Abstract
The regulatory gene aldR was identified 95 bp upstream of the ald gene encoding l-alanine dehydrogenase in Mycobacterium smegmatis. The AldR protein shows sequence similarity to the regulatory proteins of the Lrp/AsnC family. Using an aldR deletion mutant, we demonstrated that AldR serves as both activator and repressor for the regulation of ald gene expression, depending on the presence or absence of l-alanine. The purified AldR protein exists as a homodimer in the absence of l-alanine, while it adopts the quaternary structure of a homohexamer in the presence of l-alanine. The binding affinity of AldR for the ald control region was shown to be increased significantly by l-alanine. Two AldR binding sites (O1 and O2) with the consensus sequence GA-N2-ATC-N2-TC and one putative AldR binding site with the sequence GA-N2-GTT-N2-TC were identified upstream of the ald gene. Alanine and cysteine were demonstrated to be the effector molecules directly involved in the induction of ald expression. The cellular level of l-alanine was shown to be increased in M. smegmatis cells grown under hypoxic conditions, and the hypoxic induction of ald expression appears to be mediated by AldR, which senses the intracellular level of alanine.
INTRODUCTION
Alanine dehydrogenase (EC 1.4.1.1; Ald) catalyzes the reversible oxidative deamination of l-alanine to pyruvate with the concomitant reduction of oxidized nicotinamide adenine dinucleotide (NAD+) to nicotinamide adenine dinucleotide phosphate (NADH). Its forward reaction appears to be necessary for the aerobic utilization of alanine as a nitrogen source in Mycobacterium tuberculosis, Mycobacterium smegmatis, and Mycobacterium bovis BCG (1–3). The reverse reaction of Ald was proposed to play a role in recycling NADH under respiration-inhibitory conditions, such as hypoxia, by oxidizing NADH to NAD+ (2, 4, 5). Ald proteins from M. tuberculosis and M. smegmatis have glyoxylate-reductive aminase activity, which converts glyoxylate to glycine, but do not catalyze the reverse reaction, in which glycine is converted to glyoxylate by oxidative deamination (3, 6). The quaternary structure of Ald is a homohexamer consisting of three dimers, and each subunit is composed of an N-terminal catalytic domain and C-terminal NADH (NAD+) binding domain (7, 8).
It was reported that expression of the ald genes encoding Ald was upregulated in M. tuberculosis under nutrient starvation and energy-limiting conditions, as well as in Mycobacterium marinum during long-term granulomatous infection in its host (5, 9, 10). The synthesis and activity of Ald as well as expression of ald were shown to be induced when M. tuberculosis and M. smegmatis were shifted from aerobic to hypoxic growth conditions (4, 6, 11–13). Furthermore, the addition of alanine to aerobic cultures of M. tuberculosis and M. smegmatis led to a strong induction of ald expression (2, 3). Although the induction conditions of the ald gene were well known, the regulatory mechanism, which underlies upregulation of the gene under the different conditions mentioned above, remained unsolved. Here, we report that expression of the ald gene of M. smegmatis is under the control of its upstream gene product (AldR), which acts as both activator and repressor depending on the presence or absence of alanine, and that the hypoxic induction of ald is a result of increased levels of alanine in M. smegmatis cells grown under hypoxic conditions.
MATERIALS AND METHODS
Bacterial strains, plasmids, and culture conditions.
The bacterial strains and plasmids used in this study are listed in Table 1. M. smegmatis strains were grown in Middlebrook 7H9 medium (Difco, Sparks, MD) supplemented with 0.2% (wt/vol) glucose as a carbon source and 0.02% (vol/vol) Tween 80 as an anticlumping agent at 37°C. M. smegmatis strains were grown aerobically or hypoxically as described previously (14). For various stress conditions, except hypoxic conditions, M. smegmatis strains were grown to an optical density at 600 nm (OD600) of 0.5 to 0.6 on a gyratory shaker (200 rpm). Following the addition of chemicals to the cultures, the strains were further grown for 1 h. The treatment concentrations of the chemicals were 25 mM l-amino acids, 5 mM diamide, 15 mM hydrogen peroxide, and 5 mM sodium nitroprusside (SNP). For heat stress conditions, the aerobic cultures grown to an OD600 of 0.5 to 0.6 were further grown at 45°C for 1 h. Escherichia coli strains were grown in Luria-Bertani (LB) medium at 37°C. Ampicillin (100 μg/ml for E. coli), hygromycin (200 μg/ml for E. coli and 50 μg/ml for M. smegmatis), and kanamycin (50 μg/ml for E. coli and 30 μg/ml for M. smegmatis) were added to the growth medium when required.
Table 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Relevant phenotype or genotype | Reference or source |
|---|---|---|
| Strains | ||
| M. smegmatis | ||
| mc2155 | High-transformation-efficiency mutant of M. smegmatis ATCC 607 | 16 |
| ΔdevR | devR (MSMEG_5244) deletion mutant derived from M. smegmatis mc2155 | 20 |
| ΔaldR | aldR (MSMEG_2660) deletion mutant derived from M. smegmatis mc2155 | 22 |
| E. coli | ||
| DH5α | φ80dlacZΔM15 ΔlacU169 recA1 endA1 hsdR17 supE44 thi1 gyrA96 relA1 | 48 |
| BL21(DE3) | F− ompT hsdSB(rB− mB−) dcm gal λ (DE3) | Promega |
| Plasmids | ||
| pBluescript II KS + | Ampr; lacPOZ′ | 14 |
| pNC | Hygr; promoterless lacZ | 49 |
| pT7-7 | Ampr; T7 promoter, ribosome binding site, and translation start codon overlapping NdeI site | 50 |
| pNBV1 | Hygr; 5.8-kb plasmid derived from p16R1 | 51 |
| pALDLACZ | pNC with 0.52-kb XbaI-ClaI fragment containing the ald promoter region | This study |
| pBSMS2659 | pBluescript II KS+ with 0.52-kb XbaI-ClaI fragment from pALDLACZ | This study |
| pBSPM1 | pBSMS2659 with point mutations (TC→CT) in the O1 region | This study |
| pBSPM2 | pBSMS2659 with point mutations (AT→GC) in the O1 region | This study |
| pBSPM3 | pBSMS2659 with point mutations (AC→GT) in the O1 region | This study |
| pNC218bp | pNC with 0.33-kb XbaI-ClaI fragment containing 218 bp of the ald promoter region | This study |
| pNC153bp | pNC with 0.26-kb XbaI-ClaI fragment containing 153 bp of the ald promoter region | This study |
| pNC109bp | pNC with 0.22-kb XbaI-ClaI fragment containing 109 bp of the ald promoter region | This study |
| pM1 | pNC with 0.52-kb XbaI-ClaI fragment from pBSPM1 | This study |
| pM2 | pNC with 0.52-kb XbaI-ClaI fragment from pBSPM2 | This study |
| pM3 | pNC with 0.52-kb XbaI-ClaI fragment from pBSPM3 | This study |
| pT7MS2660His | pT7-7 with 0.54-kb NdeI-HindIII fragment containing aldR with 6 His codons before its stop codon | This study |
| pMV306lacZald | pMV306lacZ with 0.51-kb EcoRI-HindIII fragment containing the ald promoter region | 22 |
| pNBV1aldR | pNBV1 with 1.11-kb BamHI-HindIII fragment containing aldR of M. smegmatis mc2155 | This study |
DNA manipulation and electroporation.
Standard protocols or manufacturer's instructions were followed for recombinant DNA manipulations (15). The introduction of plasmids into M. smegmatis strains was carried out by electroporation as described elsewhere (16).
Construction of plasmids. (i) pALDLACZ, pNC218bp, pNC153bp, and pNC109bp.
pALDLACZ, pNC218bp, pNC153bp, and pNC109bp are the lacZ transcriptional fusion plasmids that contain the 5′ portion (105 bp) of ald, as well as 410-, 218-, 153-, and 109-bp DNA sequences upstream of the ald start codon, respectively. For the construction of pALDLACZ, a 530-bp DNA fragment including the ald gene upstream region was amplified with F_MS2659 ( 5′-TTATTCTAGAAGCTGCGGATCTTGCCGC-3′ ) and R_MS2659 ( 5′-ATTGATCGATGATCACCTCGTGACCTCT-3′ ), using M. smegmatis mc2155 genomic DNA as the template and Pfu DNA polymerase. The PCR product was restricted with ClaI and XbaI and cloned into the promoterless lacZ vector pNC, which had been digested with the same restriction enzymes. In order to construct the other plasmids, the ald upstream regions of the corresponding lengths were amplified by PCR using the forward primers with an XbaI restriction site and the R_MS2659 primer with a ClaI restriction site. pALDLACZ was used as the template for PCR. The PCR products were restricted with ClaI and XbaI and cloned into pNC, resulting in the plasmids pNC218bp, pNC153bp, and pNC109bp.
(ii) pT7MS2660His.
A 551-bp DNA fragment including the aldR gene and the six histidine codons immediately before its stop codon was amplified with F_MS2660_Histag ( 5′-CCCCCATATGAGTGAAGGATCATCGATCAC-3′ ) and R_MS2660_Histag ( 5′-TTTTAAGCTTTCAGTGATGGTGATGGTGATGCAGCGGCGACGCGCCCCGCA-3′ ) using M. smegmatis mc2155 genomic DNA as the template and Pfu DNA polymerase. The PCR product was restricted with NdeI and HindIII and cloned into pT7-7 digested with same restriction enzymes, yielding pT7MS2660His.
(iii) pNBV1aldR.
To construct pNBV1aldR, used for complementation, a 1,110-bp BamHI-HindIII fragment containing aldR was amplified with MSMEG_2660_F ( 5′-AACGGGATCCGACCAGACCTGGTCGGCG-3′ ) and MSMEG_2660_R ( 5′-AACGAAGCTTCCAAGCCCACGCTGACGA-3′ ) using M. smegmatis mc2155 genomic DNA as the template and Pfu DNA polymerase. The PCR product was restricted with BamHI and HindIII and cloned into pNBV1 digested with the same restriction enzymes, yielding pNBV1aldR.
Site-directed mutagenesis.
To construct pM1, pM2, and pM3, site-directed mutagenesis was performed using the plasmid pBSMS2659 as the template and the QuikChange site-directed mutagenesis kit (Stratagene, La Jolla, CA). pBSMS2659 was constructed by cloning of a 518-bp XbaI-ClaI fragment from pALDLACZ into pBluescript II KS+. Synthetic oligonucleotides 33 to 34 bases long and containing mutated nucleotides in the middle of their sequences were used to mutagenize a putative AldR binding site (O1) located upstream of ald. Mutations were verified by DNA sequencing. The 518-bp XbaI and ClaI fragment from the mutated pBSMS2659 was cloned into pNC, resulting in the plasmids pM1, pM2, and pM3, which had the same construct as pALDLACZ except for the point mutations.
