. hermsii, B. parkeri, and B. burgdorferi. The sizes of the large plasmids of B. hermsii and B. parkeri were estimated to be 174 kb and 150 kb, respectively. In another gel, the large linear plasmids of the closely related species B. parkeri and B. turicatae had identical migrations (data not shown). Accordingly, the B. hermsii and B. turicatae large plasmids were designated lp174 and lp150, respectively.
Fig 1.
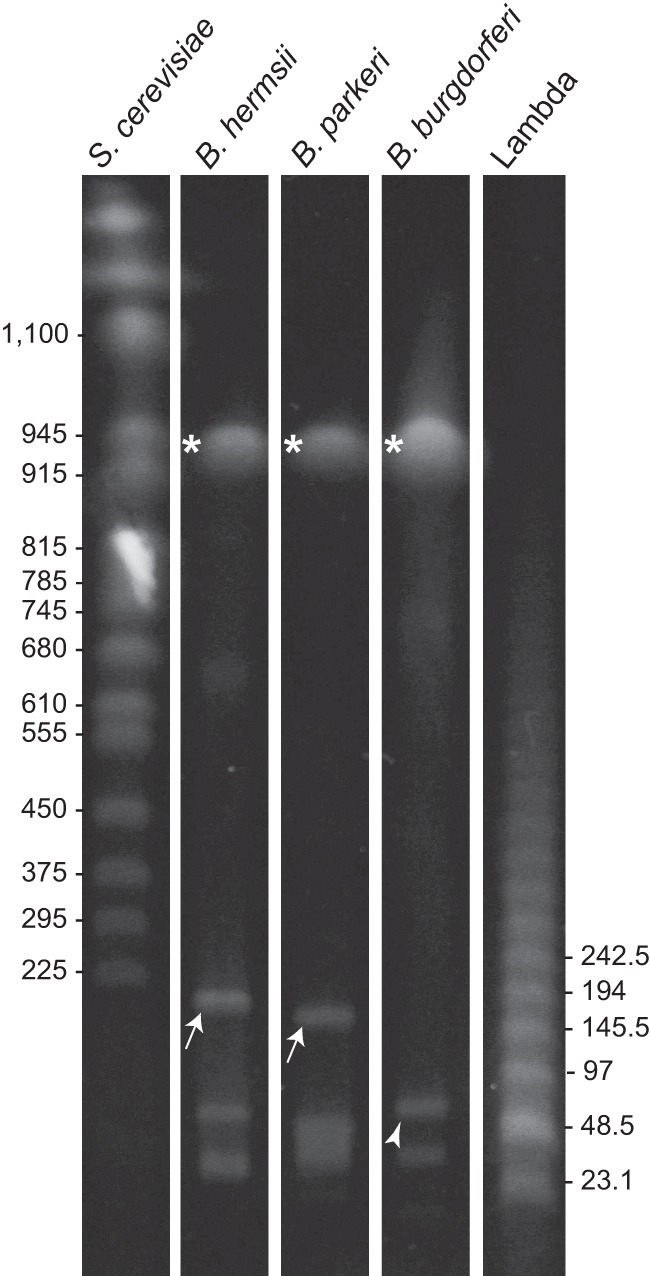
Pulsed-field gel of total DNAs of B. hermsii, B. parkeri, and B. burgdorferi. The large plasmids of B. hermsii and B. parkeri are indicated by arrows and the 54-kb linear plasmid of B. burgdorferi by an arrowhead. Asterisks indicate the linear chromosomes of all species. The sizes (in kb) of selected Saccharomyces cerevisiae chromosomes and lambda DNA concatemers are shown on the left and right, respectively.
Plasmid sequencing.
We carried out whole-genome sequencing of B. hermsii DAH and B. turicatae 91E135 by a combination of Sanger and 454 technologies, while B. hermsii HS1 was investigated with Illumina and Ion Torrent technologies. Direct Sanger sequencing of PCR products or their clones in plasmid vectors was used for gap closure, and the deduced physical map was confirmed by Southern blots of selected restriction fragments. While the sequencing of the B. hermsii large linear replicon began with strain DAH, the sequences of several other B. hermsii phylotyping genes suggested that HS1 and DAH were nearly identical or the same strain (31, 36). Our initial comparison of randomly selected homologous sequences of the large plasmids of the two isolates supported this conclusion (unpublished findings). Accordingly, large contigs accounting for a total of 163,905 bp of lp174 of strain DAH generated from Sanger and 454 technologies were used as a reference for the mapping of reads from the Illumina-based sequencing of HS1. Closure of gaps was carried out on HS1 DNA, and so that is the strain designation of record for this species' replicon described here. The completed assembly totaled 173,739 bp. The completed HS1 sequence and a draft DAH sequence were identical at 173,443 (99.8%) out of 173,739 sites compared in the alignment.
Recognizing that telomeric regions by nature of their structure may be difficult to sequence, we carried out Southern blot analysis with probes for the presumed left or right end of lp174. The probe for the left end was based on an open reading frame that was identical to the fhbA gene of B. hermsii and subsequently numbered bha008. The probe was directed against the entire bha008 gene plus an additional 100 nucleotides (nt) flanking each side. The 60-mer oligonucleotide probe for the right end was based on an open reading frame subsequently designated bha167. Table 1 gives the expected sizes of the hybridizing restriction fragments, based on the sequence assembly, and the observed sizes from the Southern blot analyses. Overall, the observed fragments with 6 different restriction enzymes were within 0.2 to 1.5% of what was predicted. Therefore, these results indicate that we either have or are very close to having the complete lp174 plasmid for HS1.
Table 1.
Expected and observed sizes of hybridizing restriction fragments
| Enzyme |
bha008 probe (left end) |
bha167 probe (right end) |
||||
|---|---|---|---|---|---|---|
| Fragment size (bp) |
End fragment | Fragment size (bp) |
End fragment | |||
| Expected | Observed | Expected | Observed | |||
| BamHI | 7,993 | 8,850 | Yes | NAa | NA | NA |
| EcoRI | 3,213 | 3,208 | No | 8,943 | 9,117 | Yes |
| EcoRV | 6,140 | 6,538 | No | 1,695 | 1,668 | Yes |
| KpnI | 11,237 | 11,037 | No | 10,802 | 10,673 | Yes |
| PstI | 10,698 | 10,218 | Yes | 10,098 | 10,218 | Yes |
| XbaI | 4,284 | 4,360 | No | 3,221 | 3,143 | Yes |
| Total size (% difference) | 43,565 | 44,211 (1.5) | 34,759 | 34,819 (0.2) | ||
NA, not applicable.
The base count for the final assembly of the B. turicatae lp150 plasmid was 114,195 bp. As detailed below, the estimated 35 kb of missing sequence was primarily at the right end of the plasmid. However, it is possible that sequence at the left end of the lp150 was also unaccounted for here. The lists of ORFs that were identified for the entire B. hermsii lp174 and for most of B. turicatae lp150 are given in Tables S2 and S3, respectively, in the supplemental material.
General features.
