Abstract
The flagellar filament enables bacteria to swim by functioning as a helical propeller. The filament is a supercoiled assembly of a single protein, flagellin, and is formed by 11 protofilaments arranged in a circle. Bacterial swimming and tumbling correlate with changes of the various helical structures, called polymorphic transformation, that are determined by the ratios of two distinct forms of protofilaments, the L and R types. The polymorphic transformation is caused by transition of the protofilament between L and R types. Elucidation of this transition mechanism has been addressed by comparing the atomic structures of L- and R-type straight filaments or using massive molecular dynamic simulation. Here, we found amino acid residues required for the transition of the protofilament using fliC-intragenic suppressor analysis. We isolated a number of revertants producing supercoiled filaments from mutants with straight filaments and identified the second-site mutations in all of the revertants. The results suggest that Asp107, Gly426, and Ser448 and Ser106, Ala416, Ala427, and Arg431 are the key residues involved in inducing supercoiled filaments from the R- and the L-type straight filaments, respectively. Considering the structures of the R- and L-type protofilaments and the relationship between the rotation of the flagellar motor and the polymorphic transformation, we propose that Gly426, Ala427, and Arg431 contribute to the first stage of the transition and that Ser106, Asp107, and Ala416 play a role in propagating the transitions along the flagellar filament.
INTRODUCTION
The bacterial flagellum is a nanomachine that makes it possible for a bacterial cell to swim freely. Bacteria change their swimming pattern when they sense stimuli, such as chemicals (1–3) and temperature changes (4, 5), and consequently move toward a more favorable environment. The stimuli are sensed by receptor proteins in the cell membrane (6) that send signals through protein phosphorylation (7, 8) to the motor that rotates the flagellar filament (9–11; for reviews, see references 12–15).
The flagellar filament of Salmonella enterica serovar Typhimurium (S. Typhimurium), composed of ∼30,000 molecules of a single protein called flagellin (494 amino acid residues), forms a tubular and left-handed supercoiled structure with a helical pitch of 2.3 μm and ∼0.4 μm in diameter (16) and is called a “normal” filament. Typically, 7 to 10 normal filaments are present on a cell. When the normal filaments rotate counterclockwise (CCW), viewed from outside the cell, the filaments form a bundle behind the cell, and the bundle rotates like a helical propeller. Rapid reversal of the motor rotation from CCW to clockwise (CW) subjects the filaments to a twisting force, so that they transform from the normal to right-handed helical forms (17, 18), and the bundle is then separated into individual filaments. Consequently, bacteria tumble during this process (17, 18) for ∼0.14 s (18). The filament then transforms from the right-handed “semicoiled” filament to the right-handed and long-pitch “curly I” filament that propels the cell in a new direction (18, 19). Finally, after flagellar rotation returns to CCW, the filament assumes its normal shape (18, 19). Normal filaments reform the bundle, and the cell resumes its initial swimming speed (18, 19). Thus, the cell swims smoothly and is able to change direction by employing this polymorphic transformation.
The bistable protofilament model (20–23) is widely accepted to explain the mechanism that drives polymorphic transformation. Electron microscopy revealed that the structure of the Salmonella filament comprises 11 protofilaments (24). If the conformation of all protofilaments is identical, filaments must be straight. In contrast, if two types of protofilaments exist, which have distinct intersubunit repeat distances and distinct inclinations, resulting curvature and twists of filaments are intrinsic. Consequently, the flagellar filament forms a supercoil. Furthermore, by the ratio of the two types of protofilaments (L or R) within 11 protofilaments, the formations of 10 distinct shapes of the supercoiled structure were predicted, e.g., 9L/2R = normal, 7L/4R = semicoiled, and 6L/5R = curly I, and two types of straight filaments were predicted, 11L/0R or 0L/11R.
Salmonella strains SJW1660 and SJW1655 carry mutations in their flagellin genes that encode G426A and A449V that lead to the formation of the L- and R-type straight filaments, respectively (25). Further, several types of helical filaments and two types of straight filaments were reconstructed by mixing various ratios of the L- and R-type flagellin monomers prepared from the L- and R-type straight filaments (26). Therefore, the bistable protofilament model is supported by much of this evidence; however, the mechanism for the polymorphic transformation at the atomic level, in particular the transition between the L and R protofilaments (L/R transition), is unknown.
The first trial to elucidate the mechanism responsible for the L/R transition at the atomic level was the massive molecular dynamics (MD) simulation conducted by Kitao et al. (27). These researchers simulated supercoiled structures from the complete atomic structure of the R-type straight filament as determined using electron cryomicroscopy (28). In the second study, Maki-Yonekura et al. determined the complete atomic structure of the L-type straight filament using electron cryomicroscopy (29) and compared it to that of the R-type straight filament (28). The third study, conducted in our laboratory, reported key residues in the L/R transition using fliC-intragenic suppressor analysis (30). As described above, the polymorphic transformation can be regarded as the interconversion between L- and R-type protofilaments. If the L-type straight filament composed of 11 L-type protofilaments is changed to a supercoiled form by a second-site mutation in flagellin molecule, the amino acid residue before substitution at the second site is likely to be significant for the L/R transition in protofilaments. In the previous work, we isolated 106 motile revertants from SJW1660 having L-type straight filaments and identified 27 second mutation sites responsible for supercoiling (30).
In the present study, we applied fliC-intragenic suppressor analysis to SJW1655, which produces the R-type straight filaments. In addition, we isolated two additional mutants (HFG180 and HFG195) that produce L-type straight filaments and attempted to identify the amino acid residues involved in the L/R transition by using the same method. By comparing the residues found in these analyses, we determined a limited number of residues that are significant for the L/R transition.
MATERIALS AND METHODS
Isolation of the straight-flagellar mutants HFG180 and HFG195.
S. Typhimurium LT2 strain SJW1103 (wild type [WT]) was grown in Luria-Bertani broth (LB) overnight, and the cells were then kept at room temperature for 4 days. The cells were streaked onto fresh LB plates and incubated at 37°C overnight. Single colonies (∼7,000) were stabbed into soft-agar plates (LB broth containing 0.3% agar), and the plates were incubated at 37°C for several hours. After incubation, we isolated 222 mutants that did not form a swarm. The mutated genes responsible for this phenotype were determined using bacteriophage P22-mediated transduction (31, 32), and five fliC mutants were identified. Two mutants lacked flagellar filaments, and one produced curly filaments. Two (designated HFG180 and HFG195) that produced straight filaments were used to perform the experiments described below.
Preparation and copolymerization of flagellin monomers.
Strains SJW1660, SJW1655, HFG180, and HFG195 were cultured in LB broth (1 liter) overnight. Polyethylene glycol 6000 (2% final concentration) and NaCl (0.5 M final concentration) were added to the cultures, and the mixtures were kept at 4°C for 2 h. Cells and filaments were centrifuged at 20,000 × g for 20 min, and the pellets were resuspended in 20 ml of 10 mM potassium phosphate buffer (pH 7.4). Flagellar filaments attached to the cells were removed using a Waring blender. The cells were harvested by centrifugation at 16,890 × g for 20 min, and the supernatant containing the flagellar filaments was centrifuged at 100,000 × g for 1 h. The pellet was suspended in 1 ml of 10 mM potassium phosphate buffer (pH 7.4), and the suspension was incubated at 65°C for 5 min to depolymerize the flagellar filaments to flagellin monomers. This preparation was centrifuged at 543,000 × g for 30 min, and the supernatant containing the flagellin monomers was collected. Ammonium sulfate (1 M final concentration) was added to the supernatant to polymerize the flagellin monomers. After 1 h, the resultant polymers (very short filaments not detected by dark-field microscopy) were harvested by centrifugation at 543,000 × g for 30 min. To remove ammonium sulfate, the pellet was washed with an appropriate volume of 10 mM potassium phosphate buffer (pH 7.4), and the wash solution was discarded after ultracentrifugation. The final preparation of flagellin polymers was resuspended in 10 mM potassium phosphate buffer (pH 7.4) and stored at 4°C.