2D electrophoresis.
The mutant M. smegmatis ΔdevR strain was grown under either hypoxic or aerobic conditions as described above, and cells were harvested. Bacterial pellets were resuspended in 2.5 ml of buffer (20 mM Tris-HCl [pH 8.0], 5 mM EDTA, and 1 mM phenylmethylsulfonyl fluoride [PMSF]), and cells were disrupted by five passages through a French pressure cell at 800 lb/in2. Following the DNase/RNase treatment (5 U DNase I and 5 U RNase I/ml in the presence of 10 mM MgCl2) for 30 min on ice, soluble fractions were obtained by ultracentrifugation at 100,000 × g for 90 min at 4°C, and proteins were precipitated by treatment with trichloroacetic acid (TCA) to a final concentration of 10% (wt/vol) for 1 h on ice. TCA was removed by acetone treatment. The sample was resuspended in 250 μl of the sample preparation solution, composed of 8 M urea, 4% (wt/vol) 3-[(3-cholamidopropyl)-dimethylammonio]-1-propanesulfonate (CHAPS), 2% (vol/vol) IPG buffer (pH 4 to 7; GE Healthcare, Piscataway, NJ), and 40 mM dithiothreitol (DTT). The protein concentration was determined by using a bicinchoninic acid (BCA) protein assay kit (Thermo, Milford, MA) and a two-dimensional (2D) Quant kit (GE Healthcare). 2D gel electrophoresis was conducted with a MultiTemp III system (GE Healthcare) according to the manufacturer's instructions. Thirteen-cm Immobiline DryStrip gels (pH 4 to 7; GE Healthcare) were rehydrated for 10 h in rehydration buffer (8 M urea, 2% [wt/vol] CHAPS, 0.002% [wt/vol] bromophenol blue) with IPG buffer at a final concentration of 0.5% (vol/vol). One hundred to 120 μg of proteins was loaded on the rehydrated gel strips, and proteins were electrofocused on an Ettan IPGphor II manifold (GE Healthcare). Isoelectrofocusing started at 100 V, and the applied voltage was increased stepwise to 8,000 V for 5 h and then maintained at 8,000 V to reach a total of 65,000 V · h, as described in the manufacturer's manual (GE Healthcare). Subsequently, the isoelectrofocused gel strips were equilibrated for 15 min in 10 ml of the equilibration solution (1% [wt/vol] DTT, 75 mM Tris-HCl [pH 8.8], 6 M urea, 29.3% [vol/vol] glycerol [87%, wt/wt], 2% [wt/vol] SDS, 0.002% [wt/vol] bromophenol blue) and for another 15 min in 10 ml of the equilibration solution with 2.5% (wt/vol) iodoacetamide (IAA). The 2D separation was performed by using an SE 600 Ruby electrophoresis system (GE Healthcare) in 12.5% (wt/vol) SDS-PAGE at 10 mA for 1 h and then at 50 mA for 4 h. Proteins were stained by Coomassie brilliant blue. The protein spots were excised from the gels and digested with trypsin for identification by matrix-assisted laser desorption ionization–time of flight mass spectroscopy (MALDI-TOF MS) as previously described (17).
RT-PCR and qRT-PCR.
RNA isolation from M. smegmatis strains, preparation of cDNA, reverse transcription-PCR (RT-PCR), and quantitative RT-PCR (qRT-PCR) were carried out as described elsewhere (14). To synthesize cDNA, the primers RT-16sr(−) ( 5′-ACAACGCTCGGACCCTAC-3′ ) for the 16S rRNA gene, Rihsp− ( 5′-CGCCCGTTGGTCTCCTTCTTC-3′ ) for the hspX gene, and MSMEG_2659_R ( 5′-TGGCCGTGGCGTCCTGATGG-3′ ) for the ald gene were used. For RT-PCR, the primers RT-16sr(+) ( 5′-CTGGGACTGAGATACGGC-3′ ) and RT-16sr(−) for the 16S rRNA gene, Rihsp+ ( 5′-GGGTCTGCCGTCGTGGGCCTC-3′ ) and Rihsp− for the hspX gene, and MSMEG_2659_F ( 5′-GAGTAGCGGGTCTCGATGCG-3′ ) and MSMEG_2659_R for the ald gene were employed.
β-Galactosidase assay and determination of the protein concentration.
The β-galactosidase activity was assayed spectrophotometrically as described previously (18). The protein concentration was determined by using a Bio-Rad protein assay kit (Bio-Rad, Hercules, CA) with bovine serum albumin as the standard protein.
Purification of AldR and determination of its molecular weight in the native form.
The E. coli BL21(DE3) strain carrying pT7MS2660His was grown aerobically at 37°C in LB medium containing 100 μg/ml ampicillin to an OD600 of 0.4 to 0.5. The aldR gene was induced by the addition of isopropyl-β-d-thiogalactopyranoside (IPTG) to a final concentration of 0.5 mM, and the cells were further grown for 4 h at 30°C. After 700 ml culture was harvested, cells were resuspended in 10 ml of buffer A (20 mM Tris-HCl [pH 8.0] containing 100 mM NaCl) and disrupted by two passages through a French pressure cell. Following DNase I treatment (10 U/ml) in the presence of 10 mM MgCl2 for 30 min on ice, cell-free crude extracts were obtained by centrifugation two times at 20,000 × g for 20 min. After addition of imidazole to a final concentration of 5 mM, 0.6 ml of 80% (vol/vol) Ni-Sepharose high-performance resin (Amersham Biosciences, Piscataway, NJ) was added to the crude extracts and mixed gently by shaking for 2 h on ice. The protein-resin mixture was loaded into a column, and the column was washed with 20 volumes of buffer A containing 5 mM imidazole, followed by 20 volumes of buffer A containing 50 mM imidazole. The His6-tagged AldR protein was eluted with buffer A containing 250 mM imidazole. Imidazole and NaCl were removed from purified AldR by means of a PD-10 desalting column (GE Healthcare) equilibrated with 20 mM Tris-HCl (pH 8.0).
Gel filtration chromatography was performed with a Superose 12 10/300 GL (GE Healthcare) column equilibrated with 20 mM Tris-HCl (pH 8.0) and calibrated with the standard proteins of known molecular masses (alcohol dehydrogenase, 150 kDa; bovine serum albumin, 66 kDa; carbonic anhydrase, 29 kDa; Sigma). When required, l-alanine or l-cysteine was added to the 20 mM Tris-HCl (pH 8.0) buffer and samples to a final concentration of 10 mM.
EMSA.
Electrophoretic mobility shift assay (EMSA) was carried out by using an EMSA kit (Invitrogen, Carlsbad, NJ) according to the manufacturer's instructions. A 243-bp DNA fragment encompassing the putative AldR binding site (O1, O2, and O3), a 216-bp DNA fragment with the deletion of O1, and a 234-bp DNA fragment with the deletion of both O1 and O2 were used in EMSA. The 243-bp DNA fragment was generated by PCR using pALDLACZ as the template and the primer sets F_EMSA ( 5′-AACCATCGATAGCATGATCGCTCCTTCAGAAG-3′ ) and R_218bp ( 5′-ATTGTCTAGATATGCGTGCATCGTCGTGCAAC-3′ ). To delete the O1 sequence, a 128-bp DNA fragment containing the upstream region of O1 was amplified by PCR using pALDLACZ as the template and the primer set R_EMSA_dI ( 5′-GAAAATTCGTTAGGATTGTGGCGTCCAAACGCCCGGTTCG-3′ ) and R_218bp ( 5′-ATTGTCTAGATATGCGTGCATCGTCGTGCAAC-3′ ). A 128-bp DNA fragment containing the downstream region of O1 was amplified by PCR using pALDLACZ as the template and the primer set F_EMSA and F_EMSA_dI ( 5′-CGAACCGGGCGTTTGGACGCCACAATCCTAACGAATTTTC-3′ ). In the secondary PCR, a 216-bp DNA fragment deleted of O1 was obtained by using both the primary PCR products as the templates and the F_EMSA and R_218bp primers. The same strategy was used for the synthesis of the 234-bp DNA fragment. A 146-bp DNA fragment containing the upstream region of O2 was amplified by PCR using pALDLACZ as the template and the primer set R_279bp ( 5′-AAAATCTAGAACGCGTCCGTGGCACGTC-3′ ) and R_EMSA_dI2 ( 5′-GAAAATTCGTTAGGATTGTGGACCTCGACGACATCGACCG-3′ ). A 128-bp DNA fragment containing the downstream region of O1 was amplified by PCR using pALDLACZ as the template and the primer set F_EMSA and F_EMSA_dI2 ( 5′-CGGTCGATGTCGTCGAGGTCCACAATCCTAACGAATTTTC-3′ ). The secondary PCR was performed with the primary PCR products and the F_EMSA and R_279bp primers, resulting in a 234-bp DNA fragment with the deletion of both O1 and O2. DNA binding reaction mixtures were composed of the appropriate amounts of DNA (2 μl), purified AldR (5 μl), 1 μl of H2O, and 2 μl of 5× binding buffer (included in the kit). l-Amino acids (alanine, serine, cysteine) were added, when necessary, to a final concentration of 20 mM. The binding reaction mixtures were incubated for 20 min at room temperature. After the addition of 2 μl of 6× loading buffer (included in the kit), the samples were subjected to the nondenaturing PAGE (6% [wt/vol] acrylamide) in buffer (pH 8.3) containing 83 mM Tris-borate and 1 mM EDTA at 60 V for 5 h at 4°C. The gels were stained with SYBR green staining solution (included in the kit). Bands were visualized by using a SYBR photographic filter (Invitrogen).
Quantification of intracellular alanine.
The cell pellets of M. smegmatis strains were resuspended in 3 ml of H2O and centrifuged to wash the cells. The cells were resuspended in 2 ml of H2O and disrupted by five passages through a French pressure cell. Cell-free crude extracts were obtained by centrifugation at 20,000 × g for 20 min. Determination of the alanine concentration in the crude extracts was conducted by measuring spectrophotometrically the reduction of NAD+ to NADH at 340 nm for 90 min in an Ald coupled enzyme assay. A 1.5-ml aliquot of the reaction mixture was composed of the crude cell extracts (100 μg of protein), 40 mM hydrazine monohydrate, 50 mM glycine, 2.67 mM NAD+, and 0.5 U/ml Ald of Bacillus subtilis (Sigma, St. Louis, MO). The obtained absorbance values were normalized to those of the controls without B. subtilis Ald. The standard curve was generated using standard solutions with different concentrations of l-alanine (0 to 120 nmol).