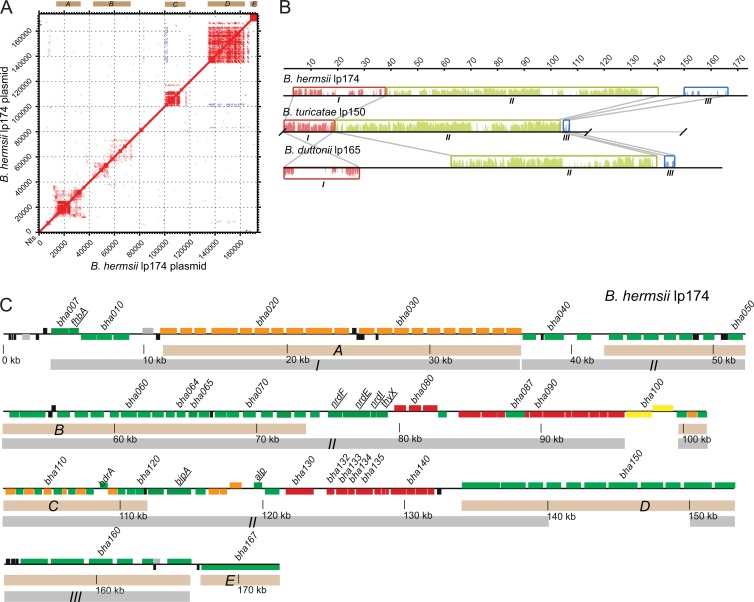
The locations and extent of repetitive DNA are indicated in the dot plot matrix of the B. hermsii lp174 sequence plotted directly against itself (Fig. 2A). Approximately 40% of the plasmid's content is repetitive DNA, and this is located mainly in regions we designated A, B, C, D, and E. Inverted repeats were infrequent and short. Region D comprises tandemly arrayed paralogous ORFs of various lengths and of unknown function. Database searches indicated that similar genes were found only in RF Borrelia species and then in various numbers. Region E at the right end is a 5,514-nt-long open reading frame, which is homologous to a partial, unmapped ORF of B. crocidurae (accession number AFI32145), and it contains multiple copies of an ∼400-nt-long sequence with 70 to 80% identity between copies. Regions A to E of B. hermsii lp174, which contain repetitive sequence, correspond to similar locational regions of repetitive sequence in the large plasmid of B. turicatae (see Fig. S1A in the supplemental material) and to the large plasmid of B. duttonii (see Fig. S1B in the supplemental material). Notably, repetitive regions A, B, C, and D appear to be locationally conserved in all three species, while region E appears to be variable relative to the species origin of the linear plasmid. Therefore, there appears to be a level of conservation of repetitive domains across the large linear plasmid from different species of RF Borrelia.
Fig 2.
Organization of repetitive DNA, colinear sequence, and individual open reading frames of relapsing fever Borrelia species. (A) Dot matrix plot of B. hermsii lp174 versus itself. Lines shown are similarities on direct strands (red) or on opposite strands (blue). Sequences were aligned using the default settings of the MAFFT alignment program. Regions of repetitive DNA are labeled A to E above the plot. (B) Schematic picture of alignment of the large linear plasmid sequences of B. hermsii, B. turicatae, and B. duttonii. Regions of contiguously homologous sequence are represented by colored local colinear blocks (LCBs I to III). An LCB is defined as a conserved segment of sequence that appears to be internally free from genome rearrangements relative to the reference sequence. Gray lines bound the orthologous LCBs. Similarity profiles (vertical lines within the LCBs) whose heights correspond to average level of sequence similarity are shown in each LCB. LCBs below the center line represent sequence in the opposite orientation relative to the reference sequence, that of B. hermsii lp174. Nucleotide positions (in kb) are indicated above the alignment. (C) Physical map of B. hermsii lp174 (accession no. HM008709) oriented left to right. Genes on the positive strand are indicated by boxes above the center line, and those on the negative strand are below the centerline. Beige shaded boxes (A to E) correspond to regions of repetitive DNA shown in panel A. Gray shaded boxes (I to III) correspond to LCBs shown in panel B. Selected genes previously mapped to the large plasmids are underlined: fhbA (bha008), nrdF (bha075), nrdE (bha076), nrdI (bha077), thyX (bha078), bipA (bha123), and alp (bha0128). Newly identified genes discussed in the text are also indicated: bha064, bha065, and the plasmid replication and partition loci (bha132 to -135). Colored ORFs represent homologs to the LD species P35 antigen (region A) or to PFam99 (orange), B. burgdorferi lp54 (red), B. burgdorferi cp26 (yellow), ORFs unique and conserved to RF species (green), or ORFs unique to a single species (black). Locus tags or gene names are indicated above the corresponding ORF.
The large plasmids of B. hermsii, B. turicatae, and B. duttonii have an approximate G+C percentage of 30%, which is within the range of G+C percentage for both the chromosome and most plasmids of Borrelia species. The coding densities of the large plasmids are at ∼75%, which is less than the coding densities of the chromosomes of these species (24, 25) (accession no. CP000048 and CP000049). This lower level of coding density is, however, similar to the coding densities of other linear plasmids of Borrelia species (22, 31, 43). Only 5 to 10% of the deduced ORFs of the 3 large plasmids defined here had identifiable homologs to proteins from organisms outside the Borrelia genus (see Tables S2 and S3 in the supplemental material). These ORFs encode products involved with nucleotide biosynthesis (thyX, nrdE, nrdF, and nrdI) (38, 50) and putative bacterial plasmid replication and partitioning (paralogous family 32 [PFam32], PFam49, PFam50, and PFam57) (16, 43). The remaining protein sequences of ORFs were found to be homologous to proteins found in LD Borrelia species as well as in RF species. In addition, homologs that were only present in RF Borrelia species were also found, and a few short ORFs that appeared to be unique to B. hermsii or B. turicatae were found (black ORFs in Fig. 2C and in Fig. S2 in the supplemental material).
In order to analyze general plasmid synteny or look for the presence of large regions of conservation or regions of inversion, an alignment and Mauve plot analysis was performed, and these results are shown in Fig. 2B. Three large colinear blocks (I, II, and III) were discovered between the B. hermsii, B. turicatae, and B. duttonii plasmids. These three colinear blocks constitute more than half the total length of each of these plasmids. Block I contains genes for two possible virulence factors of RF Borrelia species: fhbA, which encodes a factor H-binding protein (26), and bha007 and its orthologs, which encode proteins that bind complement pathway components or fibronectin (28, 51) (E. R. G. Lewis, R. A. Marcsisin, S. A. Campeau Miller, A. Phillips, D. P. AuCoin, and A. G. Barbour, submitted for publication). The longer length of B. hermsii block I relative to B. turicatae block I is attributable to a higher number of repeated “P35” (PFam54) homologs, a family of lipoproteins in LD Borrelia species (52). “P35”-like coding sequences were even fewer in the B. duttonii, as evidenced by the lack of homology (red lines) within block I (Fig. 2B) or B. recurrentis large plasmids (data not shown), and were not discernible at all in the B. crocidurae large-plasmid sequence (data not shown).
