Abstract
The aminotransferase IlvE was implicated in the acid tolerance response of Streptococcus mutans when a mutation in its gene resulted in an acid-sensitive phenotype (B. Santiago, M. MacGilvray, R. C. Faustoferri, and R. G. Quivey, Jr., J. Bacteriol. 194:2010–2019, 2012). The phenotype suggested that amino acid metabolism is important for acid adaptation, as turnover of branched-chain amino acids (bcAAs) could provide important signals to modulate expression of genes involved in the adaptive process. Previous studies have demonstrated that ilvE is regulated in response to the external pH, though the mechanism is not yet established. CodY and CcpA have been shown to regulate expression of branched-chain amino acid biosynthetic genes, suggesting that the ability to sense carbon flow and the nutritional state of the cell also plays a role in the regulation of ilvE. Electrophoretic mobility shift assays using the ilvE promoter and a purified recombinant CodY protein provided evidence of the physical interaction between CodY and ilvE. In order to elucidate the signals that contribute to ilvE regulation, cat reporter fusions were utilized. Transcriptional assays demonstrated that bcAAs are signaling molecules involved in the repression of ilvE through regulation of CodY. In a codY deletion background, ilvE transcription was elevated, indicating that CodY acts a repressor of ilvE transcription. Conversely, in a ccpA deletion background, ilvE transcription was reduced, showing that CcpA activated ilvE transcription. The effects of both regulators were directly relevant for transcription of ilvE under conditions of acid stress, demonstrating that both regulators play a role in acid adaptation.
INTRODUCTION
Streptococcus mutans, the major causative agent of dental caries, represents a challenge for effective treatment because of its acidogenic and aciduric nature. Survival under environmentally stressful conditions is possible due to an adaptive response which allows the organism to activate pathways that aid in coping with an acid challenge. Several contributing mechanisms have been identified for their roles in the acid adaptation process (1–4). However, reports have suggested other, uncharacterized routes that could contribute to the acid tolerance of S. mutans (5). Analyses of the metabolic profile of S. mutans have identified increased expression of genes involved in branched-chain amino acid (bcAA) biosynthesis under conditions of acid stress.
A strain of S. mutans GS-5 carrying an insertional mutation in a homolog of a branched-chain amino acid aminotransferase (ATase) referred to as AP-185 was impaired in both growth and sensitivity to acid challenge (6). When transcription of AP-185 was examined under various environmental stresses, expression was elevated under conditions of acid stress (6). Amino acid metabolism has been suggested as an alternative method for S. mutans to cope with acidic end products, whereby branched-chain amino acid synthesis, as an overflow pathway for pyruvate, reduces the generation of acidic end products and alleviates the acidification of the intracellular milieu (7). While potentially important for autoacidification as a result of carbohydrate metabolism, branched-chain amino acids are the most abundant amino acids in proteins and function as the precursors for several components necessary for effective cellular function. Branched-chain amino acids are also important as intracellular signals for the overall homeostasis of bacteria. The accumulation of bcAAs depends on many essential precursors, and hence, their relative abundance reflects the physiological state of the cell. Thus, as coeffectors for regulatory proteins, bcAAs act as metabolic sensors, allowing for the activation and/or repression of target genes at appropriate times (8).
CodY is a GTP-binding global transcriptional repressor that has been characterized extensively in several bacterial pathogens (9–11). CodY was first identified as a repressor in Bacillus subtilis, where an insertional mutation within the open reading frame (ORF) carrying codY resulted in derepression of its target gene, dpp, encoding a dipeptide permease (12, 13). Changes caused by nutritional limitation allow CodY to exert its regulatory function by sensing intracellular pools of branched-chain amino acids and GTP, which fluctuate when the organism enters stationary phase (14).
The ilvBHC and leuABCD operons, involved in biosynthesis of branched-chain amino acids in B. subtilis, are known to be regulated by CodY (15, 16). In Staphylococcus aureus, the ilv-leu operon has been identified as a target for CodY binding (17). CodY activation in Bacillus subtilis is dependent on direct interaction with branched-chain amino acids, which cause a conformational change in CodY upon binding that increases the affinity for its cognate motif (18). The DNA-binding region for CodY is localized upstream of promoters of the genes that it regulates, and in some cases, such as in Lactococcus lactis, a consensus sequence has been found that has a high binding affinity for CodY (19, 20). Branched-chain amino acid aminotransferases in L. lactis (bcaT) and B. subtilis (ybgE) (15, 21) have been identified as CodY targets, supporting the role of CodY in the regulation of the biosynthetic operon for amino acid synthesis. In turn, aminotransferases regulate CodY action by modulating intracellular pools of branched-chain amino acids (22).