RESULTS
Hypoxic induction of ald is independent of the DevSR two-component system.
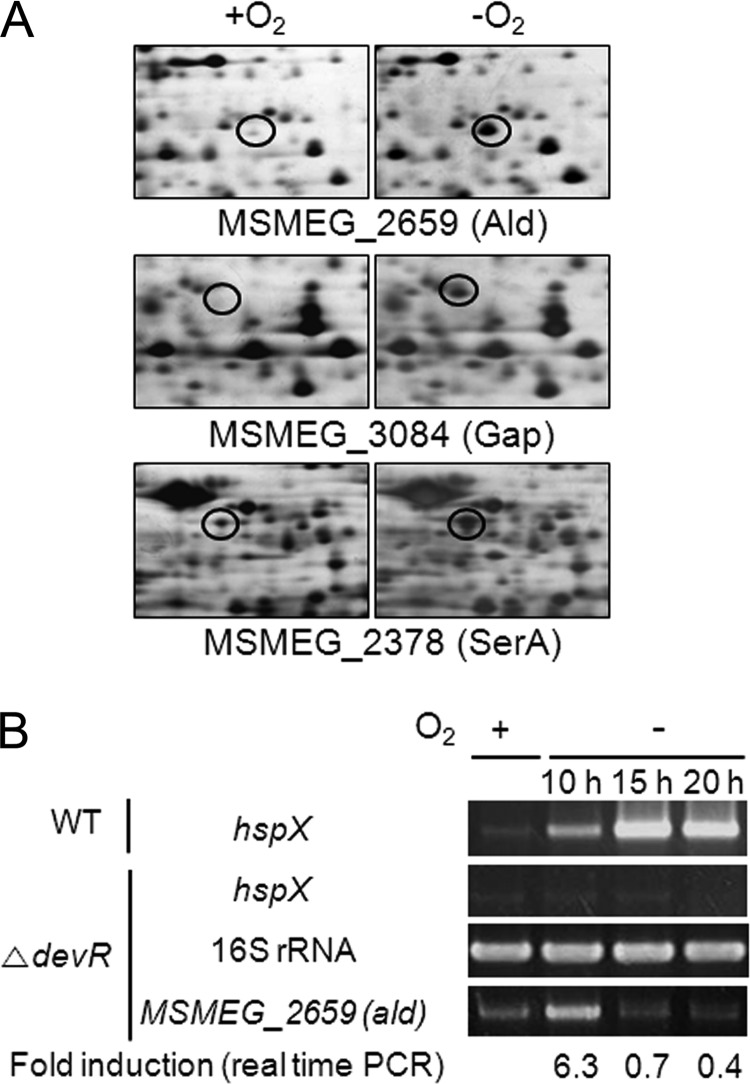
The DevSR two-component system (TCS) (DevS, MSMEG_5241; DevR, MSMEG_5244) is a major regulatory system that is responsible for hypoxic induction of gene expression in M. smegmatis (19). There is another set of genes for DevS (MSMEG_3941) and DevR (MSMEG_3944) in M. smegmatis. However, they appear not to be functional for hypoxic induction of the genes that are under the control of DevR, since the hspX gene, which is regulated by the DevSR TCS, was shown not to be induced in either MSMEG_5241 or MSMEG_5244 mutants grown under hypoxic conditions (14, 20). To identify regulatory systems other than the DevSR TCS which are involved in hypoxic induction of genes in M. smegmatis, we first looked for proteins whose levels were increased by more than 5-fold in an M. smegmatis ΔdevR (MSMEG_5244) mutant strain subjected to the gradual depletion of oxygen for 20 h (i.e., 20 h of hypoxic conditions) compared to the same strain grown aerobically. As shown in Fig. 1A, 2D electrophoresis and subsequent MALDI-TOF MS led to the identification of three proteins, l-alanine dehydrogenase, glyceraldehyde-3-phosphate dehydrogenase, and d-3-phosphoglycerate dehydrogenase, encoded by MSMEG_2659 (ald), MSMEG_3084 (gap), and MSMEG_2378 (serA), respectively. Since the synthesis of alanine dehydrogenase (Ald) was increased the most (11.8-fold) among these three proteins in the strain grown under hypoxic conditions, we further studied the regulation of ald expression. To examine the hypoxic induction of ald at the transcriptional level, the ΔdevR mutant strain of M. smegmatis was grown either aerobically or under hypoxic conditions for 10, 15, and 20 h, and the expression level of the ald gene was determined by RT-PCR (Fig. 1B). RT-PCR for the 16S rRNA gene was performed to ensure that the same amounts of total RNA were used in the experiment. The hspX gene under the control of DevR was used as the control. As expected, expression of hspX was not induced in the ΔdevR mutant strain grown under hypoxic conditions, whereas it was strongly induced in the wild-type strain grown under the same conditions (10, 15, and 20 h of hypoxic conditions). Expression of ald was strongly induced under 10 h of hypoxic conditions, and its expression level was drastically reduced thereafter. qRT-PCR showed that the expression level of ald in the ΔdevR mutant strain grown under hypoxic conditions for 10 h was 6.3-fold higher than that in the mutant grown aerobically. The discrepancy in the levels of ald mRNA and Ald protein in M. smegmatis grown under 20 h of hypoxic conditions can be explained by the slow turnover of the Ald protein.
Fig 1.
Identification of the ald gene whose expression was upregulated in a ΔdevR mutant strain of M. smegmatis grown under hypoxic conditions. (A) Comparative 2D electrophoresis analysis of soluble proteins of the M. smegmatis ΔdevR strain grown under hypoxic (−O2) or aerobic (+O2) conditions for 20 h. The spots indicated by the open circles represent three proteins whose synthesis was increased more than 5-fold under hypoxic conditions compared to their levels under aerobic conditions. By means of MALDI-TOF MS, the proteins were identified as MSMEG_2659 (Ald), with a molecular mass of 38 kDa and pI of 5.9, MSMEG_3084 (Gap), with a molecular mass of 32 kDa and pI of 5.2, and MSMEG_2378 (SerA), with a molecular mass of 51 kDa and pI of 4.9. (B) Determination of the transcription levels of ald under hypoxic conditions. The wild-type (WT) and ΔdevR strains were grown either aerobically (+O2) or under hypoxic conditions (−O2) for 10, 15, and 20 h. The expression levels of ald and 16S rRNA genes were determined by RT-PCR. The hspX gene, whose expression was known to be induced by DevR under hypoxic conditions, was included in the experiment as a control. The levels of mRNA specific for ald were also determined by qRT-PCR and normalized to those of 16S rRNA. Fold induction of ald expression indicates the level of ald mRNA in hypoxic cultures relative to that in aerobic culture and is given below the RT-PCR results.
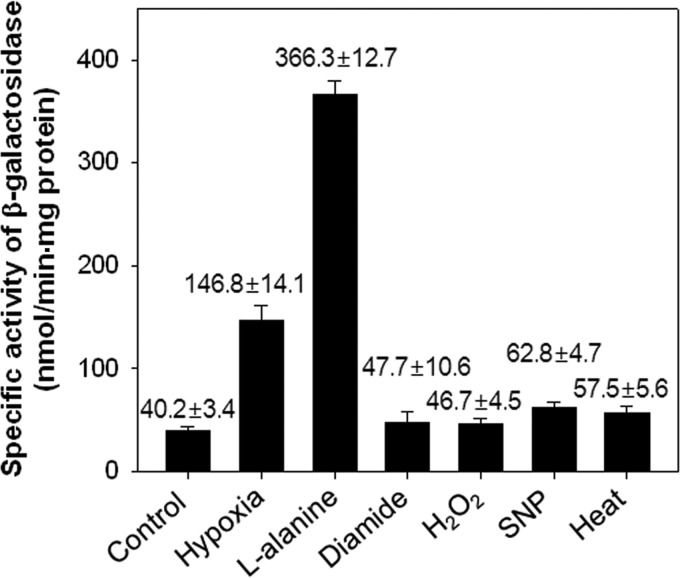
The mechanism by which expression of ald is induced in mycobacteria under hypoxic conditions remained undetermined. A possible regulatory mechanism is that an alternative sigma factor, like SigH, is involved in the regulation of ald. To examine this possibility, we measured ald expression in the M. smegmatis ΔdevR mutant exposed to various stress conditions (diamide, H2O2, SNP, and heat), as well as to hypoxic and alanine-supplemented conditions, by mean of an ald::lacZ transcriptional fusion plasmid, pALDLACZ (Fig. 2). Expression of ald in the ΔdevR mutant strain grown under hypoxia and with l-alanine was increased by 3.7- and 9.1-fold, respectively, compared to that in the strain grown aerobically (control). Expression of ald in the ΔdevR strain exposed to SNP and heat stress conditions was approximately 1.6- and 1.4-fold increased, respectively, relative to that of the control. In contrast, when the ΔdevR mutant strain was treated with diamide or H2O2, which are the signals activating expression of the SigH regulon (21), the expression level of ald was similar to that of the control, indicating that the hypoxic induction of ald expression is not caused by SigH.
Fig 2.
Expression of ald in the M. smegmatis ΔdevR strain exposed to various conditions. The M. smegmatis ΔdevR strain, carrying the ald::lacZ transcriptional fusion plasmid pALDLACZ, was grown either aerobically (control), under hypoxic conditions for 10 h (hypoxia), or exposed to various conditions as described in Materials and Methods. Cell-free crude extracts were used to determine β-galactosidase activity. The β-galactosidase activity is expressed as nanomoles per minute per milligram of protein. All values provided are the averages of the results from two independent determinations. Error bars indicate the deviations from the means.
Identification of the aldR gene.