To prepare SJW1660, SJW1655, HFG180, and HFG195 flagellin monomers for copolymerization assays, we added NaCl (0.2 M final concentration) to the flagellin polymer solutions before heat treatment (65°C for 5 min), and the flagellin monomers were collected by ultracentrifugation (543,000 × g for 30 min). The protein concentration of the resultant supernatant was adjusted to 5 mg/ml with 10 mM potassium phosphate buffer (pH 7.4) containing 0.2 M NaCl. Various combinations of mutant flagellins were mixed at various ratios, and then a short fragment of flagellar filament was added to seed polymerization. The mixtures were incubated at 4°C for 21 to 125 h. After incubation, they were diluted 20 to 30 times with 10 mM potassium phosphate buffer (pH 7.4) and observed using dark-field microscopy.
Isolation of revertants of SJW1655, HFG180, and HFG195.
A large number of colonies of SJW1655, HFG180, and HFG195 were selected from LB plates and stabbed into soft-agar plates and incubated at 37°C for 12 to 96 h. Swarms occasionally appeared. A revertant was picked from the fringe of each swarm.
Determination of the first and the second mutation sites.
The first mutation sites in HFG180 and HFG190 and the second mutation sites of all revertants were determined using DNA sequence analysis. The DNA of fliC (1,485 bp) and the sequences flanking (90 bp) both 5′ and 3′ ends of fliC were amplified using the PCR, primers, and genomic DNA templates prepared from the mutants and revertants. The nucleotide sequences of the purified PCR products were determined using a DYEnamic ET terminator cycle sequencing kit (GE Healthcare) and an ABI Prism 310 genetic analyzer (Applied Biosystems).
Swarm rate and swimming-speed assays.
Single colonies of the revertants were stabbed into the soft-agar plates and incubated at 37°C for 3 h. Cells able to swim and tumble form a swarm in soft agar. The swarm rate was measured as the diameter of the swarm after incubation. The cells on the fringe of the swarm were picked, inoculated into LB liquid medium, and incubated at 37°C overnight. A 5-μl sample of the culture was reinoculated into 5 ml of fresh LB liquid medium and further incubated at 37°C. Early-log-phase cells were kept at 25°C for 0.5 h. The cells were then observed using a CH-2 phase-contrast microscope (Olympus). The images were recorded using a complementary metal oxide semiconductor image sensor. Swimming speeds were analyzed using the two-dimensional dynamic image analysis program Move-tr/2D (Library). The swarm rate and the swimming speed of each revertant were converted to values relative to those of WT.
Observations of flagellar filaments.
We observed flagellar filaments attached or detached from cells. Each strain (SJW1655, HFG180, and HFG195), their revertants, and SJW1103 were cultured in LB broth (5 ml) overnight. Then, 10 μl of culture was diluted 10 to 20 times with fresh LB liquid medium, and filaments attached to the cells were observed under the dark-field microscopy with a customized BX50 (Olympus). One milliliter of the culture was transferred to a 1.5-ml tube and vortexed to detach the flagellar filaments from cells. After centrifugation (15,000 × g for 10 min), the filaments in the supernatant were observed under the dark-field microscopy. Filament images were recorded using an ultrasensitive black-and-white charge-coupled device camera, a CCD ROX 40B (Bel-Tex) or a WAT-902H Ultimate (Watec Co., Ltd.). Selected frames of the video are presented in Fig. 5. The helical handedness of the supercoiled filament was determined by raising and lowering the sample stage of the microscope to focus on the bottom and top of the filament on the glass slide (33). The other helical parameters of the filaments, pitch and the helical diameter, were determined from the images according to a published method (30).
Mapping the second mutation sites on flagellin subunits.
PDB files for drawing neighboring 9 (3 by 3) flagellin subunits in L- and R-type straight filaments were a gift from K. Namba, K. Yonekura, and S. Maki-Yonekura. The locations of the second mutation sites were visualized using the molecular graphics program MolMol (34).
Multiple alignments of FliC sequences.
Amino acid sequence data were obtained from the Microbial Genome Database (MBGD), provided by National Institute for Basic Biology, National Institutes of Natural Sciences, Japan. Twenty-seven FliC sequences from various genera of bacteria were selected and aligned using CLUSTAL W (35).
RESULTS
Two new mutants producing the L-type straight filament.
We isolated two mutants, HFG180 and HFG195, whose flagellar filaments appeared straight and extended from cells in all directions (Fig. 1). Detached filaments from both mutants were also straight (Fig. 1). These properties are consistent with those of the straight-filament mutants described previously (36).
Fig 1.
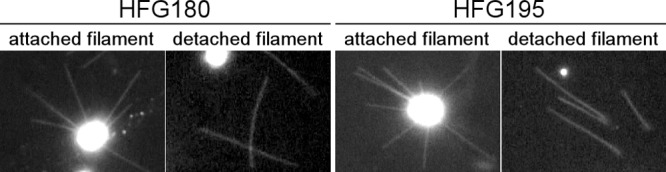
Flagellar filaments of HFG180 and HFG195. Flagellar filaments attached or detached from cells are shown using dark-field microscopy.
To determine whether these filaments are the R or L type, we performed copolymerization assays using the R and L types of flagellin monomers prepared from SJW1655 and SJW1660, respectively (Fig. 2). At first, we confirmed that polymerization of only the R-type flagellin monomers or only the L-type flagellin monomers produced straight filaments (Fig. 2a and b) and that mixing of the R- and L-type flagellin monomers produced supercoiled filaments (Fig. 2c and d). These results are consistent with the results described by Kamiya et al. (26) and confirm that supercoiled filaments were produced by copolymerization of different types of flagellin monomers.
Fig 2.