Studies of operons encoding branched-chain amino acid synthesis components in B. subtilis have elucidated a complex regulatory scheme. It is common for transcriptional regulation of biosynthetic genes to be mediated by several proteins not limited to CodY (23). Catabolite response protein A (CcpA) is a well-known regulator of carbon metabolism involved in carbon catabolite repression (CCR) (24). In the presence of a preferred carbon source, CcpA binds to and represses expression of genes involved with secondary carbon sources while activating carbon overflow pathways (8, 25). CcpA binds to cre (catabolite-responsive element) sequences in the promoter regions of its target genes. The activity of CcpA is responsive to intermediates from glycolysis, which activate HPr, a histidine protein that functions as a binding partner for regulation. CcpA is both an activator and a repressor of gene expression, and cre site positioning can indicate function, with upstream promoter sites being important for activation and downstream promoter sites being important for repression (24).
Other than regulating genes involved with carbon catabolite repression, CcpA has also been shown to regulate branched-chain amino acid biosynthetic operons, such as the ilv-leu operon (23). Insufficient expression of these genes in a ccpA deletion mutant leads to a nutritional requirement for branched-chain amino acids, indicating that CcpA functions as an activator (26). The expression of ilv genes is stimulated in the presence of glucose, a preferred carbon source, a condition where CcpA is the most active (27). In S. mutans, CcpA has been shown to be important for central metabolism and the regulation of virulence. Loss of CcpA results in increased acidogenicity and faster growth at low pH, due to dysregulation of metabolic pathways (25).
The regulation of branched-chain amino acid biosynthetic genes by CcpA and CodY has been shown to be antagonistic, with CcpA as a positive regulator and CodY as a repressor. Interplay between CcpA and CodY enables the organism to respond to its nutritional needs and to modulate gene expression per its requirements. CcpA activates gene expression for branched-chain amino acid biosynthetic genes, which eventually results in increased intracellular branched-chain amino acid pools. Amino acid accumulation enhances the DNA-binding capacity of CodY and eventually results in the repression of the target genes. Activation by CcpA has been described as self-limiting, since CodY repression becomes stronger as more branched-chain amino acids are available (16). Positive regulation by CcpA is suggested to be due to direct interaction with the RNA polymerase as well, by preventing CodY binding via blockage of access to the Cod box (23). In most cases, positive regulation of CcpA cannot overcome negative regulation by CodY, and hence, changes in amino acid pools are the signals that alter the balance between the effects these proteins have on gene expression.
SMU.1203c encodes a branched-chain amino acid ATase, IlvE, in S. mutans UA159. Our work has shown that IlvE is involved with branched-chain amino acid metabolism and plays a role in acid adaptation. Loss of ilvE results in an acid-sensitive phenotype and reduced levels of F-ATPase activity when the organism is grown at low pH (28). In several organisms, such as Streptococcus pneumoniae (11), L. lactis (29), and S. aureus (17), ilvE has been identified as a CodY-regulated gene. Hence, the acid-sensitive phenotype of the ilvE mutant and the possibility that ilvE is a target for CodY regulation led us to explore the mechanism of expression of this bcAA ATase.
In this study, we investigated the regulatory mechanisms for ilvE and the interaction of ilvE with CodY. The binding of CodY to its cognate regulatory element in the ilvE promoter was demonstrated by electrophoretic mobility shift assays (EMSAs), in which branched-chain amino acids were identified as coeffectors of binding for CodY. Similar to results for L. lactis, CodY in S. mutans did not require GTP for enhanced binding. In addition, loss of the CodY consensus binding domain in target DNA led to an inability of the protein to bind. Transcriptional reporter fusions revealed that ilvE transcription is modulated by the presence of branched-chain amino acids at acidic pH, suggesting a response to intracellular pools of bcAAs, consistent with the role of ilvE as a biosynthetic gene. Under nutrient-limiting conditions, however, the ilvE transcript was induced regardless of growth conditions, indicating that the nutritional requirements of the cell act as important signals, in combination with environmental pH. Transcription of ilvE was also examined in strains of S. mutans deficient in CcpA or CodY. The results demonstrated reduced expression of ilvE in a ΔccpA background and elevated expression in a ΔcodY background, thereby supporting the roles of the corresponding regulators as activators and repressors of transcription, respectively. Collectively, the data presented show that transcriptional regulation of ilvE is responsive to carbon flow and the nutritional state of the organism.
MATERIALS AND METHODS
Bacterial strains.