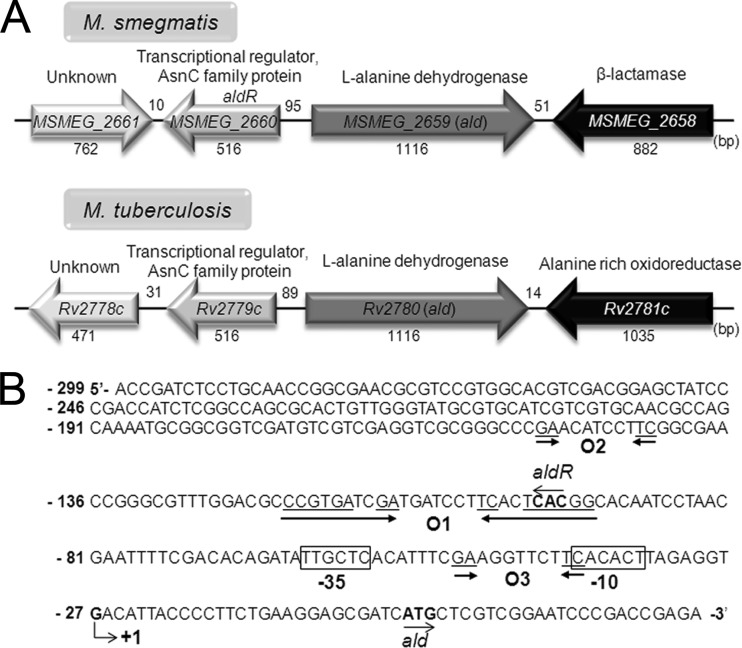
The open reading frame designated MSEMG_2660 was identified 95 bp upstream of ald and in the opposite transcriptional orientation (Fig. 3). Its deduced protein product consists of 171 amino acid residues with a calculated molecular mass of 18.6 kDa. We named MSMEG_2660 aldR (for alanine dehydrogenase regulator). AldR is a member of the family of Lrp/AsnC transcriptional regulators. The genetic organization of the genes encoding the Ald and AldR homologs is also conserved in M. tuberculosis (Fig. 3A). The amino acid sequences of Ald and AldR of M. smegmatis exhibit 80 and 75% overall identity, respectively, to their homologs in M. tuberculosis.
Fig 3.
Genetic organization of the ald locus and the putative cis-acting elements for ald expression. (A) The ald genes and their flanking genes in M. smegmatis mc2155 and M. tuberculosis H37Rv. The genes of the putative transcriptional regulators, MSMEG_2660 (aldR) and Rv2779c, are divergently located upstream of the ald genes. The lengths of genes and intergenic regions are given as the nucleotide numbers below and above their names, respectively. (B) The transcription start site (+1) of the ald gene of M. smegmatis was previously reported to be a guanosine residue that is located 27 nucleotides upstream of the start codon of ald (2). The −10 and −35 regions of the putative promoter for ald deduced from the transcription start point are boxed. The putative AldR binding site (O1), which shows an interrupted inverted repeat sequence ( CCGTGAN2GA-N7-TCN2TCACGG ), is marked by two head-facing arrows. The O2 and O3 sequences (GA-N7-TC) exhibiting partial sequence similarity to O1 are indicated by two arrows. The start codons of ald and aldR are highlighted in boldface and by the arrows indicating the transcriptional direction. The numbers on the left of the nucleotide sequence indicate the positions of the leftmost nucleotides relative to the ald gene.
AldR acts as both activator and repressor in the regulation of ald expression.
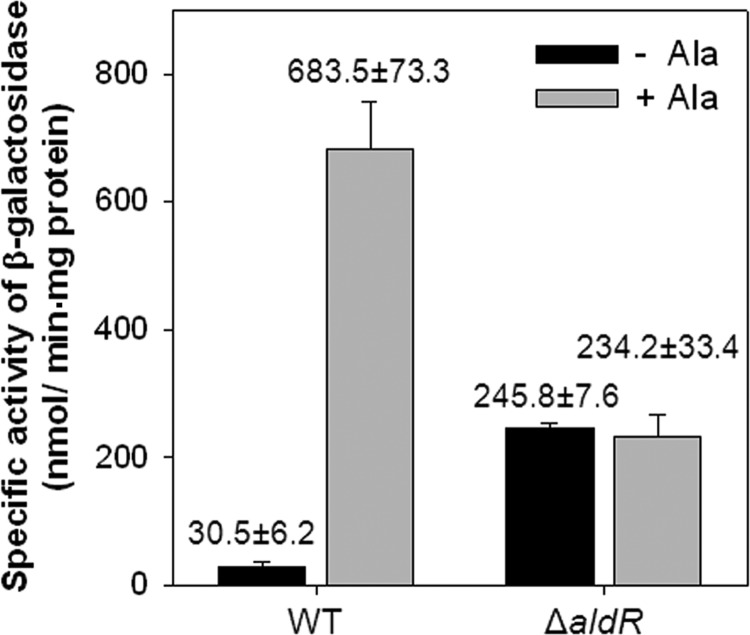
To gain further insights into the function of AldR on ald expression in the M. smegmatis strain, a mutant carrying a deletion within aldR (ΔaldR) was constructed (22). No difference in the growth rate between the wild-type and ΔaldR mutant strains of M. smegmatis was observed under aerobic conditions (data not shown). The possible role of AldR in the regulation of ald expression was investigated by determining the activity of β-galactosidase in the wild-type and ΔaldR mutant strains with pALDLACZ grown in the presence or absence of l-alanine (Fig. 4). The wild-type strain of M. smegmatis grown in the presence of l-alanine showed a significantly higher expression level of ald than the same strain grown in the absence of l-alanine, indicating that alanine is a strong inducer of ald expression. In contrast, expression of ald in the ΔaldR mutant was derepressed in the absence of l-alanine to a level corresponding to ∼35% of that in the wild type grown in the presence of l-alanine, and the addition of l-alanine to the culture did not alter the expression level of ald. When the aldR gene on pNBV1aldR was introduced into the ΔaldR mutant in trans, the regulation pattern of ald was restored to that observed in the wild type (data not shown).
Fig 4.
Expression of the ald gene in the wild-type and ΔaldR mutant strains of M. smegmatis. M. smegmatis wild-type (WT) and ΔaldR strains containing the ald::lacZ transcriptional fusion plasmid (pALDLACZ) were grown aerobically in 7H9-glucose medium to an OD600 of 0.5 to 0.6. Following the addition of 25 mM l-alanine to the cultures, the strains were further grown for 1 h (+Ala). For the control, the M. smegmatis strains were grown aerobically without the addition of l-alanine (−Ala). The ald promoter activities were measured by determining the β-galactosidase activity. All values provided are the averages of the results from two independent determinations. Error bars indicate the deviations from the means.
Identification of cis control elements upstream of the ald gene.
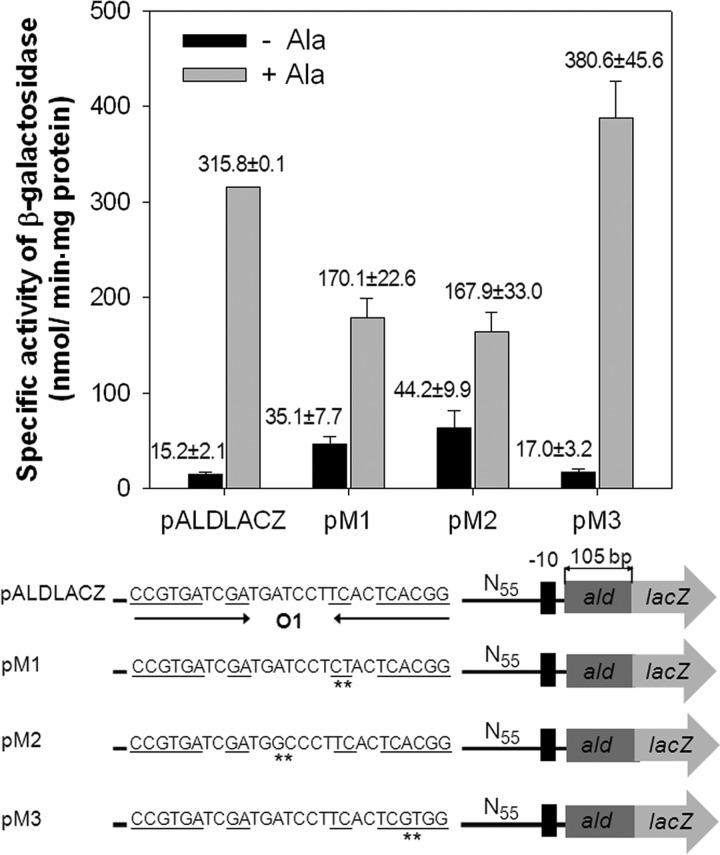
The transcriptional start point of ald was previously determined by Feng et al. (2). The putative promoter of ald resembling the mycobacterial −35 and −10 regions was identified on the basis of the position of the transcriptional start point (Fig. 3B). A conspicuous inverted repeat sequence (O1) was identified 93 bp upstream of ald, and it completely overlaps with the start codon of aldR (Fig. 3B). The O1 site contains a long, interrupted inverted repeat sequence, namely, CCGTGATCGATGATCCTTCACTCACGG (the dyad symmetry sequences are underlined, and the central sequence is indicated in boldface). Using several ald::lacZ transcriptional fusions (pM1, pM2, and pM3, which are derivatives of pALDLACZ) containing point mutations within the O1 region, we examined the role of the identified O1 region in ald expression and which sequences within the O1 region are important for the regulation of ald expression (Fig. 5). As a positive control, the wild-type strain harboring pALDLACZ was included in the experiment. As shown in Fig. 5, base substitution mutations (pM3, AC to GT) within the hexameric dyad ( CCGTGA-N15-TCACGG [the mutated nucleotides are underlined]) did not affect the regulation of ald expression, whereas those (pM1, TC to CT; pM2, AT to GC) within the internal dimeric dyad ( GA-N7-TC ) and the central sequence (ATC) led to partial deregulation of ald expression in the presence and absence of l-alanine. This result indicates that the internal dyad and the central ATC sequence ( GA-N2-ATC-N2-TC ) are important for the regulation of ald expression. The consensus DNA sequence for E. coli Lrp, Pseudomonas putida MdeR, and Pyrococcus sp. strain OT3 F11 is known as GA-N2-WWW-N2-TC (W is A or T) (23–25), which is very similar to the O1 sequence, except for the C in the central sequence. Using the criterion of GA-T7-TC and two W nucleotides in the central sequence, we identified two additional putative AldR binding sites (O2 and O3) in the region upstream of ald. The O2 sequence perfectly matches the corresponding sequence of O1 ( GA-N2-ATC-N2-TC ) and is located upstream of O1, with an interval of 40 bp between the T nucleotides of the central sequences of O1 and O2. The O3 site has the sequence GA-N2-GTT-N2-TC , and its location overlaps that of the promoter of ald (Fig. 3B).
Fig 5.