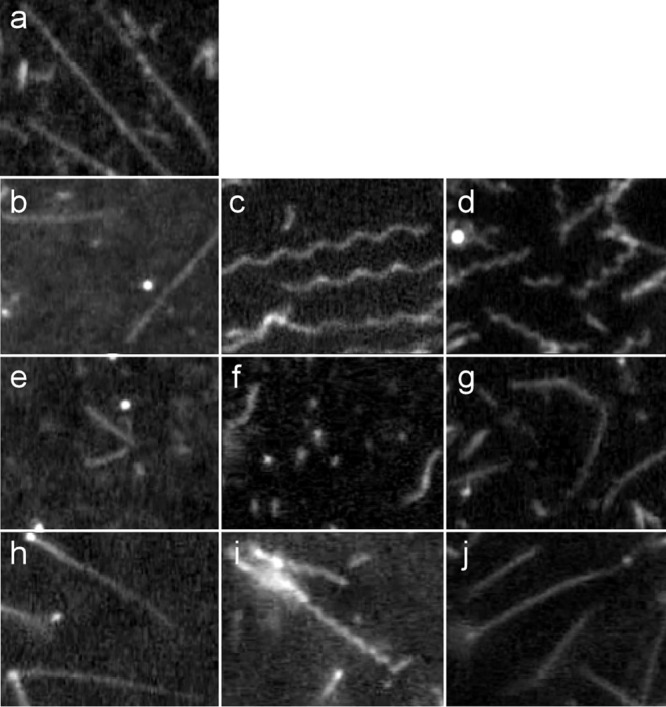
Products of copolymerization of flagellin monomers from SJW1655, SJW1660, HFG180, and HFG195 at various mixing ratios. (a) 100% SJW1655 flagellin; (b) 100% SJW1660 flagellin; (c) SJW1655:SJW1660 = 2:9; (d) SJW1655: SJW1660 = 4:7; (e) 100% HFG180 flagellin; (f) SJW1655:HFG180 = 4:7; (g) SJW1660:HFG180 = 4:7; (h) 100% HFG195 flagellin; (i) SJW1655:HFG195 = 4:7; (j) SJW1660:HFG195 = 2:9. The seeds for promoting copolymerization are as follows: SJW1655 derived (a, c, and d); SJW1660 derived (b, g, and j); HFG180 derived (e and f); and HFG195 derived (h and i).
Polymerization of HFG180 flagellin monomers alone produced straight filaments, although the polymerization efficiency was poor and their lengths were short (Fig. 2e). Copolymerization of HFG180 and SJW1655 flagellin monomers in the 7:4 ratio produced filaments similar to normal filaments, but their polymerization efficiency was still as poor as that of HFG180 flagellin monomers alone (Fig. 2f). We observed filaments in this ratio, whereas we did not observe any filaments at mixing ratios of 9:2, 4:7, and 2:9. In contrast, HFG180 and SJW1660 flagellin added together in the 7:4 ratio produced straight filaments (Fig. 2g). Polymerization of HFG195 flagellin monomers alone also produced straight filaments (Fig. 2h). Copolymerization of HFG195 and SJW1655 flagellin monomers in the 7:4 ratio produced curly I-like filaments (Fig. 2i), but at the 9:2 ratio, there was no filament observed. Copolymers of HFG195 and SJW1660 (9:2) were straight (Fig. 2j). From these results, the filaments produced by HFG180 and HFG195 were both identified as L types as SJW1660 does, but their abilities to polymerize are somewhat different.
The mutations in HFG180 and HFG195 were G70C and A80V, respectively. Gly70 and Ala80 are both located on helix 2 of the flagellin molecule (Fig. 3). Ala449 and Gly426, which are the mutations of SJW1655 and SJW1660, respectively, are located on helix 4. Thus, the two new mutations are different from the traditional two sites in terms of the helix on which they are located; however, the locations of Gly70 and Ala80 are relatively close to Gly426 in the three-dimensional structure (Fig. 3).
Fig 3.
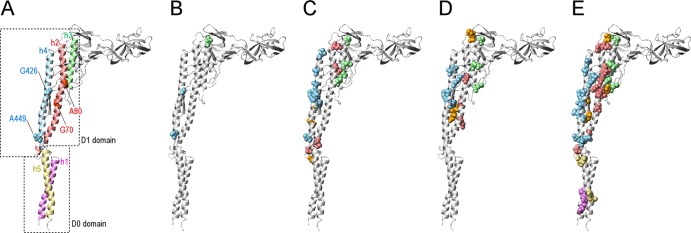
Locations of the first mutation and second mutation sites on the ribbon model of the R-type flagellin. (A) The helices 1, 2, 3, 4, and 5 are represented by violet, light coral, pale green, sky blue, and khaki, respectively. The helices 1and 5 and 2, 3, and 4 are categorized into D0 and D1 domains, respectively. The first mutation sites Gly426, Ala449, Gly70, and Ala80 are represented using a CPK model. Gly426 and Ala449 are sky blue, and Gly70 and Ala80 are light coral. (B to E) The second mutation sites of SJW1655, SJW1660, HFG180, and HFG195 revertants, respectively. The second mutation sites are represented by a CPK model. The second mutation sites located on the helices 1, 2, 3, 4, and 5 are violet, light coral, pale green, sky blue, and khaki, respectively. The residues not located on helices 1 to 5 are orange.
Revertants of SJW1655, HFG180, and HFG195.
We isolated 94, 77, and 88 revertants from SJW1655, HFG180, and HFG195, respectively, and determined the second mutation sites in all of them. The second mutation sites in the revertants from SJW1660 has previously been reported (30). All of the second mutations, including those of SJW1660, are summarized for each parental strain in Fig. S1 in the supplemental material. Notable features were observed in the SJW1655 revertants (termed R-straight revertants); it was difficult to obtain pseudorevertants (Table 1), and there were only three second-site mutations: Asp107, Ala426, and Ser448. In contrast, in the L-straight revertants from SJW1660, HFG180, and HFG195 (revertants from the L-type straight filament strains), the numbers of the second mutation sites were much higher than that of SJW1655 revertants. These results suggest that the R-type straight filament may be thermodynamically more stable than the L type.
Table 1.
Summary of parent strains, their filament types, the numbers of pseudorevertants isolated, and the numbers of second mutation sites of revertants in each parent strain
| Parent strain (mutation) | Straight type | No. of pseudorevertantsa | Yield (%)b | No. of second mutation sitesc |
|---|---|---|---|---|
| SJW1655 (A449V) | R | 52 | 1.7 | 3 |
| SJW1660 (G426A) | L | 101 | 18 | 19 |
| HFG180 (G70C) | L | 44 | 26 | 15 |
| HFG195 (A80V) | L | 81 | 56 | 49 |
“Pseudorevertants” represent only pseudorevertants having different mutation sites between the first and the second sites.
“Yield” represents the percentage of the number of the pseudorevertants acquired versus the number of single colonies stabbed on soft-agar plates.
If the second mutation occurred on the first mutation site, even if the mutation was not true revert mutation, the site was not counted as the second mutation site. For example, the first mutation of SJW1655 was A449V, and some revertants had the A449G mutation as the second mutation. In that case, Ala449 was not counted as the second mutation site.
The locations of the second mutation sites were mapped on the R-type flagellin molecule (Fig. 3). The second mutation sites of the R-straight revertants are mapped only to the D1 domain (Fig. 3B), and most of those of the L-straight revertants are also located on the D1 domain (Fig. 3C to E).
Key amino acid residues for L/R transition.
We found only four mutations—D107E, G426A, G426S, and S448F—that may be involved in transforming the R-type straight filament to supercoiled filaments. In contrast, we obtained many revertants from the L-straight mutants, HFG180 and HFG195; the numbers of the second mutation sites were 15 and 49, respectively. We have previously reported the identification of 19 from SJW1660 (30). Among these three L-straight mutants, the common second sites are Ser106, Ala416, Ala427, and Arg431.
Asp107, Gly426, and Ser448 of the R-straight revertants and Ser106, Ala416, Ala427, and Arg431 of the L-straight revertants are mapped on the R-type flagellin structures shown in Fig. 4. It is noteworthy that the second mutation sites of the R-straight revertants and those of the L-straight revertants described above do not overlap. Further, the remaining second mutation sites of the L-straight revertants are not present in the R-straight revertants. That is, the revertants from SJW1660, HFG180, and HFG195 did not have mutations at Asp107, Gly426, or Ser448 (except the mutations at Gly426 of SJW1660 revertants, because Gly426 was the first mutation site). Another interesting aspect is that Asp107 and Gly426 of the R-straight revertants and Ser106 and Ala427 of the L-straight revertants are next to each other, respectively.