Strains used in this study are detailed in Table 1. Streptococcus mutans UA159, the genomic type strain, was the parent strain used in this study (30, 31). Mutant strains of S. mutans UA159 were created by deleting the entire coding regions of codY (SMU.1824c) and ccpA (SMU.1591c), using a PCR-based, ligation-independent cloning method, LIC mutagenesis, as previously described (32–34). Flanking regions of approximately 400 bp upstream and downstream were used to mediate replacement of genomic copies of genes with a nonpolar erythromycin cassette, resulting in S. mutans ΔcodY (SMU.1824c) and ΔccpA (SMU.1591c) strains. Appropriate constructs were verified using two pairs of specific primers for each strain (Table 2).
Table 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Relevant feature | Source or reference |
|---|---|---|
| Strains | ||
| E. coli strains | ||
| DH10B | Invitrogen | |
| BL21(λDE3) | Novagen | |
| BL21(λDE3)(pET19bCodY) | CodY expression construct; Ampr | This study |
| BL21(λDE3)(pET19bVC) | pET19b vector control; Ampr | This study |
| S. mutans strains | ||
| UA159 | Genomic type strain of S. mutans | 30, 31 |
| ΔilvE strain | SMU.1203c (ΔilvE) mutant strain of UA159; Ermr | 28 |
| ΔcodY strain | SMU.1824c (ΔcodY) mutant strain of UA159; Ermr | This study |
| ΔccpA strain | SMU.1591c (ΔccpA) mutant strain of UA159; Ermr | (R. C. Faustoferri and C. Hubbard, unpublished data) |
| UR314 | codY-complemented strain (codYc); Kanr Ermr | This study |
| UR212 | UA159 carrying pJL84 (promoterless cat); Kanr | 28 |
| UR213 | UA159 carrying pJL1099 (ilvE promoter-cat); Kanr | 28 |
| UR214 | ΔcodY strain carrying pJL84; Kanr Ermr | This study |
| UR217 | ΔilvE strain carrying pJL1099; Kanr Ermr | This study |
| UR220 | ΔcodY strain carrying pJL1099; Kanr Ermr | This study |
| UR268 | ΔccpA strain carrying pJL1099; Kanr Ermr | This study |
| UR301 | ΔccpA strain carrying pJL1099CRE; Kanr Ermr | This study |
| UR302 | UA159 carrying pJL109CRE; Kanr | This study |
| UR304 | ΔcodY strain carrying pJL1099CRE; Kanr Ermr | This study |
| UR317 | ΔcodY complemented strain with pJL1099; Kanr Ermr | This study |
| Plasmids | ||
| pET19b | T7lac, N-terminal 6× His; Ampr | Novagen |
| pET19bCodY | pET19b with codY coding region; Ampr | This study |
| pJL84 | Chloramphenicol acetyltransferase gene (cat) reporter plasmid; Kanr | 28 |
| pJL1099 | pJL84 with ilvE promoter region; Kanr | 28 |
| pJlL1099NCB | pJL84 with truncated ilvE promoter, no CodY box; Kanr | This study |
| pSUGBcodY | codY complement construct used to create UR314 | This study |
Table 2.
Oligonucleotide primers used in this study
| Primer application and name | Nucleotide sequence (5′–3′)a | Restriction site |
|---|---|---|
| Primers for cloning | ||
| 1662P8DN (codY) | ATTGATGATTTTCTTGACTTAGGG | |
| 1662P7UP (codY) | ATCACAGCGGTAAAACCTCAGGCG | |
| 1442P8DN (ccpA) | AACCAAGCGGTACCATAGATAC | |
| 1442P7UP (ccpA) | AAATGGGCTATTGCTAGTGATG | |
| 1662BglUpb | CATAGATCTAAGGGCTTTCATGAAAATTGTCGA | BglII |
| 1662BglDownb | CATAGATCTGGTATAATCAATTGAAATCAGTGCTTTACTCATT | BglII |
| Primers for EMSAs and CAT constructs | ||
| 1099Fwdc | CATGAGCTCGATAACATTTCTTACAAAAAGACAGG | SacI |
| 1099Revc | CATGGATCCTTTAATACCTCTTTCCTCG | BamHI |
| 1099RevNCBc,d | CATGGATCCTTTAAAAGATAAAATGGAGCAG | BamHI |
| BglIIATPe | ATTCAACATAGATCTTTCTCCTTC | BglII |
| BRLSMp567>e | ACCTAATATTTATAAGATAAG | |
| Primers for protein expression | ||
| CodYFwd | GAAAGCCCATATGGCTAATTTATTATCG | NdeI |
| CodYRev | GAGTGACTTGGATCCTTAACTATAGTC | BamHI |
Restriction sites are underlined.
Primer used for cloning of codY complement.
Primer used for ilvE target.