Effect of base substitution mutations within the O1 site on ald expression. The ald promoter activities were determined by using the ald::lacZ transcriptional fusions with point mutations in the O1 region (pM1, pM2, and pM3), which are derivatives of pALDLACZ. Transition mutations within the O1 site are indicated by the asterisks. As the positive control, pALDLACZ was included in the experiment. M. smegmatis wild-type strains harboring the transcriptional fusion plasmids were grown aerobically in the presence (+Ala) or absence (−Ala) of l-alanine, and β-galactosidase activities were determined. All values provided are the averages of the results from two independent determinations. Error bars indicate the deviations from the means.
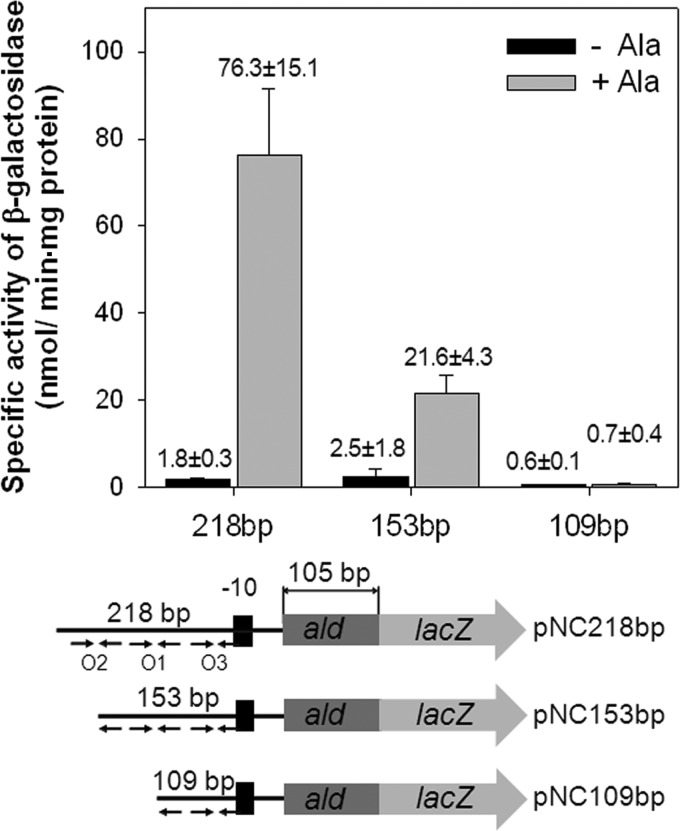
To investigate the role of O1 and O2 in the regulation of ald expression, a series of ald::lacZ transcriptional fusions with 5′-nested deletions of the ald upstream region were employed to study the expression of ald in the presence and absence of l-alanine (Fig. 6). The wild-type strain of M. smegmatis carrying pNC218bp with O1, O2, and O3 showed strong induction of ald expression in the presence of l-alanine. The deletion of the 5′ half of O2 (pNC153bp) severely affected the induction of ald expression by l-alanine, indicating that both O1 and O2 are required for the full induction of ald. When the ald upstream region was further deleted to remove the 5′ half of O1 (pNC109bp), the ald gene was no longer expressed in the presence and absence of l-alanine, indicating that the upstream region between −153 and −109 with regard to the start codon of ald is essential for ald expression.
Fig 6.
Promoter activities of the serially deleted upstream regions of ald in M. smegmatis. The ald promoter activities were determined by using ald::lacZ transcriptional fusions with 5′-nested deletions of the ald upstream region. The DNA fragments cloned into pNC consist of a shared 105-bp 5′ portion of ald and ald upstream regions in the different lengths denoted above the schematic diagrams (218, 153, and 109 bp). The relative positions and the presence or absence of the O1, O2, and O3 sites in the transcriptional fusions are indicated by the arrows. M. smegmatis wild-type strains harboring the transcriptional fusions were grown aerobically in the presence (+Ala) or absence (−Ala) of l-alanine, and β-galactosidase activities were determined. All values provided are the averages of results from two independent determinations. Error bars indicate the deviations from the means.
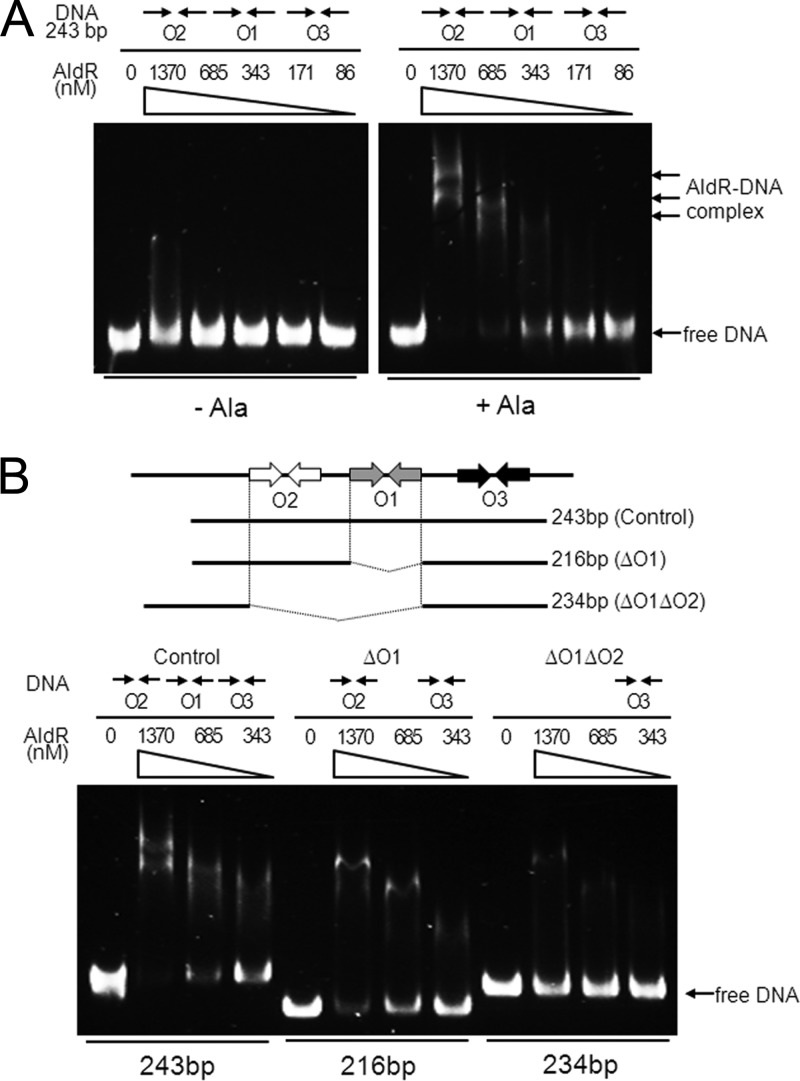
To ascertain whether the identified sites were required for AldR binding in vitro, we performed EMSA with purified AldR and the 243-bp DNA fragment containing the O1, O2, and O3 sequences in the presence or absence of l-alanine (Fig. 7A). The binding affinity of AldR for the DNA fragment was considerably enhanced by l-alanine, indicating that l-alanine serves as a positive effector for AldR. The binding was proportional to the amount of applied AldR, with higher concentrations of the protein causing multiple retarded DNA-protein complexes. The multiple retarded complexes imply that at least two AldR binding sites exist in the DNA fragment. For more detailed analysis of the role of O1, O2, and O3 in the binding of AldR, we carried out EMSA with purified AldR and three different DNA fragments (243, 216, and 234 bp) in the presence of l-alanine (Fig. 7B). The 216- and 234-bp DNA fragments contained both O2 and O3 (ΔO1) and only O3 (ΔO1O2), respectively. The 243-bp DNA fragment containing O1, O2, and O3 was included in the experiment as a control. A smaller amount of DNA was retarded when O1 was deleted from the DNA fragment (ΔO1) than with the control DNA. When both O1 and O2 were deleted (ΔO1O2), the formation of retarded bands was almost abolished. This result indicates that the O1 and O2 sites are required for AldR binding, that O1 and O2 are more favored sites for AldR binding than O3, and that any combination of two binding sites is needed for effective binding of AldR to the ald control region.
Fig 7.
Binding of purified AldR to the ald control region. (A) The 243-bp DNA fragment (10 ng, corresponding to 6.7 nM) encompassing the O1, O2, and O3 sequences was incubated with various concentrations of purified AldR in the presence (+Ala) or absence (−Ala) of 20 mM l-alanine. The concentrations of AldR used are given above the lanes. Native PAGE was run in the presence of 83 mM Tris-borate buffer (pH 8.3) containing 1 mM EDTA, and the gels were stained with SYBR green EMSA gel staining solution. The arrows indicate bands of free DNA and retarded AldR-DNA complexes. (B) Ten ng of each of the DNA fragments (10 ng corresponds to 6.7 nM for control, 7.5 nM for ΔO1, and 6.9 nM for ΔO1O2), which contain O1, O2, and O3 (control), both O2 and O3 (ΔO1), and only O3 (ΔO1O2), were incubated with various concentrations of purified AldR in the presence of 20 mM l-alanine and subjected to native PAGE.