Fig 4.
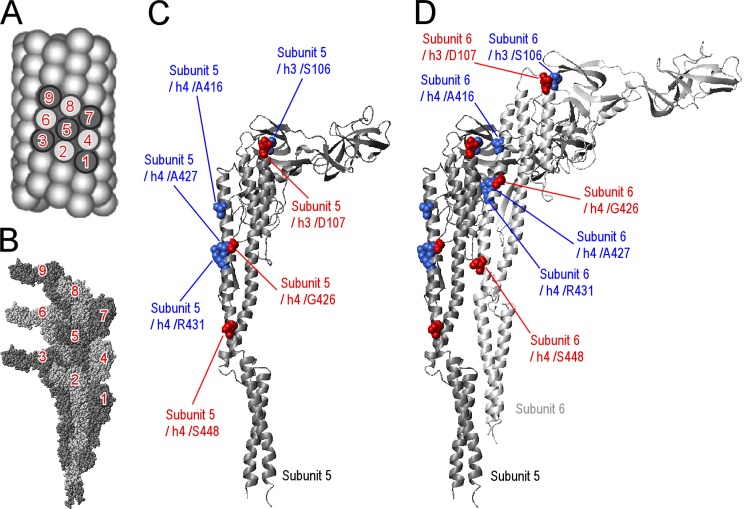
Locations of the key residues for the L/R transition. (A) The surface lattice of R-type straight filament is drawn with spheres, which represent flagellin subunits. The numbers on flagellin subunits are expediential to represent the orientation of the nine subunits. (B) The atomic structure and arrangement of nine subunits in R-type straight filament are displayed. The numbers are compatible with those of panel A. (C) The key residues are represented by a CPK model of subunit 5. The key residues determined from R-straight revertants are red, and those from L-straight revertants are blue. (D) Subunit 6 is also displayed. Panel A was modified from Quarterly Reviews of Biophysics (41) with permission of the publisher. The present subunit numbers, 1 to 9, correspond to the −16, −11, −6, −5, 0, 5, 6, 11, and 16 in a previous report (27). The latter numbering is based on the helical symmetry of the flagellar filament.
These findings indicate that the residues that are preferred in the L-type and R-type protofilaments are completely different but that they are adjacent in the three-dimensional flagellin structure in certain cases. Although Ala416 is distant from Asp107 and Ser106 in the flagellin monomer, considering subunits 5 and 6 (Fig. 4A and B), Ala416 is located at the interface of these subunits and forms a cluster with Ser106 and Asp107 (Fig. 4D). These results suggest that two clusters, one containing Ser106, Asp107, and A416 and the other containing Gly426, Ala427, and Arg431, mainly contribute to L/R transitions.
Swimming and swarming of the revertants with mutations of the key residues.
We investigated the swimming speed in the LB liquid medium and the swarming rate on the soft-agar gels of the revertants with mutations at the key sites. The L-straight revertants swam at >60% relative speed to the WT, indicating that the second mutations induced supercoiling in their filaments. In contrast, the swarms of many of the revertants were smaller than 60% of WT. These results suggest that their abilities to undergo polymorphic transformation were partly affected.
Among the R-straight revertants, SJW1655-D73(A449V, G426A) swam at about half the speed of the WT, whereas the rests swam very poorly (Table 2). Interestingly, SJW1655-D42(A449V, G426S) did not swim after 3 h in culture but did swim after a 21-h culture at a rate that was 84.8% of the WT.
Table 2.
Summary of the second mutations, relative swimming speeds, and relative swarm rates of representatives of the revertants
| Parent strain | Revertant no. | Second mutation | Mean relative value ± SD |
|
|---|---|---|---|---|
| Swimming speed | Swarming rate | |||
| SJW1655 | D1340 | D107E | 0.08 ± 0.09 | 0.05 ± 0.01 |
| D42 | G426S | 0.08 ± 0.09 | 0.37 ± 0.00 | |
| D73 | G426A | 0.55 ± 0.27 | 0.23 ± 0.06 | |
| D1824 | S448F | 0.06 ± 0.07 | 0.07 ± 0.00 | |
| SJW1660 | D561 | S106Y | 0.93 ± 0.24 | 0.29 ± 0.02 |
| D570 | A416V | 0.69 ± 0.22 | 0.17 ± 0.00 | |
| D455 | A427G | 1.00 ± 0.27 | 0.07 ± 0.00 | |
| D489 | A427V | 0.88 ± 0.32 | 0.22 ± 0.00 | |
| D520 | A427T | 1.01 ± 0.34 | 0.17 ± 0.00 | |
| D253 | R431S | 0.61 ± 0.19 | 0.11 ± 0.00 | |
| D276 | R431C | 0.91 ± 0.29 | 0.09 ± 0.00 | |
| D541 | R431H | 0.68 ± 0.21 | 0.11 ± 0.03 | |
| HFG195 | D59 | S106F | 0.67 ± 0.21 | 0.40 ± 0.00 |
| D144 | A416T | 1.08 ± 0.30 | 0.39 ± 0.07 | |
| D87 | A427E | 0.82 ± 0.32 | 0.42 ± 0.03 | |
| D132 | A427T | 1.12 ± 0.31 | 0.35 ± 0.04 | |
| D135 | A427V | 1.17 ± 0.31 | 0.67 ± 0.04 | |
| D130 | R431C | 1.01 ± 0.24 | 0.49 ± 0.00 | |
| HFG180 | D68 | S106Y | 0.94 ± 0.32 | 0.71 ± 0.04 |
| D77 | S106F | 0.68 ± 0.24 | 0.44 ± 0.00 | |
| D121 | A416V | 0.82 ± 0.29 | 0.59 ± 0.00 | |
| D29 | A427V | 0.73 ± 0.28 | 0.78 ± 0.03 | |
| D63 | A427S | 0.83 ± 0.28 | 0.73 ± 0.00 | |
| D147 | A427T | 0.92 ± 0.29 | 0.57 ± 0.11 | |
| D14 | R431C | 0.60 ± 0.22 | 0.60 ± 0.09 | |
Values of relative swimming speed and relative swarming rate represent means of triplicate assays. The swimming speed and the swarming rate of the wild type were 25.8 ± 3.3 µm/s and 18.1 ± 3.8 mm, respectively. The data for the revertants from SJW1660 were from a previous report (30).
Filament morphology.
We determined the shapes of flagellar filaments detached from cells and other properties during swimming using dark-field microscopy (Fig. 5). The detached filament shape is most stable thermodynamically, whereas the attached filament shape during swimming is subjected to torsion by rotation of the motor. Here, we define the shapes of the detached and attached filaments as “static filament shape” and “dynamic filament shape,” respectively.
Fig 5.