Primer used to construct a truncated ilvE promoter without the CodY consensus binding sequence.
Primer used for ATPase control probe SMU.1534c (3).
Generation of CodY-complemented strain.
A genetic complement strain of S. mutans was created using primers (Table 2) to amplify the codY coding and preceding intergenic regions from UA159 chromosomal DNA. A codY complement plasmid, pSUGBcodY, was constructed as previously described (28, 35). The ΔcodY mutant strain was transformed with the integration vector pSUGBcodY, and colonies were selected on brain heart infusion (BHI) agar containing kanamycin (1,000 μg/ml). Positive transformants were identified by colony PCR. One strain, containing the codY coding region and promoter integrated into the gtfA locus (SMU.0881), was named UR314.
Growth conditions.
Escherichia coli strains were maintained on Luria-Bertani (LB) agar plates with appropriate selection, where necessary, as detailed below. S. mutans strains were maintained on BHI (Becton, Dickinson/Difco, Franklin Lakes, NJ) agar plates with appropriate antibiotic selection (see below). For chloramphenicol acetyltransferase (CAT) assays (described below), S. mutans batch cultures were grown in BHI broth or FMC minimal medium (36) plus 1% (wt/vol) glucose in a 95% (vol/vol) air–5% CO2 atmosphere. Nutritional supplements were exogenously supplied amino acids or fatty acids. Final concentrations of additives were as follows: 1 mM leucine (L), 1 mM isoleucine (I), and 1 mM valine (V), individually or mixed (at a 1 mM concentration of each) (ILV). Overnight cultures were diluted 1:20 (in triplicate) in 50 ml of fresh medium titrated to pH 5 or buffered to pH 7, with or without additives. The optical density at 600 nm (OD600) was followed until the cell density was between 0.4 and 0.6. Cells were harvested, washed with 10 mM Tris-Cl, pH 7.8, pelleted, and stored frozen until assayed for chloramphenicol acetyltransferase activity.
ilvE promoter-cat fusion construction.
For cat transcriptional fusions, full-length ilvE promoter-cat fusions were cloned as previously described (28). In addition, a truncated ilvE promoter-cat construct (NCB [no Cod box]) in which the predicted native CodY-binding domain was absent was created (Fig. 1). Briefly, truncated promoters were cloned into a promoterless chloramphenicol acetyltransferase reporter plasmid, pJL84 (28), using primers described in Table 2. Transformants were selected on LB agar plates containing kanamycin (50 μg/ml) and were screened by colony PCR. Correct constructs were named pJL1099CRE and pJL1099NCB (Table 1). The S. mutans parent strain (UA159) and the ΔcodY and ΔccpA mutant strains were transformed with pJL1099 to generate single-copy, integrated reporter fusions.
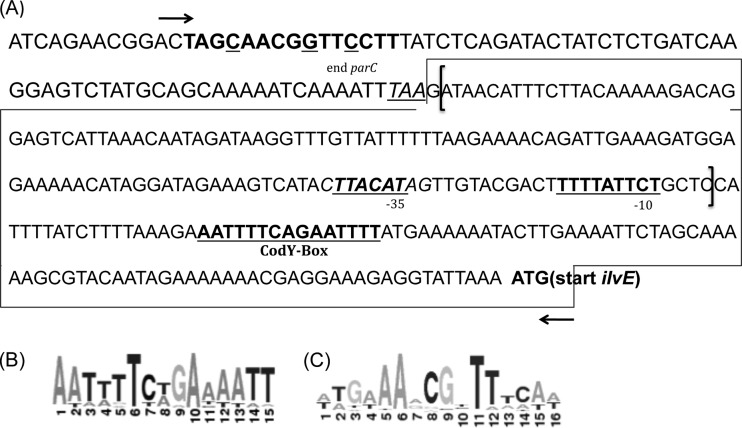
Fig 1.
Intergenic region preceding ilvE. (A) Putative −10 and −35 sequences for ilvE determined by BPROM (Softberry) promoter prediction. The boxed region represents the ilvE promoter region. Brackets represent the truncated ilvE promoter region (NCB; no CodY box). Arrows indicate an extended ilvE promoter region that contains a cre site. The CodY-binding recognition site is shown in bold and underlined. The hypothetical cre site is shown in bold, with mismatched bases underlined. Primer binding sites are denoted by arrows. (B) Streptococcal CodY consensus sequence. (C) CcpA consensus sequence. The consensus sequences were predicted by using the RegPrecise (http://regprecise.lbl.gov/RegPrecise/) database.