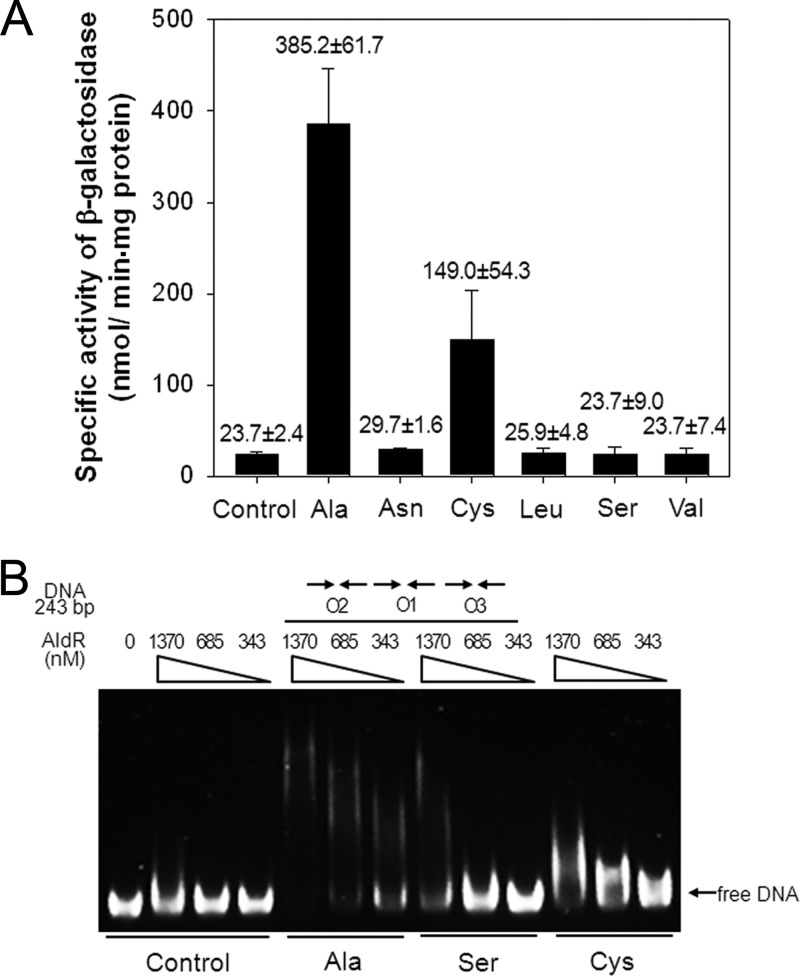
Some Lrp/AsnC family regulators, like E. coli Lrp and M. tuberculosis LrpA, are known to recognize a broad range of amino acids as effector molecules (26, 27). This fact led us to determine the effect of various amino acids on both ald expression and binding of AldR to the ald control region (Fig. 8). In addition to l-leucine and l-asparagine, we examined the l-amino acids (cysteine, serine, and valine) that have structures close to that of alanine. As a positive control, l-alanine was included in the experiment. Consistent with previous results, the addition of l-alanine to the culture of M. smegmatis led to a significant increase in ald expression compared to that of the control without amino acid treatment. Interestingly, expression of ald was 6.3-fold induced by l-cysteine relative to that of the control, while the addition of l-serine did not induce ald expression. The addition of neither l-leucine nor l-asparagine affected expression of ald. Furthermore, EMSA was performed to investigate the effect of l-alanine, l-serine, and l-cysteine on the binding of AldR to the 243-bp DNA fragment containing O1, O2, and O3 (Fig. 8B). Interestingly, the presence of l-cysteine resulted in the formation of AldR-DNA complexes. However, the band pattern of the AldR-DNA complexes in the presence of l-cysteine was different from that in the presence of l-alanine. When l-serine was used in the experiment, the DNA fragment was retarded only with a high concentration of AldR, indicating that l-serine can bind to AldR and alter the binding affinity of AldR for the ald control region, and that the binding affinity or conformation of the serine-bound form of AldR is not enough to induce ald expression in vivo.
Fig 8.
Effect of various amino acids on ald expression and binding of AldR to the ald control region. (A) M. smegmatis wild-type strain containing the ald::lacZ transcriptional fusion plasmid pALDLACZ was grown aerobically in 7H9 glucose medium to an OD600 of 0.5 to 0.6. Following the addition of 25 mM l-amino acids, the strain was further grown for an additional 1 h. The ald promoter activities were measured by determining the β-galactosidase activity. All values provided are the averages of results from two independent determinations. Error bars indicate the deviations from the means. (B) EMSA with the 243-bp DNA fragment (6.7 nM) encompassing O1, O2, and O3 and purified AldR in the presence of 20 mM l-alanine (Ala), l-serine (Ser), or l-cysteine (Cys). The concentrations of AldR used are given above the lanes. Native PAGE was run in the presence of 83 mM Tris-borate buffer (pH 8.3) containing 1 mM EDTA, and the gels were stained with SYBR green EMSA gel staining solution.
Quaternary structure of AldR.
Changes in the quaternary structure of AldR by l-alanine and l-cysteine were examined by means of gel filtration chromatography (see Fig. S1 in the supplemental material). Although the theoretical molecular mass of the N-terminally His6-tagged AldR protein is 19.4 kDa, the molecular mass of purified AldR was found to be 17 kDa in SDS-PAGE (see Fig. S1A). On the basis of the elution volumes in gel filtration chromatography, the molecular mass of native AldR in the absence and presence of l-alanine was estimated to be 41 and 107 kDa, respectively. Considering the molecular mass of the AldR monomer (19.4 kDa), AldR exists as a homodimer in amino acid-free solution. The presence of l-alanine appears to change the oligomerization state of AldR from homodimer to homohexamer. In the presence of l-cysteine, the AldR protein was eluted at the position corresponding to 90 kDa, implying that AldR dimers assemble into homotetramers.
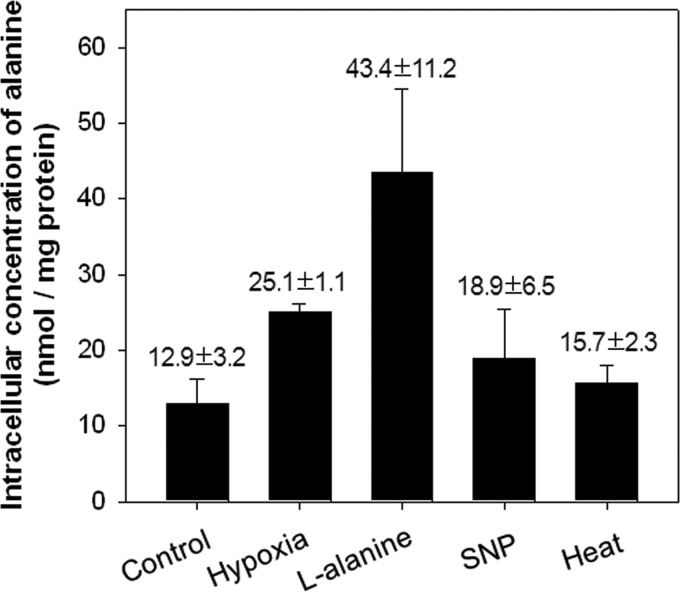
Intracellular concentration of alanine is increased under hypoxic conditions.
The fact that expression of ald was increased under various stress conditions, such as hypoxic, SNP, and heat stress conditions, led us to speculate that the intracellular concentration of alanine is increased under these conditions, thereby resulting in the induction of ald expression by AldR. To explore this hypothesis, we determined the intracellular concentration of l-alanine in the M. smegmatis ΔdevR strain grown under various conditions (hypoxia, l-alanine, SNP, and heat stress). The same strain grown aerobically without any treatment was included in the experiment as a control. As shown in Fig. 9, the M. smegmatis ΔdevR strain grown with the supplementation of l-alanine showed the highest intracellular concentration of l-alanine. The strain grown under hypoxic conditions exhibited an intermediate value between those of the control and l-alanine-supplemented strains. A slightly increased level of l-alanine was detected in the strains grown under SNP and heat conditions. The concentration of l-alanine under various conditions correlated well with the expression levels of ald under the corresponding conditions (Fig. 2), implying that induction of ald expression under hypoxic, SNP, and heat conditions is the result of increased levels of alanine under these conditions.
Fig 9.
Determination of the intracellular alanine concentration in the M. smegmatis ΔdevR mutant strain grown under various conditions. To measure the intracellular alanine levels, the M. smegmatis ΔdevR mutant strain was grown either aerobically (control), under hypoxic conditions for 10 h (hypoxia), or exposed to various conditions (l-alanine, SNP, and 45°C heat) as described in Materials and Methods. The concentration of alanine in M. smegmatis ΔdevR cells was determined by spectrophotometrically measuring the amounts of NADH produced in a B. subtilis l-alanine dehydrogenase-linked enzyme assay. All values provided are the averages of the results from two independent determinations. Error bars indicate deviations from the means.
DISCUSSION
Regulation of ald expression in M. smegmatis: trans- and cis-regulatory elements.
The hspX gene, which is under the control of the DevSR TCS, was strongly upregulated in the wild-type strain of M. smegmatis subjected to the gradual depletion of oxygen for 15 and 20 h (15 and 20 h of hypoxic conditions), while the three identified genes (ald, gap, and serA) that are regulated in a DevSR-independent way were strongly induced under 10 or 15 h of hypoxic conditions, and their expression levels were reduced thereafter (data not shown for gap and serA). This result indicates that expression of ald, gap, and serA is induced under early hypoxic conditions and is distinct from that of the genes under the control of DevR. A common feature of the enzymes encoded by ald, gap, and serA is that they are NADH/NAD+-dependent dehydrogenases, implying that the change in the ratio of NADH to NAD+ in the cells under early hypoxic conditions is a direct or indirect signal for the upregulation of these genes.
Since the hypoxic induction of ald was the strongest of the three genes and a gene (MSMEG_2660; aldR) encoding an Lrp/AsnC family regulator is located immediately upstream of ald, we first chose the ald gene for further study of gene regulation. Expression of ald was far more induced by exogenous l-alanine than by hypoxic stress, and the intracellular level of l-alanine was increased in the cells grown under hypoxic conditions relative to those grown aerobically. These findings led us to hypothesize that the hypoxic induction of ald in M. smegmatis is mediated through an increase in alanine levels in the cells under hypoxic conditions.
A BLAST search revealed that the aldR gene product is a member of the Lrp/AsnC family of regulators. Like other members of this family, AldR is a small DNA binding protein (18.6 kDa) composed of two domains, the N-terminal DNA binding domain (amino acids 41 to 71) containing a helix-turn-helix (HTH) motif and the C-terminal ligand binding domain (amino acids 92 to 163), called RAM (regulation of amino acid metabolism) (28, 29). The C-terminal domain is also known to be involved in the oligomerization of the proteins in this family (29). AldR shows more than 75% sequence identity to its homologs in mycobacteria, and the genetic organization in the gene loci encoding the AldR homologs is well conserved, i.e., the aldR gene is located upstream of and adjacent to ald. AldR also exhibits an overall amino acid sequence identity of 22 to 28% to other Lrp/AsnC family members, e.g., 27% to Lrp of E. coli, 22% to AsnC of E. coli, 22% to LrpC of B. subtilis, 23% to FL11 of Pyrococcus sp. strain OT3, 24% to LrpA of Pyrococcus furiosus, 25% to LrpA of M. tuberculosis H37Rv, and 28% to MdeR of P. putida. Since most Lrp/AsnC family regulators are known to be involved in the regulation of the genes with related amino acid metabolism (30), we assumed the involvement of AldR in ald regulation. Using a ΔaldR mutant of M. smegmatis, we demonstrated that induction of ald expression by exogenous l-alanine and hypoxic stress was abolished in the mutant (data not shown for hypoxic stress). Intriguingly, the ald gene was partially derepressed in the mutant in the absence of l-alanine compared to the wild type and was constitutively expressed in the mutant regardless of the presence or absence of l-alanine. This regulation pattern provided evidence for a dual role of AldR in the control of ald. AldR serves as both an activator for ald expression in the presence of l-alanine and a repressor in the absence of l-alanine. Furthermore, this finding indicates that the ald gene has a promoter that drives a basal level of transcription without help of trans-acting regulatory protein(s). This novel type of positive and negative regulation by a single regulator is known for the regulation of the dadAX operon by Lrp of E. coli in the Lrp/AsnC family (31), and it has been well studied for AraC, which regulates the araBAD operon of E. coli (32). The FdsR regulator belonging to the LysR-type transcriptional regulator family was also reported to act as both activator and repressor to regulate expression of the fdsGBACD operon encoding the soluble formate dehydrogenase of Ralstonia eutropha (33).