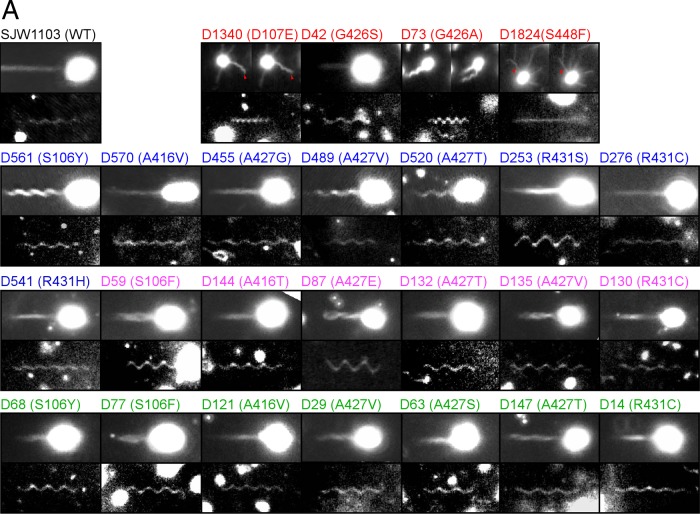
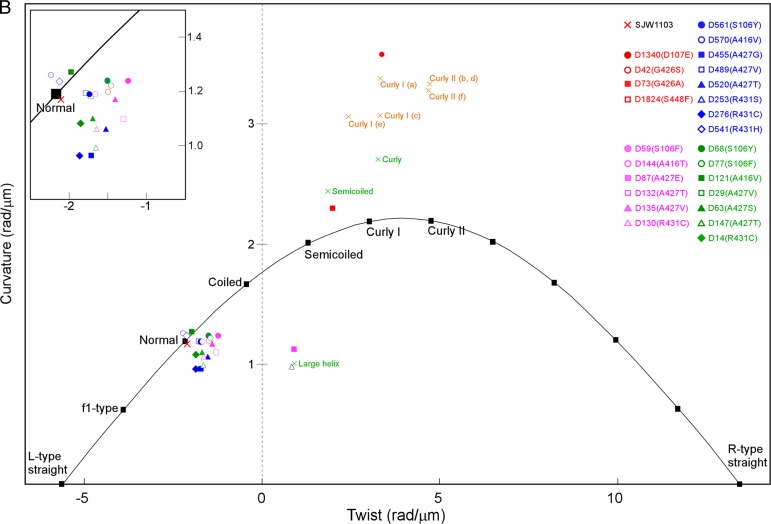
Shapes of flagellar filaments. (A) Images of the dynamic filament (upper pictures) and static filament shapes (lower) from WT and the 25 revertants. Dxxx (x represents a number) and parenthetical letters represent revertant number and amino acid substitution, respectively. The numbers in red, blue, magenta, and green represent SJW1655, SJW1660, HFG195, and HFG180 revertants, respectively. Two images are displayed to illustrate the dynamic filaments shape in D1340, D73, and D1824. In D1340 and D1824, the left and the right images are sequential snap shots (the right was taken 0.03 s after the left). In D73, the right was taken 0.27 s after the left. Because the two pictures are arranged together, their aspect ratios are different from that of the others. The red arrowheads indicate significant filaments. (B) The helical parameters of the static filaments shapes of WT and the 25 revertants are plotted on a twist-versus-curvature graph (23). The plotted positions of the colored symbols represent averages of the calculated values from five independent filaments of each strain. The standard deviations of X and Y of WT were 0.07 and 0.10 are represented as WT (0.07, 0.10). Similarly, the standard deviations for the revertants were D1340 (0.13, 0.28), D42 (0.08, 0.09), D73 (0.26, 0.13), D561 (0.11, 0.11), D570 (0.13, 0.12), D455 (0.05, 0.10), D489 (0.09, 0.05), D520 (0.06, 0.03), D253 (0.06, 0.03), D276 (0.13, 0.09), D541 (0.10, 0.14), D59 (0.06, 0.03), D144 (0.05, 0.08), D87 (0.28, 0.31), D132 (0.11, 0.08), D135 (0.09, 0.09), D130 (0.05, 0.11), D68 (0.10, 0.07), D77 (0.08, 0.10), D121 (0.13, 0.07), D29 (0.08, 0.13), D63 (0.07, 0.10), D147 (0.13, 0.12), and D14 (0.09, 0.08). The parameters of D1824 were not determined because of its straight appearance. The orange crosses show the helical parameters calculated from pitch length and diameter data reported by Kamiya and Asakura (37). (a) Salmonella strain SJ30 (pH 7.0); (b) SJ30 (pH 7.0) and 0.2 M KCl; (c) SJ670 (pH 12.0) and 0.1 M KCl; (d) SJ670 (pH 12.4) and 0.4 M KCl; (e and f) SJ25 (pH 12.4) and 0.5 M KCl. The green crosses show the helical parameters calculated from pitch length and diameter data of Salmonella SJ25 strain under steady flow of a viscous solution reported in Hotani (39). The closed black squares represent the helical parameters of various types of flagellar filaments estimated from simulation data (44). The extended figure around “Normal” is inserted.
The static and dynamic filament shapes of all R-straight-revertants were highly diverse (Fig. 5 and see Fig. S3 in the supplemental material). As shown in Fig. 5, the static filament shape of SJW1655-D1340(A449V, D107E) was similar to that of curly I or II (37). As known curly mutants, this mutant swam poorly and the filaments extended in all directions from the cell (Fig. 5A). However, there was a difference in their movement. In the known curly mutants, their filaments sometimes swung similarly to the motion of a windshield wiper without changing their shapes, whereas the filaments of SJW1655-D1340(A449V, D107E) did not swing. Instead, they sometimes changed to apparently wavy shapes, and notably, the D107E mutation changed the R-type straight filament to a flexible filament with a curly-like shape.
The filament shapes of SJW1655-D42(A449V, G426S) cultured for 21 h looked similar to those of WT (Fig. 5), indicating that the G426S mutation changed the R-type straight filament to a normal-like filament. In contrast, it was difficult to find filaments in cells cultured for 3 h, and the few filaments found were short (data not shown). As described above, cells cultured for 3 h swam poorly, whereas those cultured for 21 h swam well. These results indicate that SJW1655-D42(A449V, G426S) filaments grew slowly but that, when grown, the filaments were able to propel the cells similarly to the normal filaments.
The static filament shape of SJW1655-D73(A449V, G426A) was curly I or semicoiled (Fig. 5 and see Fig. S3 in the supplemental material), whereas its dynamic filament showed completely different morphology from that of any known filaments. The dynamic filament shapes occurred in several forms, such as a WT-like thick bundle, a thick corkscrew, and others, and vigorously converted between them, presumably when the motor reversed. The static filament shape of SJW1655-D1824(A449V, S448F) appeared to be straight (Fig. 5A), but this revertant swam poorly. Microscopy revealed that the filaments extended in all directions from the cells, as did the known straight mutants. Nevertheless, the filament shapes were not uniform, and most filaments appeared to be wavy with very small amplitudes. Some of them bent up and down at their midpoint (Fig. 5A), and a few occasionally formed irregular waves.
In contrast to R-straight revertants showing multimodal static and dynamic filament properties, most of the L-straight revertants had flagellar filaments with normal-like static shapes and WT-like dynamic shapes (Fig. 5). An exception was that the filaments of SJW1660-D253(G426A, R431S) and HFG195-D87(A80V, A427E) showed a larger helical radius than the coiled shape with a longer pitch than the normal and a right-handed shape.
DISCUSSION
Key residues involved in transformation.
In the present study, 62 of 72 sites of the different parental strains are located within the D1 domain of the flagellin monomer. Vonderviszt et al. suggested that the D1 domain is involved in the polymorphic transformation of the filament (38). Our present results demonstrate that fliC-intragenic suppressor analysis is a reasonable approach to determine amino acid residues involved in the polymorphic transformation of flagellar filaments.