UR314, the codY-complemented strain, was transformed with pJL1099 (33), and transformants were selected on BHI plates containing both kanamycin (1,000 μg/ml) and erythromycin (5 μg/ml). Clones were screened for proper integration into the chromosome by PCR. Integration of the promoter-cat construct occurs within the intergenic regions between mtlA (SMU.1185c)-glmS (SMU.1187c) and mtlD (SMU.1182c)-phnA (SMU.1180c). Transformants for each construct are described in Table 1.
CodY protein expression.
CodY protein expression and purification were performed as previously described (11, 18), with modifications. The S. mutans codY gene was PCR amplified from UA159 chromosomal DNA by using the primer pair CodYFwd and CodYRev (Table 2) and then cloned into the Zero Blunt vector (Invitrogen, Carlsbad, CA). The codY coding sequence was confirmed by sequencing, and a positive clone was digested at engineered NdeI and BamHI sites to release the fragment. The gel-purified insert was cloned into NdeI/BamHI sites of pET19b (Novagen, Rockland, MA) to generate an N-terminally His-tagged recombinant CodY protein, and a correct construct was named pET19CodY. E. coli BL21(λDE3) was transformed with pET19b (as a negative expression control) or pET19CodY for protein expression. The BL21 strain carrying pET19b or pET19CodY was grown in LB medium containing ampicillin at 100 μg/ml, at room temperature, until the OD600 reached approximately 0.4 to 0.6. IPTG (isopropyl-β-d-thiogalactopyranoside; 1 mM) was then added to induce protein expression, and growth was continued for 3 h. Cells were harvested by centrifugation, and cell pellets were lysed using a mini-bead beater (BioSpec Products, Bartlesville, OK). His-tagged CodY protein was purified under native conditions, using Ni-nitrilotriacetic acid (Ni-NTA) agarose (Qiagen, Valencia, CA). Purified CodY protein was analyzed by matrix-assisted laser desorption ionization–time of flight (MALDI-TOF) (UR Proteomics Core Facility) and verified to be S. mutans CodY. Total protein concentration was estimated by the method of Bradford (Bio-Rad, Hercules, CA) (37). Protein samples were mixed with 5× SDS-PAGE loading buffer, boiled for 5 min, and loaded onto a NuPAGE Novex 4 to 12% Bis-Tris gel (Invitrogen). Proteins were electrotransferred to a polyvinylidene difluoride (PVDF) membrane for Western blotting, using a Milliblot (Millipore, Billerica, MA) system, and then were immunoblotted (38). The presence of the 6×-His tag was verified by Western blot analysis using an anti-His IgG primary antibody (GE Healthcare, Piscataway, NJ).
CAT assay.
CAT assays were performed following a previously described protocol (3, 28, 39). Briefly, 50-ml cell pellets were resuspended in 1 ml 10 mM Tris-Cl, pH 7.8. Cells were lysed using 0.1-mm glass beads in a mini-bead beater (BioSpec Products, Bartlesville, OK). Cell debris was removed by centrifugation at 10,000 rpm for 10 min. Lysates were removed and used for total protein quantification (37) and CAT assays. Each CAT assay reaction mixture consisted of 50 μl whole-cell lysate, 100 mM Tris-Cl, pH 7.8, 0.1 mM acetyl-coenzyme A (acetyl-CoA), and 0.4 mg/ml 5,5′-dithio-bis(2-nitrobenzoic acid) (DTNB) in a total volume of 1 ml. Reactions were initiated by addition of 0.1 mM chloramphenicol. Optical density measurements at 412 nm were monitored over 3 min. Reaction rate and total protein concentration were used to determine CAT activity, and values are presented as μmol chloramphenicol acetylated/min/mg of total protein.
Electrophoretic mobility shift assay.
Electrophoretic mobility shift assays were performed as described previously (11, 40). Briefly, DNA fragments containing ilvE (SMU.1203c) and ATPase (SMU.1534c/c subunit ATPase) promoter regions were amplified by PCR using primers shown in Table 2. DNA products were purified and end labeled with T4 polynucleotide kinase (PNK) and [γ-32P]ATP (PerkinElmer, Waltham, MA) for 10 min at 37°C. Quantification of labeled DNA was performed by scintillation counting. A representative binding reaction mixture (25 μl) consisted of the following: binding buffer (20 mM Tris-HCl, pH 8; 8.7% [vol/vol] glycerol; 1 mM EDTA; 5 mM MgCl2; 250 mM KCl; 0.5 mM dithiothreitol [DTT]; 2 μg bovine serum albumin [BSA]), radiolabeled DNA probe (3,000 cpm), purified CodY (1,500 nM), and the branched-chain amino acids isoleucine, leucine, and valine, separately or in combination, to a final concentration of 3 mM or 10 mM, as indicated. GTP was added to a final concentration of 3 mM. Binding reaction mixtures were incubated at 37°C for 45 min. CodY-DNA binding was assessed using an 8% nondenaturing gel in 1× Tris-glycine buffer or a 6% DNA retardation gel (Invitrogen). DnaK protein (1,500 nM) and the ATPase promoter were used as negative controls for protein binding and probe specificity, respectively. The gels were exposed to a phosphorimager screen, and binding was detected with a Molecular FX phosphorimager and Bio-Rad Quantity One software (Bio-Rad, Hercules, CA).