The basic assembly unit of AldR appears to be the homodimer, as in other members of the Lrp/AsnC family. The purified AldR protein exists as a homodimer in the absence of l-alanine and appears to assemble into a higher-order structure of homohexamer in the presence of l-alanine, unlike the ring-like octamer structure consisting of four dimers that most members of the Lrp/AsnC family adopt (24, 25, 27, 29, 34–40). Among the Lrp/AsnC regulators, the oligomerization state of the FL11 protein of Pyrococcus sp. strain OT3, LrpA of M. tuberculosis, and Lrp of E. coli was reported to be affected by their cognate ligands, lysine, phenylalanine, and leucine, respectively (25, 27, 38). In the case of FL11, the presence of lysine brings about the assembly of FL11 dimer into homooctamer, while LrpA and Lrp change their quaternary structure from hexadecamer to octamer in the presence of their ligands. Purified Grp of Sulfolobus tokodaii and AsnC of Neisseria meningitides were demonstrated to exist as homooctamers irrespective of the presence or absence of their ligands (glutamine for Grp and leucine for AsnC), but the association of the ligands with the proteins was shown to increase the stability for their octamer formation (37, 39). The increased binding affinity of AldR for the control region of ald and homohexamer formation of AldR in the presence of l-alanine indicate that the higher order form of AldR has a higher binding affinity for the ald control region than the dimeric form. The fact that AldR serves as the repressor for ald expression in the absence of l-alanine indicates that the dimer form of AldR can also bind to the ald control region with a low binding affinity in a manner that represses expression of ald. In this study, we identified two AldR binding sites (O1 and O2) and one putative AldR binding site (O3) upstream of ald (Fig. 3B). The O1 and O2 sites have a consensus sequence of GA-N2-ATC-N-2-TC and are located upstream of the ald promoter. The O1 and O2 sites are separated by 40 bp between the T nucleotides of the central sequences of O1 and O2. The O3 site has the sequence of GA-N2-GTT-N2-TC , and its location overlaps that of the ald promoter. The location of a cis-regulatory DNA sequence determines its function in many cases. Transcriptional activators bind predominantly to positions between −80 and −30 relative to the transcriptional start points in the case of σ70-dependent promoters to recruit RNA polymerase or to promote open complex formation, while many repressor binding sites overlap the promoters to occlude the binding of RNA polymerase to the promoters (41, 42). In this regard, we assume that O1 and O2 are involved in activation of ald expression in the presence of l-alanine, whereas O3 might be responsible for repression of ald expression in the absence of l-alanine. If AldR dimers are arranged in a 3-fold axis to form a ring-like hexamer with the HTH DNA binding domains facing out, the rotational angle between two neighboring dimers would be 120°. It was predicted that the two adjacent dimers bind to the target DNA sequences that are separated by ∼30 bp between the centers of dyad symmetry in the case of the Lrp octamer with an angle of 90°, and that the two neighboring dimers with an angle of 122.5° simultaneously bind to the target DNA sequences separated by ∼40 bp (35, 43), which implies that a molecule of AldR hexamer simultaneously binds to O1 and O2, which are separated by 40 bp, instead of binding to O1 and O3, which are separated by 61 bp. Since the simultaneous binding of AldR to both O1 and O2 seems to be thermodynamically more stable than the binding of AldR to O3 alone, binding of AldR hexamer to O1 and O2 might be favored. When O1 and O2 are occupied by the AldR hexamer and DNA bending occurs as a result, the access of another AldR hexamer to O3 might be occluded by steric hindrance. The AldR hexamer bound to O1 and O2 in the presence of l-alanine results in the recruitment of RNA polymerase to the ald promoter or the promotion of open complex formation at the promoter to activate the transcription of ald. In the absence of l-alanine, each AldR dimer might bind to O1, O2, and O3, but with a much less binding affinity than the AldR hexamer. When the O3 site is occupied by the AldR dimer, ald transcription is repressed. Binding of the AldR dimer to the O1, O2, and O3 binding sites might be cooperative, like many Lrp/AsnC regulators binding to the multiple target sites (44). Therefore, it is possible that mutations in O1 led to partial derepression of ald in the absence of l-alanine through compromised binding of AldR dimer to O3 (Fig. 5).
Exogenous l-cysteine, which has an additional sulfhydryl group at the side chain of l-alanine, also induced the expression of ald in vivo, although it caused a substantially lower level of induction than l-alanine. Furthermore, l-cysteine appears to increase the binding affinity of AldR for the target DNA (Fig. 8). Intriguingly, EMSA analysis and gel filtration chromatography revealed that the quaternary structure of AldR in the presence of l-cysteine (tetramer) is different from that in the presence of l-alanine (Fig. 8; also see Fig. S1 in the supplemental material). It is currently unknown whether l-cysteine plays a role in the regulation of ald in vivo. Further study is required to address this and to confirm the quaternary structure of AldR in the presence of l-cysteine.
Proposed model for hypoxic induction of ald.
Hypoxic induction of ald expression is mediated by AldR, which recognizes the cellular level of l-alanine. When the culture of M. smegmatis undergoes a gradual transition from aerobic conditions to hypoxic conditions, the depletion of oxygen leads to a reduced functionality of the respiratory electron transport chain (ETC). As a result, the redox state of the NADH/NAD+ pool in the cell is shifted toward the more reduced state, and the cellular metabolism producing NADH might be slowed. Under these circumstances, the reductive amination reaction by Ald converting pyruvate to alanine, with the concomitant oxidation of NADH to NAD+, might be accelerated due to an increase in the reactants, thereby producing more alanine, which in turn promotes the transcription of the ald gene through AldR. This positive feedback of ald gene expression helps mycobacteria regenerate NAD+ to maintain the redox state of NADH/NAD+ under hypoxic conditions. In good agreement with this model, it was reported that the growth of an ald knockout mutant of M. smegmatis was compromised under hypoxic conditions (2). The treatment of M. smegmatis cells with SNP, a generator of NO and CN− (45), led to increased expression of ald. We also observed that expression of ald was increased by 27% relative to that of the control when M. smegmatis cells were treated with 500 μM CN− (data not shown). There are two functional terminal oxidases in the ETC of M. smegmatis, aa3 cytochrome c oxidase and bd quinol oxidase (46, 47). The activity of aa3 cytochrome c oxidase is inhibited by NO and CN−, while bd-type quinol oxidase is insensitive to CN− (46). The induction of ald expression by SNP and heat might be a consequence of partial inhibition of the respiratory ETC. Since the intracellular level of alanine reflects the functional state of the respiratory ETC, AldR can recognize not only alanine directly but also respiration-inhibitory signals, such as hypoxia, SNP, heat, etc.
Supplementary Material
ACKNOWLEDGMENT
This work was supported by a Bio-Scientific Research Grant funded by Pusan National University (PNU; Bio-Scientific Research Grant 2010-061-32110001).
Footnotes
Published ahead of print 7 June 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JB.00482-13.