In the published sequence for the large plasmid of B. duttonii (NC011247) block I is inverted, involving approximately 27 kb of sequence, with respect to its orientation in the New World species (Fig. 2B). We noted a similar inversion for the block I region in B. crocidurae (NC017778). To verify that this inversion was not the result of an assembly error, we carried out PCR on total DNAs isolated from both B. hermsii and B. duttonii, using 6 pairs of primers that spanned the junction of blocks I and II for each of the two possible orientations. Each of the six PCRs yielded appropriately sized fragments that support the current assembly (see Table S4 in the supplemental material). Therefore, these results suggest that there was an inversion of this region when this location in New World RF species B. hermsii and B. turicatae is compared to that in Old World RF species B. duttonii and B. crocidurae.
Comparison with LD Borrelia plasmids.
Figure 2C shows a graphical representation of the 167 ORFs, including three pseudogenes, identified in the B. hermsii lp174 plasmid. A comparable map of the 108 ORFs in B. turicatae lp150 is provided in Fig. S2 in the supplemental material. These maps distinguish ORFs that are unique to B. hermsii and at least one other RF Borrelia species (green) from those that appear to be at least genus-wide (orange, red, or yellow) and those unique to a specific species (black) in their distribution. Among the genus-wide group, we further distinguish between ORFs whose closest orthologs are in the lp54 linear plasmid of B. burgdorferi and related species and ORFs in common with genes carried on other B. burgdorferi plasmids.
Of the 76 ORFs described for the lp54 plasmid (AE000790), 28 of these ORFs were found to have orthologs on the RF species large plasmids. These 28 orthologs appear predominantly in two extended regions of B. hermsii lp174, namely, ca. kb 79 to kb 96 and kb 121 to kb 132 (Fig. 2C), which also are largely syntenic with the lp54 linear plasmid of B. burgdorferi. In the region from kb 79 to 96, there are orthologs of the following ORFs of lp54: bba38 to -43, bba45 to -49, and bba51 to -52. In lp54, bba50 resides between the genes bba49 and bba51; however, in B. hermsii lp174 a 1,424-bp-long ORF, designated bha087, that is unique to RF species resides in this location. In addition, for the large linear plasmids of B. turicatae and B. duttonii, genes homologous to bha087 designated bta062 and bdu1086, respectively, both of which are unique to RF Borrelia species, also reside in this location. Further to the right and present in B. hermsii lp174, but not in the large plasmids of B. turicatae, B. duttonii, B. recurrentis, or B. crocidurae, are bha099 to -102 (yellow ORFs in Fig. 2C), which are orthologs of 4 genes of B. burgdorferi circular plasmid 26 (cp26). These genes are three chitobiose transport genes (chbABC) and a gene for an integrin-binding protein (53–55). These results suggest that lp174 contains orthologs that reside on LD species cp26 and lp54.
Continuing further downstream on the HS1 plasmid, we encountered the repetitive sequence region C (Fig. 2A), which resides from ca. kb 100 to kb 112, and the genes bha103 to bha129 (Fig. 2C). There were repeats of two alternating sequences, one of which was unique to RF species (bha107, -109, -111, -113, -115, -118, -120, and -126) and the other of which is homologous to the PFam99 family of B. burgdorferi (bha104, -106, -108, -110, -114, -117, -119, and -125). In the midst of these repeats is the bdrA gene (bha116) of B. hermsii (56). Paralogous genes were also arranged as tandem repeats in repetitive sequence region C of B. turicatae lp150 (see Fig. S2 in the supplemental material) but not in the large plasmids of the Old World species. This repetitive sequence region C in B. hermsii (kb 112 to kb 121) and B. turicatae (kb 71 to kb 92) was followed by a series of ORFs that were either unique to RF species, such as the alp gene (bha128 or bta096) for the arthropod-associated lipoprotein (30), or common to both RF and LD groups, including 3 ORFs (bha125 to -127 or bta092 to -094) with homology to ORFs restricted to the lp38 plasmid among B. burgdorferi plasmids.
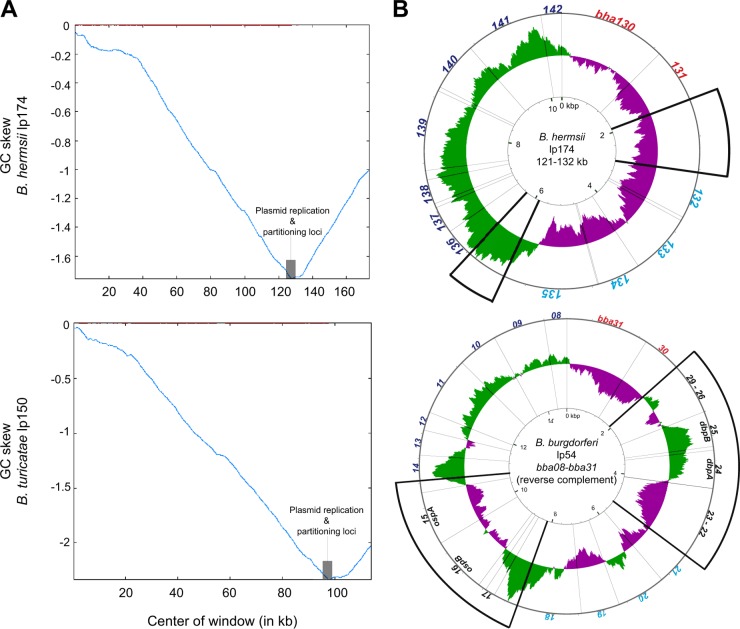
From kb 121 to kb 132 of B. hermsii lp174, there is sequence that is partially syntenic with lp54. Included here are four genes, bha132 to -135 (encoding PFam32, PFam49, PFam50, and PFam57), constituting a plasmid maintenance locus (16, 43). All Borrelia plasmids characterized to date carry a subset of genes from PFam32, -49, -50, and either -57 or -62, where PFam32 is homologous to the ParA protein of other bacteria (57). Skew shifting allows identification of the origin of replication in an otherwise cryptic plasmid. Total GC skew calculations and plots for B. hermsii lp174 and B. turicatae lp150 show that the skew shifted at the site where this plasmid maintenance gene set is located and, more specifically, within the PFam57 (bha134) gene (Fig. 3A), suggesting that this locus may be the origin of replication for the large plasmids (58).
Fig 3.
GC skew analysis of Borrelia plasmid sequences. (A) GC skew diagrams of B. hermsii lp174 and B. turicatae partial lp150 sequences. Shaded gray boxes indicate the plasmid replication and partitioning loci. Red lines at the top indicate the window and the point at which the skew shifts. Total GC skew was calculated with a sliding window size of 500 bp and a step size of 50 bp. (B) Circular G-C skew representation of orthologous regions from B. hermsii lp174 and B. burgdorferi lp54. Deviation from the average GC skew of the entire sequence is represented by green lines (above average) or purple lines (below average). Radiating lines originating from the center circle indicate ORF boundaries. Orthologous ORFs are noted by the same font colors (red, cyan, or blue), with thick diagonal lines enclosing regions of nonhomologous sequence (ORFs indicated in black). GC skew was calculated with a 500-bp window and a step size of 1 bp (75).
Partition locus genes of lp54 and RF species large linear plasmids share an origin.