RESULTS
CodY interacts with the ilvE promoter region.
A consensus CodY-binding sequence in the putative promoter region of ilvE in S. mutans was previously identified by Lemos et al. (20), suggesting that ilvE could be regulated by CodY. To determine whether the CodY protein was capable of direct interaction with the ilvE promoter, we evaluated DNA-protein binding via EMSAs. A recombinant CodY protein was purified and verified by mass spectrometry prior to performing the binding assays (data not shown). The ilvE promoter region containing the CodY consensus sequence (Fig. 1) was used as the target. In the presence of CodY, migration of the ilvE promoter target was hindered, indicating DNA-protein complex formation (Fig. 2, lane 4, and Fig. 3, lane 6). No shift was detected in the presence of control DNA (Fig. 2, lane 1, and Fig. 3, lanes 1 and 2) or with the use of a control protein, DnaK (Fig. 2, lane 3), indicating that the interaction between ilvE and CodY was specific. The EMSA data demonstrated that the ilvE promoter region, containing a predicted CodY consensus sequence, is capable of binding CodY.
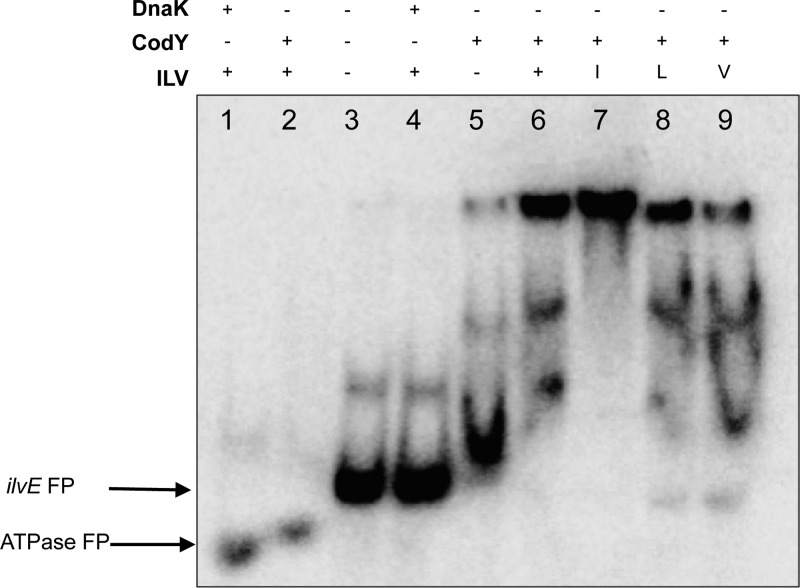
Fig 2.
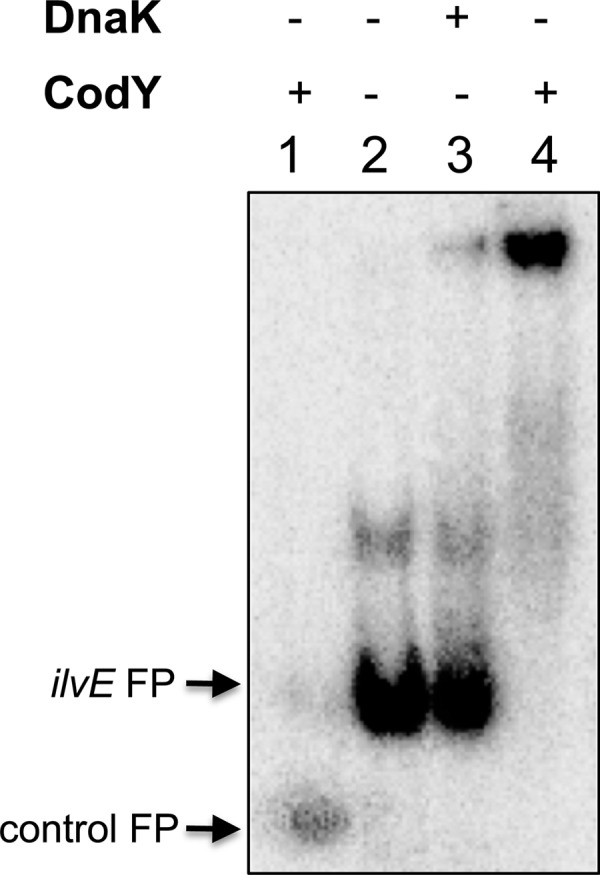
CodY interaction with ilvE promoter. Radiolabeled DNA probes were incubated with 1,500 nM CodY or DnaK, as indicated. Promoter regions used for binding assays were the ATPase promoter (lane 1) and the ilvE promoter region (lanes 2 to 4). No branched-chain amino acids were added to binding reaction mixtures. Arrows at left indicate migration of free probes (FP).