Among the R-straight-revertant mutations, D107E and G426A were also identified as the mutations to produce the L-type straight filaments in SJW1663 and SJW1660, respectively (25). Because these mutations transform the R-type straight filament to a supercoiled filament and the normal type filament to the L-type straight filament, we suggest that Glu107 and Ala426 tend to induce the formation of the L-type protofilament. Conversely, Asp107 and Gly426 tend to form the R-type protofilament. Although S448F has not been identified as the mutation to produce the L-type straight filament, Ser448 should also prefer the R-type protofilament based on the same logic. Ser106, Ala416, Ala427, and Arg431 are also thought to tend to form the L-type protofilament.
Large helical filaments.
The static filament shapes of SJW1660-D253(G426A, R431S) and HFG195-D87(A80V, A427E) have a larger helical radius than the coiled form, a longer pitch than the normal form, and are right-handed (Fig. 5). A similar shape has been reported as an unstable large helical form under a steady flow of a viscous solution of methylcellulose (39). The large helical form has been described to appear upon interconversion between the normal form and either the curly or semicoiled forms. The large helical shapes shown in the present study were also similar to what was proposed as an intermediate using massive MD simulations (27). Considering these findings, we suggest that the large helical conformation may represent an intermediate in the transformation from the normal to the semicoiled forms and be somehow stabilized by an unknown mechanism. If it is true, the cluster consisting of Arg431 and Ala427, the second mutation sites of SJW1660-D253(G426A, R431S) and HFG195-D87(A80V, A427E), respectively, is likely involved in the transformation of the normal to the semicoiled shape. Although further investigations are required to understand the formation of large helical filaments, it is true that the second mutation sites, Ala427 and Arg431, are involved in helix formation of the flagellar filament, that is, these second mutation sites are involved in L/R transitions.
Flagellar filaments possessing both mutations responsible for R- and L-type straight filaments.
The static filament shapes of SJW1655-D73(A449V, G426A) were curly I (L6/R5) or semicoiled (L7/R4) (Fig. 5 and see Fig. S3 in the supplemental material). From these results, we suggest that the filament shape is a consequence of a tug-of-war between the effects of G426A and A449V. A similar idea proposed by Calladine et al. (40) asserts the absence of a simple addition of effects of two mutations. In the case of SJW1655-D1340(A449V, D107E), there is a similar situation between the two mutations. The D107E mutation transforms the normal to the L-type straight shape, as does G426A. Addition of A449V to D107E increases the number of the R-type protofilaments and results in the appearance of the curly I (L6/R5) or II (L5/R6) filament (Fig. 5 and see Fig. S3 in the supplemental material). Although G426A and D107E share properties inducing the L-type straight filament structure and showing curly-like filaments with A449V, there are some distinct differences in the exact static and dynamic filament shapes between SJW1655-D73(A449V, G426A) and SJW1655-D1340(A449V, D107E), suggesting that the roles of Gly426 and Asp107 in the polymorphic transformation are different.
Differences between L- and R-type straight filaments in the local structures surrounding Ser106, Asp107, and Ala416 and Gly426, Ala427, and Arg431.
To consider the roles of Ser106, Asp107, Ala416, Gly426, Ala427, and Arg431 in the polymorphic transformation, we compared the atomic structures of L- and R-type straight filaments around Gly426, Ala427, and Arg431 (Fig. 6A to C) and around Ser106, Asp107, and Ala416 (Fig. 6D to F). Because the two structures were built based on the three-dimensional maps at 4- to 5-Å resolution, some ambiguities remain at the side chain level. However, the two structures are still useful for considering the roles of the six residues in the polymorphic transformation.
Fig 6.
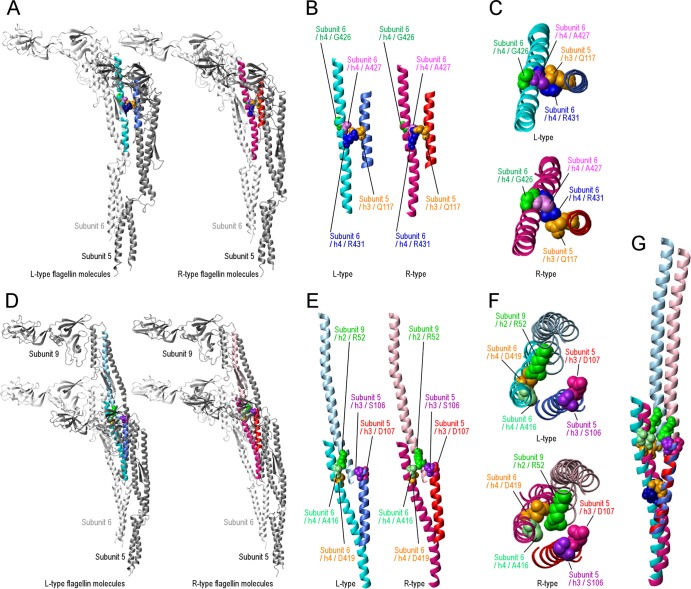
Differences in structure between L- and R-type flagellin subunits. (A) The left and right panels present ribbon models of subunits 5 and 6 of L- and R-type flagellin structures, respectively. The helix 4 of subunit 6 and the helix 3 of subunit 5 are cyan and royal blue in the L-type straight filament and deep pink and red in the R-type straight filament, respectively. The four residues are depicted in different colors, and the color coordinates are the same in both L and R types. (B) The colored region in panel A is cropped and magnified. (C) The structures shown in panel B are viewed from top. The upper and lower panels present L and R types, respectively. (D) The left and right panels present the ribbon models of subunits 5, 6, and 9 of L- and R-type flagellin structures, respectively. The helix 4 of subunit 6, the helix 3 of subunit 5, and the helix 2 of subunit 9 are cyan, royal blue, and light blue in the L-type straight filament and deep pink, red, and pink in the R-type straight filament, respectively. The five residues are depicted in different colors, and the color coordinates are the same in both L and R types. (E) The colored region in panel D is cropped and magnified. (F) The structures shown in panel F are viewed from top. The upper and lower panels present L and R types, respectively. (G) Helices 4, 3, and 2 of the subunits 6, 5, and 9, respectively, and Arg431, Gln117, Ala416, Asp419, Ser106, and Asp107 are shown. The partial structures of the L- and R-type straight filaments are superimposed as the short helices, helix 3, blue and red in L and R, respectively, overlapped.
First, we examined the region around Gly426, Ala427, and Arg431 on helix 4 of subunit 6 (Fig. 6A to C). Two of them, Ala427 and Arg431, are at the boundary surface to the neighboring subunit, subunit 5 (Fig. 6A and B). Looking at the positional relationship between Arg431 of subunit 6 and Gln117 on the helix 3 of subunit 5, there is an obvious difference between L- and R-type straight-filament structures (Fig. 6B and C). Thus, Arg431 might switch its position by “jumping over” Gln117 during the polymorphic transformation. Considering that the polymorphic transformation is triggered by the sliding between the neighboring protofilaments as previously described (41), subunits 5 and 6 are in such a relationship. Further, it is likely that Gly426 and Ala427 on a subunit control the jumping-switch of Arg431on the subunit, because they surround the switching residues.