The 4 gene products in the partition locus have 46 to 80% amino acid identity with their counterparts on the lp54 plasmid. Phylogenetic analysis of Pfam32 genes from LD species linear and circular plasmids and the linear chromosomes of both RF and LD Borrelia species indicates that the orthologs of RF species large-plasmid homologs are more closely related to B. burgdorferi lp54 genes than they are to those of any other replicon, including other replicons in their own species (Fig. 4). The same phylogenetic relationships were inferred from the sequences of other genes in the plasmid maintenance locus (data not shown). Therefore, plasmid maintenance genes show sequence bias toward the replicon structure that encodes them across species, rather than across replicons within a single species or isolate.
Fig 4.
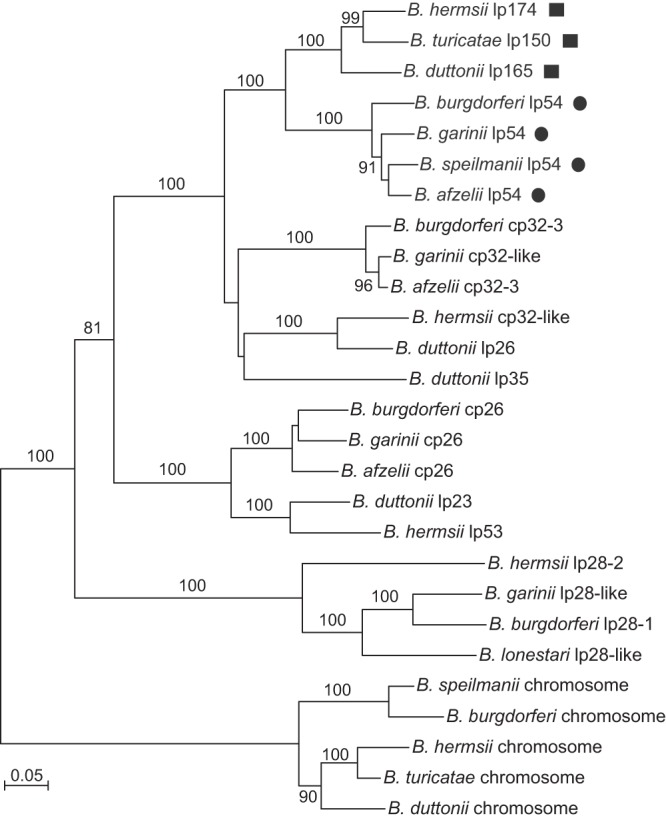
Phylogram of codon-aligned nucleotide sequences for selected PFam32 orthologs from different Borrelia replicons. Genes from the RF species large plasmids (squares) and LD species lp54 plasmids (circles) are indicated. The replicons and accession numbers used for each species and strain are provided in Materials and Methods. Nodes with bootstrap values of >70% support by neighbor-joining distance criteria from 10,000 replicates are indicated. The scale bar represents nucleotide substitutions per site.
We noticed that in these regions of B. hermsii lp174 (kb 122 to kb 133) and B. turicatae lp150 (kb 92 to kb 103) that had partial synteny to lp54, two sets of lp54 ORFs, namely, bba22 to -29 and bba14 to -17, were absent in RF Borrelia species and without replacements by other ORFs. Were these ORFs lost from the lineage leading to B. hermsii and other RF species but not from the LD species lineage, or were they gained by the LD Borrelia lineage after the split from a common ancestor? Taking advantage of the plasmid's sharp shift in GC skew in the midst of the plasmid maintenance locus of lp174 (Fig. 3A), we compared the GC skew patterns of these syntenic regions in each replicon. We reasoned that if all or some of the ORFs unique to LD species were relatively recent acquisitions, this might be apparent in the GC skew pattern (59, 60). Indeed, as analysis of kb 121 to 132 of B. hermsii lp174 and the corresponding span of ORFs bba08 to -31 of B. burgdorferi lp54 reveals (Fig. 3B), the GC skew around the operons for outer surface proteins A and B (bba15 and bba16) and for decorin-binding proteins A and B (bba24 and bba25) is opposite to that of the surrounding sequence that is common to RF and LD species. This is an indication that these operons, which are unique to LD species, were likely acquired horizontally in the LD clade after the descent from an ancestral taxon, as appears to be the case for other operons (50, 61).
Regions unique to RF Borrelia species.
Part of local colinear block II, specifically from ca. kb 36 to 75 of B. hermsii lp174 (Fig. 2C), is just left or upstream of a functional pyrimidine biosynthesis locus containing thyX (bha078), which encodes a thymidylate synthase (50), and the ribonucleotide reductase subunit cluster nrdFEI (bha075 to -077) (38, 50). The majority of the coding sequences in this region were on the minus strand (Fig. 2C), and the polypeptide sizes range from 200 to 300 residues. The proteins in this region appeared to be paralogs with considerable divergence between members in the same species and between orthologs in other colinear block-containing species (data not shown). The exceptions were a pair of ORFs, bha064 and bha065 in B. hermsii (Fig. 2C), bta037 and bta038 in B. turicatae (see Fig. S3 in the supplemental material), and bdu1066 and bdu1067 in B. duttonii (not shown), as well as their counterparts in B. recurrentis and B. crocidurae (not shown). BHA064 and BHA065 likely are paralogs duplicated prior to the divergence of RF and LD lineages, with greater identity between corresponding loci of the three species than between pair members of the same species (see Fig. S4 in the supplemental material). For example, BHA065 of B. hermsii and BTA039 of B. turicatae were 78% identical in protein sequence, while the identity between BHA065 and BHA064 was only 67%.
Expression and characterization of bha064.
There are near-consensus ribosomal binding sequences (GGAGA) positioned 9 nt in front of the start codons for both bha64 and bha65 with an intergenic distance of 174 bp. Corresponding distances in other RF species were 228 bp between bta037 and bta038 of B. turicatae and 170 bp between bdu1066 and bdu1067 of B. duttonii. Accordingly, we assessed the transcription of bha064 by quantitative reverse transcriptase PCR with 5 replicates each for cells growing in culture medium at different temperatures (23 versus 37°C) and pHs (7.6 versus 8.0) or in the blood of mice. Results of each replicate were normalized by the values for copies of the constitutively and abundantly expressed flaB gene for the same replicate. Expression for bha064 was significantly higher under conditions that mimicked the tick environment, i.e., 23°C and pH 8.0 (62, 63), than with culture medium at 37°C at both pHs or in mouse blood (Fig. 5). By linear regression analysis, the combination of temperature and pH accounted for the expression values in vitro with a coefficient of 4.1 (95% confidence interval, 3.3 to 4.9) and a coefficient of determination (r2) of 0.77 (P < 0.0001). Normalized expression of bha064 was also higher in vitro at 23°C at pH 8.0 (5.40 [3.24 to 7.55]) than in mouse blood (2.02 [1.46 to 2.59]) (Mann-Whitney U test, P = 0.009). These findings suggest that bha064 is potentially preferentially expressed in the tick vector relative to the mammalian host environment, but this remains to be determined.
Fig 5.