REFERENCES
- 1. Chen JM, Alexander DC, Behr MA, Liu J. 2003. Mycobacterium bovis BCG vaccines exhibit defects in alanine and serine catabolism. Infect. Immun. 71: 708– 716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Feng Z, Caceres NE, Sarath G, Barletta RG. 2002. Mycobacterium smegmatis L-alanine dehydrogenase (Ald) is required for proficient utilization of alanine as a sole nitrogen source and sustained anaerobic growth. J. Bacteriol. 184: 5001– 5010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Giffin MM, Modesti L, Raab RW, Wayne LG, Sohaskey CD. 2012. ald of Mycobacterium tuberculosis encodes both the alanine dehydrogenase and the putative glycine dehydrogenase. J. Bacteriol. 194: 1045– 1054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Hutter B, Dick T. 1998. Increased alanine dehydrogenase activity during dormancy in Mycobacterium smegmatis. FEMS Microbiol. Lett. 167: 7– 11 [DOI] [PubMed] [Google Scholar]
- 5. Betts JC, Lukey PT, Robb LC, McAdam RA, Duncan K. 2002. Evaluation of a nutrient starvation model of Mycobacterium tuberculosis persistence by gene and protein expression profiling. Mol. Microbiol. 43: 717– 731 [DOI] [PubMed] [Google Scholar]
- 6. Usha V, Jayaraman R, Toro JC, Hoffner SE, Das KS. 2002. Glycine and alanine dehydrogenase activities are catalyzed by the same protein in Mycobacterium smegmatis: upregulation of both activities under microaerophilic adaptation. Can. J. Microbiol. 48: 7– 13 [DOI] [PubMed] [Google Scholar]
- 7. Agren D, Stehr M, Berthold CL, Kapoor S, Oehlmann W, Singh M, Schneider G. 2008. Three-dimensional structures of apo- and holo-L-alanine dehydrogenase from Mycobacterium tuberculosis reveal conformational changes upon coenzyme binding. J. Mol. Biol. 377: 1161– 1173 [DOI] [PubMed] [Google Scholar]
- 8. Tripathi SM, Ramachandran R. 2008. Crystal structures of the Mycobacterium tuberculosis secretory antigen alanine dehydrogenase (Rv2780) in apo and ternary complex forms captures “open” and “closed” enzyme conformations. Proteins 72: 1089– 1095 [DOI] [PubMed] [Google Scholar]
- 9. Berney M, Cook GM. 2010. Unique flexibility in energy metabolism allows mycobacteria to combat starvation and hypoxia. PLoS One 5: e8614. 10.1371/journal.pone.0008614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Chan K, Knaak T, Satkamp L, Humbert O, Falkow S, Ramakrishnan L. 2002. Complex pattern of Mycobacterium marinum gene expression during long-term granulomatous infection. Proc. Natl. Acad. Sci. U. S. A. 99:3920– 3925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Rosenkrands I, Slayden RA, Crawford J, Aagaard C, Barry CE, III, Andersen P. 2002. Hypoxic response of Mycobacterium tuberculosis studied by metabolic labeling and proteome analysis of cellular and extracellular proteins. J. Bacteriol. 184: 3485– 3491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Sherman DR, Voskuil M, Schnappinger D, Liao R, Harrell MI, Schoolnik GK. 2001. Regulation of the Mycobacterium tuberculosis hypoxic response gene encoding alpha-crystallin. Proc. Natl. Acad. Sci. U. S. A. 98: 7534– 7539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Raynaud C, Etienne G, Peyron P, Laneelle MA, Daffe M. 1998. Extracellular enzyme activities potentially involved in the pathogenicity of Mycobacterium tuberculosis. Microbiology 144: 577– 587 [DOI] [PubMed] [Google Scholar]
- 14. Kim MJ, Park KJ, Ko IJ, Kim YM, Oh JI. 2010. Different roles of DosS and DosT in the hypoxic adaptation of Mycobacteria. J. Bacteriol. 192: 4868– 4875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Sambrook J, Russell D. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 16. Snapper SB, Melton RE, Mustafa S, Kieser T, Jacobs WR., Jr 1990. Isolation and characterization of efficient plasmid transformation mutants of Mycobacterium smegmatis. Mol. Microbiol. 4: 1911– 1919 [DOI] [PubMed] [Google Scholar]
- 17. Starck J, Kallenius G, Marklund BI, Andersson DI, Akerlund T. 2004. Comparative proteome analysis of Mycobacterium tuberculosis grown under aerobic and anaerobic conditions. Microbiology 150: 3821– 3829 [DOI] [PubMed] [Google Scholar]
- 18. Oh JI, Kaplan S. 1999. The cbb3 terminal oxidase of Rhodobacter sphaeroides 2.4.1: structural and functional implications for the regulation of spectral complex formation. Biochemistry 38: 2688– 2696 [DOI] [PubMed] [Google Scholar]
- 19. Mayuri Bagchi G, Das TK, Tyagi JS. 2002. Molecular analysis of the dormancy response in Mycobacterium smegmatis: expression analysis of genes encoding the DevR-DevS two-component system, Rv3134c and chaperone alpha-crystallin homologues. FEMS Microbiol. Lett. 211: 231– 237 [DOI] [PubMed] [Google Scholar]
- 20. Lee HN, Lee NO, Ko IJ, Kim SW, Kang BS, Oh JI. 2013. Involvement of the catalytically important Asp54 residue of Mycobacterium smegmatis DevR in protein-protein interactions between DevR and DevS. FEMS Microbiol. Lett. 343: 26– 33 [DOI] [PubMed] [Google Scholar]
- 21. Raman S, Song T, Puyang X, Bardarov S, Jacobs WR, Jr, Husson RN. 2001. The alternative sigma factor SigH regulates major components of oxidative and heat stress responses in Mycobacterium tuberculosis. J. Bacteriol. 183: 6119– 6125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Jeong JA, Lee HN, Ko IJ, Oh JI. 2013. Development of new vector systems as genetic tools applicable to mycobacteria. J. Life Sci. 23: 290– 298 [Google Scholar]
- 23. Cui Y, Wang Q, Stormo GD, Calvo JM. 1995. A consensus sequence for binding of Lrp to DNA. J. Bacteriol. 177: 4872– 4880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Koike H, Ishijima SA, Clowney L, Suzuki M. 2004. The archaeal feast/famine regulatory protein: potential roles of its assembly forms for regulating transcription. Proc. Natl. Acad. Sci. U. S. A. 101: 2840– 2845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Yokoyama K, Ishijima SA, Koike H, Kurihara C, Shimowasa A, Kabasawa M, Kawashima T, Suzuki M. 2007. Feast/famine regulation by transcription factor FL11 for the survival of the hyperthermophilic archaeon Pyrococcus OT3. Structure 15: 1542– 1554 [DOI] [PubMed] [Google Scholar]
- 26. Hart BR, Blumenthal RM. 2011. Unexpected coregulator range for the global regulator Lrp of Escherichia coli and Proteus mirabilis. J. Bacteriol. 193: 1054– 1064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Shrivastava T, Ramachandran R. 2007. Mechanistic insights from the crystal structures of a feast/famine regulatory protein from Mycobacterium tuberculosis H37Rv. Nucleic Acids Res. 35: 7324– 7335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Ettema TJ, Brinkman AB, Tani TH, Rafferty JB, van Der Oost J. 2002. A novel ligand-binding domain involved in regulation of amino acid metabolism in prokaryotes. J. Biol. Chem. 277: 37464– 37468 [DOI] [PubMed] [Google Scholar]
- 29. Leonard PM, Smits SH, Sedelnikova SE, Brinkman AB, de Vos WM, van der Oost J, Rice DW, Rafferty JB. 2001. Crystal structure of the Lrp-like transcriptional regulator from the archaeon Pyrococcus furiosus. EMBO J. 20: 990– 997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Calvo JM, Matthews RG. 1994. The leucine-responsive regulatory protein, a global regulator of metabolism in Escherichia coli. Microbiol. Rev. 58: 466– 490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Zhi J, Mathew E, Freundlich M. 1999. Lrp binds to two regions in the dadAX promoter region of Escherichia coli to repress and activate transcription directly. Mol. Microbiol. 32: 29– 40 [DOI] [PubMed] [Google Scholar]
- 32. Schleif R. 1996. Two positive regulated systems, ara and mal, p 1300–1309 In Curtiss IR, Ingraham JL, Lin ECC, Low KB, Magasanik B, Reznikoff WS. (ed), Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed. ASM Press, Washington, DC [Google Scholar]
- 33. Oh JI, Bowien B. 1999. Dual control by regulatory gene fdsR of the fds operon encoding the NAD+-linked formate dehydrogenase of Ralstonia eutropha. Mol. Microbiol. 34: 365– 376 [DOI] [PubMed] [Google Scholar]
- 34. Thaw P, Sedelnikova SE, Muranova T, Wiese S, Ayora S, Alonso JC, Brinkman AB, Akerboom J, van der Oost J, Rafferty JB. 2006. Structural insight into gene transcriptional regulation and effector binding by the Lrp/AsnC family. Nucleic Acids Res. 34: 1439– 1449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. De los Rios S, Perona JJ. 2007. Structure of the Escherichia coli leucine-responsive regulatory protein Lrp reveals a novel octameric assembly. J. Mol. Biol. 366: 1589– 1602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Reddy MC, Gokulan K, Jacobs WR, Jr, Ioerger TR, Sacchettini JC. 2008. Crystal structure of Mycobacterium tuberculosis LrpA, a leucine-responsive global regulator associated with starvation response. Protein Sci. 17: 159– 170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Ren J, Sainsbury S, Combs SE, Capper RG, Jordan PW, Berrow NS, Stammers DK, Saunders NJ, Owens RJ. 2007. The structure and transcriptional analysis of a global regulator from Neisseria meningitidis. J. Biol. Chem. 282: 14655– 14664 [DOI] [PubMed] [Google Scholar]
- 38. Chen S, Calvo JM. 2002. Leucine-induced dissociation of Escherichia coli Lrp hexadecamers to octamers. J. Mol. Biol. 318: 1031– 1042 [DOI] [PubMed] [Google Scholar]
- 39. Kumarevel T, Nakano N, Ponnuraj K, Gopinath SC, Sakamoto K, Shinkai A, Kumar PK, Yokoyama S. 2008. Crystal structure of glutamine receptor protein from Sulfolobus tokodaii strain 7 in complex with its effector L-glutamine: implications of effector binding in molecular association and DNA binding. Nucleic Acids Res. 36: 4808– 4820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Yamada M, Ishijima SA, Suzuki M. 2009. Interactions between the archaeal transcription repressor FL11 and its coregulators lysine and arginine. Proteins 74: 520– 525 [DOI] [PubMed] [Google Scholar]
- 41. Raibaud O, Schwartz M. 1984. Positive control of transcription initiation in bacteria. Annu. Rev. Genet. 18: 173– 206 [DOI] [PubMed] [Google Scholar]
- 42. Collado-Vides J, Magasanik B, Gralla JD. 1991. Control site location and transcriptional regulation in Escherichia coli. Microbiol. Rev. 55: 371– 394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Shrivastava T, Dey A, Ramachandran R. 2009. Ligand-induced structural transitions, mutational analysis, and ‘open’ quaternary structure of the M. tuberculosis feast/famine regulatory protein (Rv3291c). J. Mol. Biol. 392: 1007– 1019 [DOI] [PubMed] [Google Scholar]
- 44. Brinkman AB, Ettema TJ, de Vos WM, van der Oost J. 2003. The Lrp family of transcriptional regulators. Mol. Microbiol. 48: 287– 294 [DOI] [PubMed] [Google Scholar]
- 45. Friederich JA, Butterworth JF. 1995. Sodium nitroprusside: twenty years and counting. Anesth. Analg. 81: 152– 162 [DOI] [PubMed] [Google Scholar]
- 46. Kana BD, Weinstein EA, Avarbock D, Dawes SS, Rubin H, Mizrahi V. 2001. Characterization of the cydAB-encoded cytochrome bd oxidase from Mycobacterium smegmatis. J. Bacteriol. 183: 7076– 7086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Megehee JA, Hosler JP, Lundrigan MD. 2006. Evidence for a cytochrome bcc-aa3 interaction in the respiratory chain of Mycobacterium smegmatis. Microbiology 152: 823– 829 [DOI] [PubMed] [Google Scholar]
- 48. Jessee J. 1986. New subcloning efficiency competent cell: >1×106 transformants/μg. Focus 8: 9 [Google Scholar]
- 49. Oh JI, Park SJ, Shin SJ, Ko IJ, Han SJ, Park SW, Song T, Kim YM. 2010. Identification of trans- and cis-control elements involved in regulation of the carbon monoxide dehydrogenase genes in Mycobacterium sp. strain JC1 DSM 3803. J. Bacteriol. 192:3925– 3933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Tabor S, Richardson CC. 1985. A bacteriophage T7 RNA polymerase/promoter system for controlled exclusive expression of specific genes. Proc. Natl. Acad. Sci. U. S. A. 82: 1074– 1078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Howard NS, Gomez JE, Ko C, Bishai WR. 1995. Color selection with a hygromycin-resistance-based Escherichia coli-mycobacterial shuttle vector. Gene 166: 181– 182 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.