Second, in the three-dimensional structure of flagellin, Ala416 of subunit 6 is not close enough to interact with Ser106 and Asp107 of subunit 5 (Fig. 6E and F). Regarding the relationships among Asp107 of subunit 5, Asp419 of subunit 6, and Arg52 of subunit 9, there seems to be a significant difference between the L- and R-type straight filaments, such as exchanging the interacting partner of Arg52 (Fig. 6E and F). Arg52 interacts with Asp419 in L-type straight filaments, whereas Arg52 interacts with Asp107 in R-type straight filaments. Such switching is very significant because the longitudinal interactions are supposed to be essential for propagating the transformation generated by sliding between adjacent subunits. In this model, Ser106 and Ala416 would control the propagation through the positions of Asp107 and Asp419, respectively.
Conserved amino acid residues revealed by comparisons of FliC sequences.
The contributions of Gln117, Arg52, and Asp419 to the polymorphic transformation are revealed by the structural comparison between the two kinds of straight filaments. To confirm their contributions, we performed multiple sequence alignments between 27 flagellin molecules of different bacterial species (see Fig. S2 in the supplemental material). Gln117 is conserved in 16 species (59%), and when substitutions to the polar residues Ser and Asn were taken into account, the number increases to 21 (78%). Arg52 is conserved in 18 species (67%) and in 22 species (81%) when substitution by Lys is considered. Asp419 is conserved in 12 species (44%) or 20 species (74%) when conserved substitutions to polar amino acids, such as Ser, Asn, and Thr are included. Beatson et al. also performed multiple alignment of the sequences of flagellin homologues of 20 species of bacteria (42). Even though 15 of them are different from the bacteria used in the present study, their results are in general agreement with ours.
In the seven key residues, Gly426 and Ala427 were conserved in all 27 species, and Asp107 and Arg431 were conserved in 25 and 24 species (93 and 89%), respectively. As described above, Gly426 and Ala427 may control the jumping-switch of Arg431 based on the analyses of atomic structure. Considering the high degree of conservation of Gly426 and Ala427, these small amino acid residues likely play an important role in strictly controlling the switch of position of Arg431. In contrast, Ser106 and Ala416 are conserved in 13 and 10 species (48 and 37%), respectively, suggesting that the control of the positioning of Asp117 and Asp419 by Ser106 and Ala416, respectively, is less stringent than that of Arg431. On the 448th residue, Ser was not well conserved (15%), but Ala was preferred (56%).
Ser450 and Asp456 are conserved in all of the alignments, and many other residues are highly conserved. There are two possible reasons why these residues were not identified as second mutation sites. First, the suppressor analysis used here is limited by amino acid substitutions, and second, highly conserved residues play an important role in other processes, such as type III flagellin export and/or self-assembly of flagellar filaments.
Mechanism for polymorphic transformation at the atomic level.
The relationships between switching the direction of motor rotation and transformation of the filament can be expressed as three steps as follows (43): (i) rapid reversal from CCW to CW induces the normal to the semicoiled; (ii) subsequently, during CW rotation, the semicoiled transforms to curly I; and (iii) switching from CW to CCW induces curly I to transform to the normal. Here, we propose the transformation mechanism described in step i. Because the normal and the semicoiled filaments are L9/R2 and L7/R4, respectively, step i is regarded as the L-to-R transition of two protofilaments. Then, we assume the L-type structure in Fig. 6 to be the initial state preceding step i. The transformation mechanism proceeds presumably as follows. (i) A rapid-reverse rotation in the motor applies twisting forces to the flagellar filament, and the forces induce sliding between neighboring protofilaments (41). In detail, the sliding occurs between the helix 4 of subunit 6 and the helix 3 of subunit 5. (ii) Based on our interpretation stated in the first section of Discussion, Arg431 seems to prefer the L-type structure, so that under such twisting forces, Arg431 changes its position to the R type from one side to the other (Fig. 6B and C). (iii) After this repositioning, helix 4 of subunit 6 moves toward helix 2 of subunit 9 and then helix 2 of subunit 9 moves toward helix 3 of subunit 5 (Fig. 6E to G). (iv) The two movements disrupt interactions between helix 4 of subunit 6 and helix 2 of subunit 9, and then new interactions occur between helix 2 of subunit 9 and helix 3 of subunit 5 (Fig. 6E to G), resulting in the conversion of a protofilament of the R type. Thus, we suggest that the cluster containing Arg431 contributes to the first action of the transition and that another cluster propagates the transitions along the flagellar filament. In this model, Ser448 and Ala449 do not participate but may play a role in steps ii and iii.
Recently, two reports considered the mechanism for the polymorphic transformation at atomic level. The first study reported the massive MD simulation of L-type straight through supercoiled filaments from the R-type straight filaments (27). Comparison between the L- and R-type flagellin molecules in the simulation identified 32 residues that are different between both types of flagellins. Among them, Arg52, Asp107, Asp419, and Arg431 were identified in the present study. The second report compared the atomic structures of R-type and L-type straight filaments (29). Phe53, Phe131, and Ala449, which participate in hydrophobic interactions, were identified as candidates for the conformational switching between the L and R type. However, Phe53 and Phe131 were not identified in either the present or MD simulation study. Perhaps Phe53 and Phe131 are involved in the Ala449-centered mechanism for polymorphic transformation in either step ii and/or iii.
The fliC-intragenic suppressor analysis is useful for determining the key residues for the transition between L- and R-type protofilaments. By taking advantage of this method, we determined the key residues using the straight-filament mutants as parental strains and proposed a mechanism for step i at the atomic level based on the residues identified. However, the results of our present study did not allow us to obtain clues to the mechanisms of transition in steps ii and iii. To identify the key residues and mechanism involved in step iii, in particular, we believe that fliC-intragenic suppressor analysis using curly-filament mutants as parental strains will serve as an appropriate approach.
Supplementary Material
ACKNOWLEDGMENTS
We thank Chris R. Calladine and Ben F. Luisi for discussing each of our prepublication results. We also thank Keiichi Namba, Koji Yonekura, and Saori Maki-Yonekura for kindly providing PDB files for drawing nine flagellin subunits in L- and R-type straight filaments.
Footnotes
Published ahead of print 31 May 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JB.02091-12.