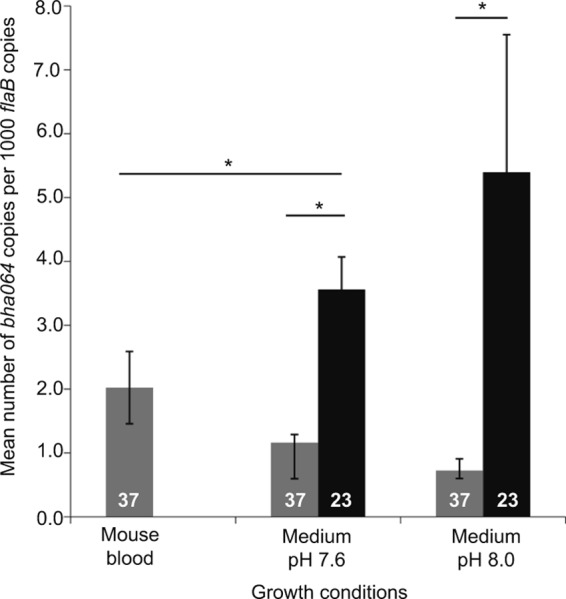
Gene expression analysis of bha064. Quantitative real-time PCR analysis of bha064 cDNA copies normalized to copies of the constitutively expressed flaB for B. hermsii grown at 37°C (gray bars) in the blood of immunodeficient mice or in culture or grown at 23°C (black bars) in culture is shown. Ninety-five percent confidence intervals are shown for each condition. Asterisks above lines indicate significant differences as measured by the t test (P < 0.001).
Another clue about the possible function of BHA064 and BHA065 was provided by prediction of their tertiary structures through virtual protein folding and threading of their amino acid sequences against known protein structures in the database. Predictions for BHA064 and BHA065 of B. hermsii, as well as their orthologs in B. turicatae and B. duttonii, had the same top alignment hit, Protein Data Bank (PDB) structure 1W33. The E value scores for this call were <10−2, with the next best scores >1. 1W33 is the reference for the crystal structure of the B. burgdorferi CspA protein, formerly known as the complement regulator-acquiring surface protein or CRASP-1 (64) (see Fig. S5 and S6 in the supplemental material). This largely α-helical protein is encoded by gene bba68 on B. burgdorferi lp54 and is a member of the PFam54 family. The closest relative to BBA68 (CspA) is BBA69, encoded by a neighboring gene on lp54, with 59% identity. Thus, LD species have tandem pairs of paralogs such as BHA064/065 of B. hermsii, BTA037/038 of B. turicatae, and BDU1066/1067 of B. duttonii.
DISCUSSION
The large linear plasmids of five RF Borrelia species are fairly homologous, as demonstrated by (i) the extensive regions of colinearity between plasmids from different species, (ii) the possession of common biosynthesis genes, such as those for ribonucleotide reductase, not present elsewhere in the genome, and (iii) the relatedness of their plasmid maintenance genes. Although there is a set of core genes shared among the large plasmids and an overall similarity in architecture, there is also considerable intermolecular variability, especially at the plasmids' ends, as was noted regarding the linear replicons of LD Borrelia species (16). Unique gene content was present in the large linear plasmid of one species but not in the large linear plasmids of other species. An example is that on B. hermsii lp174, there is a cluster of large linear plasmid ORFs that are orthologous to genes associated with cp26 of B. burgdorferi (Fig. 2C). However, such instances were few. Differences in lengths of the plasmids were largely attributable to variation in the number of repeats within the extensive regions of repetitive DNA. In some cases, such as repetitive region A with its tandemly arrayed ORFs of “P35”-like sequences (Fig. 2), the repeated DNA was homologous to sequences found in LD Borrelia species lp54, lp28-4, and lp38 replicons (accession numbers AE000790, AE000789, and AE000787). However, in other regions, such as region D, the repetitive ORFs appeared to be unique to RF species.
The length of the shortest of the large linear plasmids was reported as 124 kb, for B. recurrentis (24). However, there is evidence that the deposited plasmid sequence for B. recurrentis was not complete. Grosskinsky et al. demonstrated that B. recurrentis has a gene that is an ortholog to the cihC gene of B. duttonii, bha007 of B. hermsii, and bta001 of B. turicatae, all of which are near the left termini of their respective plasmids (28). This gene and adjoining genes, such as fhbA and the clusters of “P35”-like sequences, were not included in the sequence of “lp124” of B. recurrentis. In one study, a portion of the left end of the large plasmid, including the fhbA gene, was spontaneously lost in a few strains of B. hermsii and in one strain of B. turicatae after 100 serial passes in vitro, yet the strains remained viable in culture (54). It is possible that this phenomenon occurred in the B. recurrentis strain used for sequencing; however, pulsed-field gel electrophoresis showed that the large plasmid of B. recurrentis was about the same size as the B. duttonii plasmid, at 160 to 170 kb (24, 28). Therefore, the publically available B. recurrentis linear plasmid is most likely missing 36 to 46 kb of sequence.
A prominent feature of the large linear plasmids' sequences was the ∼60% proportion of repetitive DNA, much of which was colinear or locationally similar between replicons. Some of the repeated ORFs, such as the “P35”-like coding sequences, were homologous to sequences in LD Borrelia species, while others, such as the repeats in region D, appeared to be unique to RF species. It is possible that amplification and diversification of these proteins could have occurred as the spirochetes adapted to different hosts, arthropod or vertebrate, and their associated immune or environmental selective pressures. Gene duplications in other pathogens can result in an increase of functional genes used, for example, to adapt to or exploit novel environmental niches or function in a gene dosage effect (65, 66).
Our inference about a common ancestry for the large linear plasmids of RF Borrelia species was anticipated by prior findings of the nrdEFI genes on large plasmids across species (50). A less expected finding was the extent of shared heritage for the large linear plasmids of RF Borrelia species and in particular the lp54-type linear plasmids of LD species. Lescot et al. observed syntenic regions between B. duttonii lp165 and B. burgdorferi lp54, but orthologous genes between the two plasmids were not described (24). Totaling ∼22 kb, two regions containing lp54 orthologs were found in similar locations on each of the RF species large plasmids and the more conserved interior portions of these plasmids. All LD Borrelia species and strains whose genome sequences are public have had an lp54-like plasmid of similar size. Recognizing the phylogenetic evidence that we present here regarding the PFam32 genes and the 3 other genes of the plasmid maintenance locus, which are borne by the large plasmids and those of lp54-type plasmids, we propose assigning the RF species large plasmids to the type A compatibility group for Borrelia plasmids (67, 68) and, accordingly, using “a” as the third letter after “bh,” “bt,” etc., for coding sequence designations.
The lp54 plasmids bear genes whose expression is regulated during the infectious cycle and that in their respective presence or absence distinguish LD species from RF species. These include OspA (bba15) and OspB (bba16) (69, 70) and DbpA (bba24) and DbpB (bba25) (71). GC skew analysis indicates that the ospAB and dbpAB operons were acquired horizontally by the LD lineage after the putative split from the lineage leading to RF species. Can we identify lp174 genes that distinguish RF species from LD species? Three examples are fhbA, bipA, and alp (26, 29, 30). Two other lp174 genes that first appeared to be unique to RF species on the basis of standard blast search criteria were bha064 and bha065. However, a search for structural similarities revealed what may be homology of these proteins to BBA68, the CspA (formerly CRASP-1) protein and a PFam54 family member. A different “CRASP-1” protein was named in B. hermsii (72), but this protein is the same as FhbA, encoded by bha008. Surrounding bha064 and bha065 are other ORFs that are more divergent in sequence from this pair but possibly are paralogs and, as such, additional candidates for virulence determinants. To the right and downstream of bha064 and bha065 is the thyX gene, bha076 (Fig. 2C). A thyX pseudogene occupies a position similar to that of the PFam54 genes on lp54 (50). If one also includes these in the list of shared genes, then an even larger proportion of lp174 has common ancestry with the lp54-type plasmids.