Fig 3.
CodY interaction with the ilvE promoter is enhanced in the presence of isoleucine. Radiolabeled DNA probes were incubated with 1,500 nM purified recombinant DnaK (control) or CodY. Branched-chain amino acids added were isoleucine (I), leucine (L), and valine (V), individually or in combination (ILV), to a final concentration of 3 mM (each). Promoter regions used for binding assays were the ATPase promoter (lanes 1 and 2) and the ilvE promoter (lanes 3 to 9). Arrows at left indicate migration of free probes (FP).
Interaction between CodY and the ilvE promoter occurs in the presence of bcAAs and not GTP.
The interaction between a target DNA and the CodY protein has been shown to be mediated by either GTP in conjunction with bcAAs or bcAAs alone (41). In S. mutans, the use of decoyinine, an analogue of adenosine that inhibits GMP synthase and causes a decrease in GTP pools, did not affect CodY-mediated repression of ilvE, indicating that in S. mutans, CodY activity is not responsive to GTP (20). To confirm these findings and to demonstrate that bcAAs are coeffectors of CodY, we tested CodY binding to the ilvE promoter in the presence of bcAAs as well as GTP. When 3 mM GTP was added to the binding reaction mixture, CodY binding to the ilvE promoter was not enhanced (Fig. 4, lane 6) and appeared to be similar to protein binding without coeffectors. However, in the presence of bcAAs, a significant shift was observed (Fig. 3, lane 5 versus lane 6), indicating that CodY binding to the ilvE promoter was not dependent on GTP but on bcAAs alone.
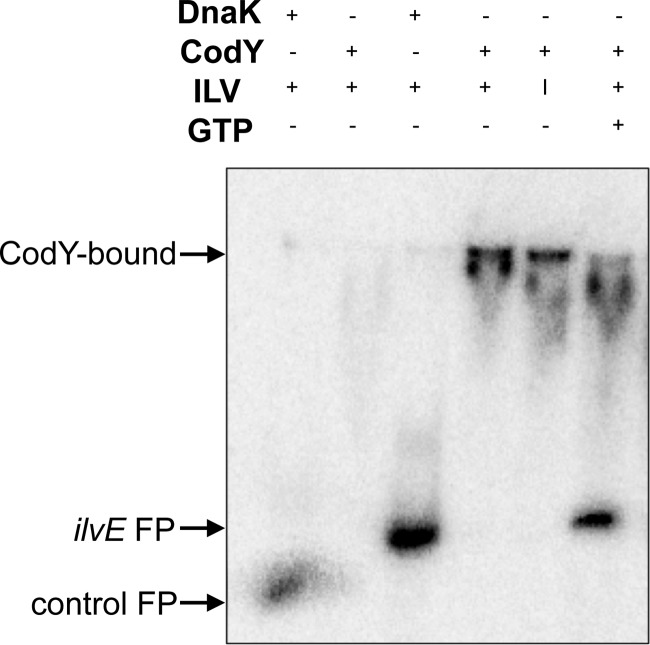
Fig 4.
CodY binding is not enhanced by GTP in S. mutans. Radiolabeled DNA probes were incubated with 1,500 nM purified recombinant DnaK (control) or CodY. Branched-chain amino acids added were isoleucine, leucine, and valine (ILV) or isoleucine alone (I), to a final concentration of 10 mM (each). GTP was added to a final concentration of 3 mM. Promoter regions used for binding assays were the ATPase promoter (lanes 1 and 2) and the ilvE promoter (lanes 3 to 6).
CodY binding to the ilvE promoter is enhanced in the presence of isoleucine.