REFERENCES
- 1.Mesibov R, Adler J. 1972. Chemotaxis toward amino acids in Escherichia coli. J. Bacteriol. 112:315–326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Seymour FWK, Doetsch RN. 1973. Chemotactic responses by motile bacteria. J. Gen. Microbiol. 78:287–296 [DOI] [PubMed] [Google Scholar]
- 3.Tso WW, Adler J. 1974. Negative chemotaxis in Escherichia coli. J. Bacteriol. 118:560–576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Maeda K, Imae Y, Shioi JI, Oosawa F. 1976. Effect of temperature on motility and chemotaxis of Escherichia coli. J. Bacteriol. 127:1039–1046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Imae Y. 1985. Molecular mechanism of thermosensing in bacteria, p 73–81 In Eisenbach M, Balaban M. (ed), Sensing and response in microorganisms. Elsevier Science Publishers, Amsterdam, Netherlands [Google Scholar]
- 6.Kort EN, Goy MF, Larsen SH, Adler J. 1975. Methylation of a membrane protein involved in bacterial chemotaxis. Proc. Natl. Acad. Sci. U. S. A. 72:3939–3943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hess JF, Oosawa K, Kaplan N, Simon MI. 1988. Phosphorylation of 3 proteins in the signaling pathway of bacterial chemotaxis. Cell 53:79–87 [DOI] [PubMed] [Google Scholar]
- 8.Oosawa K, Hess JF, Simon MI. 1988. Mutants defective in bacterial chemotaxis show modified protein phosphorylation. Cell 53:89–96 [DOI] [PubMed] [Google Scholar]
- 9.Berg HC, Brown DA. 1972. Chemotaxis in Escherichia coli analyzed by three-dimensional tracking. Nature 239:500–504 [DOI] [PubMed] [Google Scholar]
- 10.Berg HC, Anderson RA. 1973. Bacteria swim by rotating their flagellar filaments. Nature 245:380–382 [DOI] [PubMed] [Google Scholar]
- 11.Silverman MR, Simon MI. 1974. Flagellar rotation and mechanism of bacterial motility. Nature 249:73–74 [DOI] [PubMed] [Google Scholar]
- 12.Blair DF. 1995. How bacteria sense and swim. Annu. Rev. Microbiol. 49:489–522 [DOI] [PubMed] [Google Scholar]
- 13.Falke JJ, Bass RB, Butler SL, Chervitz SA, Danielson MA. 1997. The two-component signaling pathway of bacterial chemotaxis: a molecular view of signal transduction by receptors, kinases, and adaptation enzymes. Annu. Rev. Cell Dev. Biol. 13:457–512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bourret RB, Stock AM. 2002. Molecular information processing: lessons from bacterial chemotaxis. J. Biol. Chem. 277:9625–9628 [DOI] [PubMed] [Google Scholar]
- 15.Sourjik V. 2004. Receptor clustering and signal processing in Escherichia coli chemotaxis. Trends Microbiol. 12:569–576 [DOI] [PubMed] [Google Scholar]
- 16.Kamiya R, Asakura S. 1976. Helical transformations of Salmonella flagella in vitro. J. Mol. Biol. 106:167–186 [DOI] [PubMed] [Google Scholar]
- 17.Macnab RM, Ornston MK. 1977. Normal to curly flagellar transitions and their role in bacterial tumbling: stabilization of an alternative quaternary structure by mechanical force. J. Mol. Biol. 112:1–30 [DOI] [PubMed] [Google Scholar]
- 18.Turner L, Ryu WS, Berg HC. 2000. Real-time imaging of fluorescent flagellar filaments. J. Bacteriol. 182:2793–2801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Darnton NC, Turner L, Rojevsky S, Berg HC. 2007. On torque and tumbling in swimming Escherichia coli. J. Bacteriol. 189:1756–1764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Asakura S. 1970. Polymerization of flagellin and polymorphism of flagella. Adv. Biophys. 1:99–155 [PubMed] [Google Scholar]
- 21.Calladine CR. 1975. Construction of bacterial flagella. Nature 255:121–124 [DOI] [PubMed] [Google Scholar]
- 22.Calladine CR. 1976. Design requirements for construction of bacterial flagella. J. Theor. Biol. 57:469–489 [DOI] [PubMed] [Google Scholar]
- 23.Calladine CR. 1978. Change of waveform in bacterial flagella: role of mechanics at molecular level. J. Mol. Biol. 118:457–479 [Google Scholar]
- 24.O'Brien EJ, Bennett PM. 1972. Structure of straight flagella from a mutant Salmonella. J. Mol. Biol. 70:133–152 [DOI] [PubMed] [Google Scholar]
- 25.Kanto S, Okino H, Aizawa SI, Yamaguchi S. 1991. Amino acids responsible for flagellar shape are distributed in terminal regions of flagellin. J. Mol. Biol. 219:471–480 [DOI] [PubMed] [Google Scholar]
- 26.Kamiya R, Asakura S, Yamaguchi S. 1980. Formation of helical filaments by copolymerization of 2 types of straight flagellins. Nature 286:628–630 [DOI] [PubMed] [Google Scholar]
- 27.Kitao A, Yonekura K, Maki-Yonekura S, Samatey FA, Imada K, Namba K, Go N. 2006. Switch interactions control energy frustration and multiple flagellar filament structures. Proc. Natl. Acad. Sci. U. S. A. 103:4894–4899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yonekura K, Maki-Yonekura S, Namba K. 2003. Complete atomic model of the bacterial flagellar filament by electron cryomicroscopy. Nature 424:643–650 [DOI] [PubMed] [Google Scholar]
- 29.Maki-Yonekura S, Yonekura K, Namba K. 2010. Conformational change of flagellin for polymorphic supercoiling of the flagellar filament. Nat. Struct. Mol. Biol. 17:417–422 [DOI] [PubMed] [Google Scholar]
- 30.Hayashi F, Tomaru H, Oosawa K. 2010. The analysis of the flagellar filaments of the Salmonella pseudorevertants. Kobunshi Ronbunshu 67:668–678 [Google Scholar]
- 31.Yamaguchi S, Aizawa SI, Kihara M, Isomura M, Jones CJ, Macnab RM. 1986. Genetic evidence for a switching and energy-transducing complex in the flagellar motor of Salmonella typhimurium. J. Bacteriol. 168:1172–1179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yamaguchi S, Fujita H, Ishihara A, Aizawa SI, Macnab RM. 1986. Subdivision of flagellar genes of Salmonella typhimurium into regions responsible for assembly, rotation, and switching. J. Bacteriol. 166:187–193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shimada K, Kamiya R, Asakura S. 1975. Left-handed to right-handed helix conversion in Salmonella flagella. Nature 254:332–334 [DOI] [PubMed] [Google Scholar]
- 34.Koradi R, Billeter M, Wuthrich K. 1996. MOLMOL: a program for display and analysis of macromolecular structures. J. Mol. Graph. 14:51–55 [DOI] [PubMed] [Google Scholar]
- 35.Thompson JD, Higgins DG, Gibson TJ. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673–4680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Matsuura S, Kamiya R, Asakura S. 1978. Transformation of straight flagella and recovery of motility in a mutant Escherichia coli. J. Mol. Biol. 118:431–440 [DOI] [PubMed] [Google Scholar]
- 37.Kamiya R, Asakura S. 1977. Flagellar transformations at alkaline pH. J. Mol. Biol. 108:513–518 [DOI] [PubMed] [Google Scholar]
- 38.Vonderviszt F, Aizawa S-I, Namba K. 1991. Role of the disordered terminal regions of flagellin in filament formation and stability. J. Mol. Biol. 221:1461–1474 [DOI] [PubMed] [Google Scholar]
- 39.Hotani H. 1982. Micro-video study of moving bacterial flagellar filaments. 3. Cyclic transformation induced by mechanical force. J. Mol. Biol. 156:791–806 [DOI] [PubMed] [Google Scholar]
- 40.Calladine CR, Luisi BF, Pratap JV. 2013. A “mechanistic” explanation of the multiple helical forms adopted by bacterial flagellar filaments. J. Mol. Biol. 425:914–928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Namba K, Vonderviszt F. 1997. Molecular architecture of bacterial flagellum. Q. Rev. Biophys. 30:1–65 [DOI] [PubMed] [Google Scholar]
- 42.Beatson SA, Minamino T, Pallen MJ. 2006. Variation in bacterial flagellins: from sequence to structure. Trends Microbiol. 14:151–155 [DOI] [PubMed] [Google Scholar]
- 43.Berg HC. 2004. Escherichia coli in motion. Springer-Verlag, New York, NY [Google Scholar]
- 44.Hasegawa K, Yamashita I, Namba K. 1998. Quasi- and nonequivalence in the structure of bacterial flagellar filament. Biophys. J. 74:569–575 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.