While the functions of BHA064 and BHA065 in B. hermsii and orthologous ORFs in other species remain to be determined, the higher expression of bha064 under in vitro conditions that mimic the environment in the arthropod suggests to us a vector-associated role in the RF species life cycle. If BHA064, as well as its orthologs in four other Borrelia species, is homologous as well as structurally similar to CspA of LD species, how does the experimental expression data for bha064 compare with a proposed function of CspA, i.e., providing serum resistance (73)? Somewhat at odds with the serum resistance claim is the report that CspA expression is increased in the tick during feeding but rapidly decreases once the spirochetes are transmitted to the mammal (74). Whatever activity BHA064 and the other ORFs turn out to actually have, the present study provides a sharper-resolution view of regions of largely unknown function on these unique large linear plasmids and provides for insights about their evolution.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by NIH grant AI24424 and the intramural program of the National Institute of Allergy and Infectious Diseases.
We thank Sherwood Casjens for advice and Fong Hue and Renee Marcsisin for technical assistance.
Footnotes
Published ahead of print 7 June 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JB.00347-13.
REFERENCES
- 1.Levin BR, Bergstrom CT. 2000. Bacteria are different: observations, interpretations, speculations, and opinions about the mechanisms of adaptive evolution in prokaryotes. Proc. Natl. Acad. Sci. U. S. A. 97:6981–6985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ferdows MS, Barbour AG. 1989. Megabase-sized linear DNA in the bacterium Borrelia burgdorferi, the Lyme disease agent. Proc. Natl. Acad. Sci. U. S. A. 86:5969–5973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Casjens S, Huang WM. 1993. Linear chromosomal physical and genetic map of Borrelia burgdorferi, the Lyme disease agent. Mol. Microbiol. 8:967–980 [DOI] [PubMed] [Google Scholar]
- 4.Plasterk RH, Simon MI, Barbour AG. 1985. Transposition of structural genes to an expression sequence on a linear plasmid causes antigenic variation in the bacterium Borrelia hermsii. Nature 318:257–263 [DOI] [PubMed] [Google Scholar]
- 5.Crespi M, Messens E, Caplan AB, van Montagu M, Desomer J. 1992. Fasciation induction by the phytopathogen Rhodococcus fascians depends upon a linear plasmid encoding a cytokinin synthase gene. EMBO J. 11:795–804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hirochika H, Nakamura K, Sakaguchi K. 1984. A linear DNA plasmid from Streptomyces rochei with an inverted terminal repetition of 614 base pairs. EMBO J. 3:761–766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Medema MH, Trefzer A, Kovalchuk A, van den Berg M, Muller U, Heijne W, Wu L, Alam MT, Ronning CM, Nierman WC, Bovenberg RA, Breitling R, Takano E. 2010. The sequence of a 1.8-mb bacterial linear plasmid reveals a rich evolutionary reservoir of secondary metabolic pathways. Genome Biol. Evol. 2:212–224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Francis I, De Keyser A, De Backer P, Simon-Mateo C, Kalkus J, Pertry I, Ardiles-Diaz W, De Rycke R, Vandeputte OM, El Jaziri M, Holsters M, Vereecke D. 2012. pFiD188, the linear virulence plasmid of Rhodococcus fascians D188. Mol. Plant Microbe Int. 25:637–647 [DOI] [PubMed] [Google Scholar]
- 9.Barbour AG. 1988. Plasmid analysis of Borrelia burgdorferi, the Lyme disease agent. J. Clin. Microbiol. 26:475–478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hinnebusch J, Barbour AG. 1992. Linear- and circular-plasmid copy numbers in Borrelia burgdorferi. J. Bacteriol. 174:5251–5257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kitten T, Barbour AG. 1992. The relapsing fever agent Borrelia hermsii has multiple copies of its chromosome and linear plasmids. Genetics 132:311–324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hinnebusch J, Barbour AG. 1991. Linear plasmids of Borrelia burgdorferi have a telomeric structure and sequence similar to those of a eukaryotic virus. J. Bacteriol. 173:7233–7239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kobryn K, Chaconas G. 2002. ResT, a telomere resolvase encoded by the Lyme disease spirochete. Mol. Cell 9:195–201 [DOI] [PubMed] [Google Scholar]
- 14.Barbour AG, Carter CJ, Bundoc V, Hinnebusch J. 1996. The nucleotide sequence of a linear plasmid of Borrelia burgdorferi reveals similarities to those of circular plasmids of other prokaryotes. J. Bacteriol. 178:6635–6639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zuckert WR, Meyer J. 1996. Circular and linear plasmids of Lyme disease spirochetes have extensive homology: characterization of a repeated DNA element. J. Bacteriol. 178:2287–2298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Casjens S, Palmer N, van Vugt R, Huang WM, Stevenson B, Rosa P, Lathigra R, Sutton G, Peterson J, Dodson RJ, Haft D, Hickey E, Gwinn M, White O, Fraser CM. 2000. A bacterial genome in flux: the twelve linear and nine circular extrachromosomal DNAs in an infectious isolate of the Lyme disease spirochete Borrelia burgdorferi. Mol. Microbiol. 35:490–516 [DOI] [PubMed] [Google Scholar]
- 17.Ferdows MS, Serwer P, Griess GA, Norris SJ, Barbour AG. 1996. Conversion of a linear to a circular plasmid in the relapsing fever agent Borrelia hermsii. J. Bacteriol. 178:793–800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Steere AC, Coburn J, Glickstein L. 2005. Lyme borreliosis, p 176–206 In Goodman JL, Dennis DT, Sonenshine DE. (ed), Tick-borne diseases of humans. ASM Press, Washington, DC [Google Scholar]
- 19.Piesman J, Schwan T. 2010. Ecology of borreliae and their arthropod vectors, p 251–278 In Samuels DS, Radolf JD. (ed). Borrelia molecular biology, host interaction, and pathogenesis. Caister Academic Press, Norfolk, United Kingdom [Google Scholar]
- 20.Casjens SR, Fraser-Liggett CM, Mongodin EF, Qiu WG, Dunn JJ, Luft BJ, Schutzer SE. 2011. Whole genome sequence of an unusual Borrelia burgdorferi sensu lato isolate. J. Bacteriol. 193:1489–1490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Casjens SR, Mongodin EF, Qiu WG, Dunn JJ, Luft BJ, Fraser-Liggett CM, Schutzer SE. 2011. Whole-genome sequences of two Borrelia afzelii and two Borrelia garinii Lyme disease agent isolates. J. Bacteriol. 193:6995–6996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schutzer SE, Fraser-Liggett CM, Casjens SR, Qiu WG, Dunn JJ, Mongodin EF, Luft BJ. 2011. Whole-genome sequences of thirteen isolates of Borrelia burgdorferi. J. Bacteriol. 193:1018–1020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schutzer SE, Fraser-Liggett CM, Qiu WG, Kraiczy P, Mongodin EF, Dunn JJ, Luft BJ, Casjens SR. 2012. Whole-genome sequences of Borrelia bissettii, Borrelia valaisiana, and Borrelia spielmanii. J. Bacteriol. 194:545–546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lescot M, Audic S, Robert C, Nguyen TT, Blanc G, Cutler SJ, Wincker P, Couloux A, Claverie JM, Raoult D, Drancourt M. 2008. The genome of Borrelia recurrentis, the agent of deadly louse-borne relapsing fever, is a degraded subset of tick-borne Borrelia duttonii. PLoS Genet. 4:e1000185. 10.1371/journal.pgen.1000185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Elbir H, Gimenez G, Robert C, Bergstrom S, Cutler S, Raoult D, Drancourt M. 2012. Complete genome sequence of Borrelia crocidurae. J. Bacteriol. 194:3723–3724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hovis KM, McDowell JV, Griffin L, Marconi RT. 2004. Identification and characterization of a linear-plasmid-encoded factor H-binding protein (FhbA) of the relapsing fever spirochete Borrelia hermsii. J. Bacteriol. 186:2612–2618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhong J, Barbour AG. 2004. Cross-species hybridization of a Borrelia burgdorferi DNA array reveals infection- and culture-associated genes of the unsequenced genome of the relapsing fever agent Borrelia hermsii. Mol. Microbiol. 51:729–748 [DOI] [PubMed] [Google Scholar]
- 28.Grosskinsky S, Schott M, Brenner C, Cutler SJ, Simon MM, Wallich R. 2010. Human complement regulators C4b-binding protein and C1 esterase inhibitor interact with a novel outer surface protein of Borrelia recurrentis. PLoS Negl. Trop. Dis. 4:e698. 10.1371/journal.pntd.0000698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lopez JE, Schrumpf ME, Nagarajan V, Raffel SJ, McCoy BN, Schwan TG. 2010. A novel surface antigen of relapsing fever spirochetes can discriminate between relapsing fever and Lyme borreliosis. Clin. Vaccine Immunol. 17:564–571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Marcsisin RA, Campeau SA, Lopez JE, Barbour AG. 2012. Alp, an arthropod-associated outer membrane protein of Borrelia species that cause relapsing fever. Infect. Immun. 80:1881–1890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dai Q, Restrepo BI, Porcella SF, Raffel SJ, Schwan TG, Barbour AG. 2006. Antigenic variation by Borrelia hermsii occurs through recombination between extragenic repetitive elements on linear plasmids. Mol. Microbiol. 60:1329–1343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schwan TG, Schrumpf ME, Hinnebusch BJ, Anderson DE, Jr, Konkel ME. 1996. GlpQ: an antigen for serological discrimination between relapsing fever and Lyme borreliosis. J. Clin. Microbiol. 34:2483–2492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Taylor J, Moore G, Cheek J. 1991. Outbreak of relapsing fever masquerading as Lyme borreliosis, p 317 Abstr. 30th Intersci. Conf. Antimicrob. Agents Chemother [Google Scholar]
- 34.Cadavid D, Thomas DD, Crawley R, Barbour AG. 1994. Variability of a bacterial surface protein and disease expression in a possible mouse model of systemic Lyme borreliosis. J. Exp. Med. 179:631–642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hinnebusch BJ, Barbour AG, Restrepo BI, Schwan TG. 1998. Population structure of the relapsing fever spirochete Borrelia hermsii as indicated by polymorphism of two multigene families that encode immunogenic outer surface lipoproteins. Infect. Immun. 66:432–440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Porcella SF, Raffel SJ, Anderson DE, Jr, Gilk SD, Bono JL, Schrumpf ME, Schwan TG. 2005. Variable tick protein in two genomic groups of the relapsing fever spirochete Borrelia hermsii in western North America. Infect. Immun. 73:6647–6658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Barbour AG. 1984. Isolation and cultivation of Lyme disease spirochetes. Yale J. Biol. Med. 57:521–525 [PMC free article] [PubMed] [Google Scholar]
- 38.Pettersson J, Schrumpf ME, Raffel SJ, Porcella SF, Guyard C, Lawrence K, Gherardini FC, Schwan TG. 2007. Purine salvage pathways among Borrelia species. Infect. Immun. 75:3877–3884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Delcher AL, Harmon D, Kasif S, White O, Salzberg SL. 1999. Improved microbial gene identification with GLIMMER. Nucleic Acids Res. 27:4636–4641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bocs S, Cruveiller S, Vallenet D, Nuel G, Medigue C. 2003. AMIGene: Annotation of MIcrobial Genes. Nucleic Acids Res. 31:3723–3726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389–3402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wywial E, Haven J, Casjens SR, Hernandez YA, Singh S, Mongodin EF, Fraser-Liggett CM, Luft BJ, Schutzer SE, Qiu WG. 2009. Fast, adaptive evolution at a bacterial host-resistance locus: the PFam54 gene array in Borrelia burgdorferi. Gene 445:26–37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fraser CM, Casjens S, Huang WM, Sutton GG, Clayton R, Lathigra R, White O, Ketchum KA, Dodson R, Hickey EK, Gwinn M, Dougherty B, Tomb JF, Fleischmann RD, Richardson D, Peterson J, Kerlavage AR, Quackenbush J, Salzberg S, Hanson M, van Vugt R, Palmer N, Adams MD, Gocayne J, Weidman J, Utterback T, Watthey L, McDonald L, Artiach P, Bowman C, Garland S, Fuji C, Cotton MD, Horst K, Roberts K, Hatch B, Smith HO, Venter JC. 1997. Genomic sequence of a Lyme disease spirochaete, Borrelia burgdorferi. Nature 390:580–586 [DOI] [PubMed] [Google Scholar]
- 44.Finn RD, Mistry J, Tate J, Coggill P, Heger A, Pollington JE, Gavin OL, Gunasekaran P, Ceric G, Forslund K, Holm L, Sonnhammer EL, Eddy SR, Bateman A. 2010. The Pfam protein families database. Nucleic Acids Res. 38:D211–D222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Juncker AS, Willenbrock H, Von Heijne G, Brunak S, Nielsen H, Krogh A. 2003. Prediction of lipoprotein signal peptides in Gram-negative bacteria. Protein Sci. 12:1652–1662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gouy M, Guindon S, Gascuel O. 2010. SeaView version 4: a multiplatform graphical user interface for sequence alignment and phylogenetic tree building. Mol. Biol. Evol. 27:221–224 [DOI] [PubMed] [Google Scholar]
- 47.Darling AE, Mau B, Perna NT. 2010. progressiveMauve: multiple genome alignment with gene gain, loss and rearrangement. PLoS One 5:e11147. 10.1371/journal.pone.0011147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Frith MC, Hamada M, Horton P. 2010. Parameters for accurate genome alignment. BMC Bioinformatics 11:80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Karplus K. 2009. SAM-T08, HMM-based protein structure prediction. Nucleic Acids Res. 37:W492–W497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhong J, Skouloubris S, Dai Q, Myllykallio H, Barbour AG. 2006. Function and evolution of plasmid-borne genes for pyrimidine biosynthesis in Borrelia spp. J. Bacteriol. 188:909–918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Grosskinsky S, Schott M, Brenner C, Cutler SJ, Kraiczy P, Zipfel PF, Simon MM, Wallich R. 2009. Borrelia recurrentis employs a novel multifunctional surface protein with anti-complement, anti-opsonic and invasive potential to escape innate immunity. PLoS One 4:e4858. 10.1371/journal.pone.0004858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Skare JT, Foley DM, Hernandez SR, Moore DC, Blanco DR, Miller JN, Lovett MA. 1999. Cloning and molecular characterization of plasmid-encoded antigens of Borrelia burgdorferi. Infect. Immun. 67:4407–4417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tilly K, Elias AF, Errett J, Fischer E, Iyer R, Schwartz I, Bono JL, Rosa P. 2001. Genetics and regulation of chitobiose utilization in Borrelia burgdorferi. J. Bacteriol. 183:5544–5553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lopez JE, Schrumpf ME, Raffel SJ, Policastro PF, Porcella SF, Schwan TG. 2008. Relapsing fever spirochetes retain infectivity after prolonged in vitro cultivation. Vector Borne Zoonotic Dis. 8:813–820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Behera AK, Durand E, Cugini C, Antonara S, Bourassa L, Hildebrand E, Hu LT, Coburn J. 2008. Borrelia burgdorferi BBB07 interaction with integrin alpha3beta1 stimulates production of pro-inflammatory mediators in primary human chondrocytes. Cell. Microbiol. 10:320–331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zuckert WR, Barbour AG. 2000. Stability of Borrelia burgdorferi bdr loci in vitro and in vivo. Infect. Immun. 68:1727–1730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bignell C, Thomas CM. 2001. The bacterial ParA-ParB partitioning proteins. J. Biotechnol. 91:1–34 [DOI] [PubMed] [Google Scholar]
- 58.Beaurepaire C, Chaconas G. 2005. Mapping of essential replication functions of the linear plasmid lp17 of B. burgdorferi by targeted deletion walking. Mol. Microbiol. 57:132–142 [DOI] [PubMed] [Google Scholar]
- 59.Gal-Mor O, Finlay BB. 2006. Pathogenicity islands: a molecular toolbox for bacterial virulence. Cell. Microbiol. 8:1707–1719 [DOI] [PubMed] [Google Scholar]
- 60.Gao Q, Jin K, Ying SH, Zhang Y, Xiao G, Shang Y, Duan Z, Hu X, Xie XQ, Zhou G, Peng G, Luo Z, Huang W, Wang B, Fang W, Wang S, Zhong Y, Ma LJ, St Leger RJ, Zhao GP, Pei Y, Feng MG, Xia Y, Wang C. 2011. Genome sequencing and comparative transcriptomics of the model entomopathogenic fungi Metarhizium anisopliae and M. acridum. PLoS Genet. 7:e1001264. 10.1371/journal.pgen.1001264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Barbour AG, Putteet-Driver AD, Bunikis J. 2005. Horizontally acquired genes for purine salvage in Borrelia spp. causing relapsing fever. Infect. Immun. 73:6165–6168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Carroll JA, Garon CF, Schwan TG. 1999. Effects of environmental pH on membrane proteins in Borrelia burgdorferi. Infect. Immun. 67:3181–3187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Bykowski T, Woodman ME, Cooley AE, Brissette CA, Brade V, Wallich R, Kraiczy P, Stevenson B. 2007. Coordinated expression of Borrelia burgdorferi complement regulator-acquiring surface proteins during the Lyme disease spirochete's mammal-tick infection cycle. Infect. Immun. 75:4227–4236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Cordes FS, Roversi P, Kraiczy P, Simon MM, Brade V, Jahraus O, Wallis R, Skerka C, Zipfel PF, Wallich R, Lea SM. 2005. A novel fold for the factor H-binding protein BbCRASP-1 of Borrelia burgdorferi. Nat. Struct. Mol. Biol. 12:276–277 [DOI] [PubMed] [Google Scholar]
- 65.Hooper SD, Berg OG. 2003. On the nature of gene innovation: duplication patterns in microbial genomes. Mol. Biol. Evol. 20:945–954 [DOI] [PubMed] [Google Scholar]
- 66.Bratlie MS, Johansen J, Sherman BT, Huang da W, Lempicki RA, Drablos F. 2010. Gene duplications in prokaryotes can be associated with environmental adaptation. BMC Genomics 11:588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Casjens S, Eggers CH, Schwartz I. 2010. Borrelia genomics: chromosome, plasmids, bacteriophages and genetic variation, p 27–53 In Samuels DS, Radolf JD. (ed), Borrelia molecular biology, host interaction and pathogenesis. Caister Academic Press, Norfolk, United Kingdom [Google Scholar]
- 68.Stevenson B, Miller JC. 2003. Intra- and interbacterial genetic exchange of Lyme disease spirochete erp genes generates sequence identity amidst diversity. J. Mol. Evol. 57:309–324 [DOI] [PubMed] [Google Scholar]
- 69.Schwan TG, Piesman J, Golde WT, Dolan MC, Rosa PA. 1995. Induction of an outer surface protein on Borrelia burgdorferi during tick feeding. Proc. Natl. Acad. Sci. U. S. A. 92:2909–2913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Pal U, de Silva AM, Montgomery RR, Fish D, Anguita J, Anderson JF, Lobet Y, Fikrig E. 2000. Attachment of Borrelia burgdorferi within Ixodes scapularis mediated by outer surface protein A. J. Clin. Invest. 106:561–569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Fischer JR, Parveen N, Magoun L, Leong JM. 2003. Decorin-binding proteins A and B confer distinct mammalian cell type-specific attachment by Borrelia burgdorferi, the Lyme disease spirochete. Proc. Natl. Acad. Sci. U. S. A. 100:7307–7312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Rossmann E, Kraiczy P, Herzberger P, Skerka C, Kirschfink M, Simon MM, Zipfel PF, Wallich R. 2007. Dual binding specificity of a Borrelia hermsii-associated complement regulator-acquiring surface protein for factor H and plasminogen discloses a putative virulence factor of relapsing fever spirochetes. J. Immunol. 178:7292–7301 [DOI] [PubMed] [Google Scholar]
- 73.Kraiczy P, Hellwage J, Skerka C, Becker H, Kirschfink M, Simon MM, Brade V, Zipfel PF, Wallich R. 2004. Complement resistance of Borrelia burgdorferi correlates with the expression of BbCRASP-1, a novel linear plasmid-encoded surface protein that interacts with human factor H and FHL-1 and is unrelated to Erp proteins. J. Biol. Chem. 279:2421–2429 [DOI] [PubMed] [Google Scholar]
- 74.Bykowski T, Woodman ME, Cooley AE, Brissette CA, Wallich R, Brade V, Kraiczy P, Stevenson B. 2008. Borrelia burgdorferi complement regulator-acquiring surface proteins (BbCRASPs): expression patterns during the mammal-tick infection cycle. Int. J. Med. Microbiol. 298(Suppl. 1):249–256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Stothard P, Wishart DS. 2005. Circular genome visualization and exploration using CGView. Bioinformatics 21:537–539 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.