The branched-chain amino acids isoleucine, leucine, and valine are metabolite effector molecules for CodY binding in other systems (42). In L. lactis, isoleucine has been shown to be the stronger of the signals that enhance binding affinity for CodY (29). To determine if CodY binding in S. mutans exhibited a similar bias for specific bcAAs, we performed binding assays in the presence of individual bcAAs. Although CodY was able to bind the ilvE promoter without added bcAAs, the presence of bcAAs enhanced the binding of CodY, as shown in Fig. 3, lanes 6 to 9. In the presence of isoleucine, CodY was able to bind and completely shift the ilvE target in a manner similar to that seen when a mixture of all three amino acids was present (Fig. 3, lanes 6 and 7), indicating that isoleucine increased binding to a greater extent than that with any of the other amino acids added individually. These data demonstrate that of all the bcAAs tested, isoleucine is the most effective coeffector for CodY binding to the ilvE promoter in S. mutans.
Absence of the CodY consensus binding sequence abrogates the ability of CodY to bind the ilvE promoter.
To test whether the absence of a CodY consensus binding sequence would inhibit protein-DNA binding and to demonstrate that this interaction was CodY specific, an ilvE promoter sequence lacking the CodY consensus motif was used as a target in EMSAs (Fig. 1). Unlike our control reaction, where a visible shift occurred with the full-length ilvE probe, in the absence of a CodY-binding sequence, the protein was unable to bind the ilvE promoter, as shown in Fig. 5, lane 4. In the presence of an anti-His IgG antibody, no shift was detected using the ilvE promoter lacking the Cod box (data not shown), in contrast to results with the full-length ilvE promoter, containing a Cod box and CodY (data not shown). Taken together, these data indicate that the CodY consensus sequence is necessary for protein-DNA interaction between CodY and ilvE.
Fig 5.
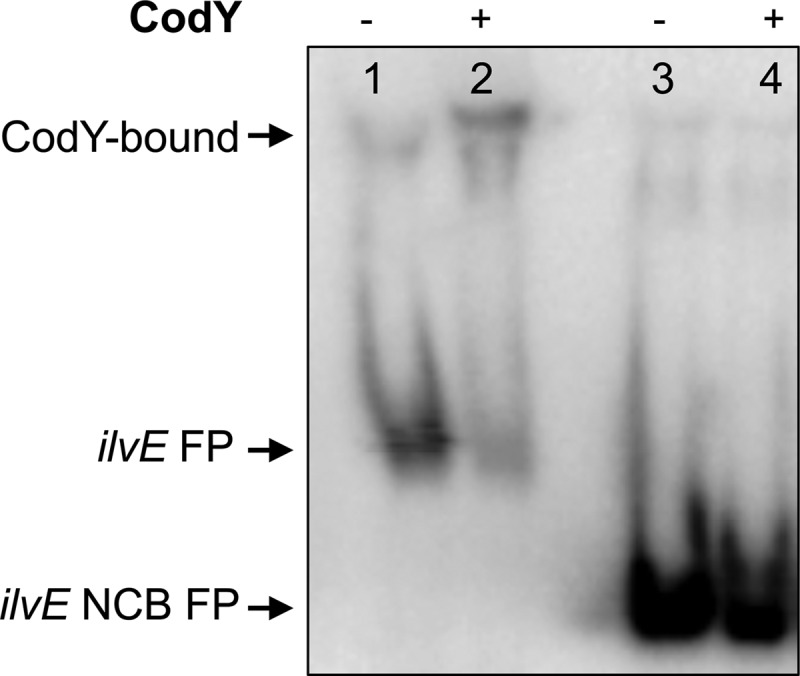
The CodY box is required for binding of CodY to the ilvE promoter in S. mutans. CodY binding assays for full-length and truncated (NCB) ilvE promoter regions (described in Materials and Methods) were performed with isoleucine (10 mM) and purified recombinant CodY (1,500 nM). Lanes 1 and 2, ilvE promoter; lanes 3 and 4, truncated ilvE promoter (NCB).
Transcription of ilvE is responsive to bcAAs.
Our binding assays provided evidence that CodY binding to ilvE was enhanced in the presence of branched-chain amino acids. Branched-chain amino acids have also been shown to be important in the regulation of biosynthetic genes as end products of the biosynthetic pathway. To assess the effects of the presence of branched-chain amino acids on expression of ilvE in vivo, an ilvE promoter-cat fusion strain, UR213, was created. When the fusion-bearing strain was grown in the presence of isoleucine or leucine, the transcription of ilvE was reduced in comparison to expression in unsupplemented medium (Fig. 6A). The reduced expression in the presence of leucine also suggests that, like other biosynthetic genes, ilvE could be subject to regulation by the level of leucine. The observable effect on the transcriptional activity occurred at a low pH, which we had previously shown is a condition that triggers an increase in ilvE transcription (28). The results suggest that signaling by bcAAs can modulate transcription of ilvE, demonstrating that both pH and the nutritional state of the cell are signals that regulate ilvE transcription.
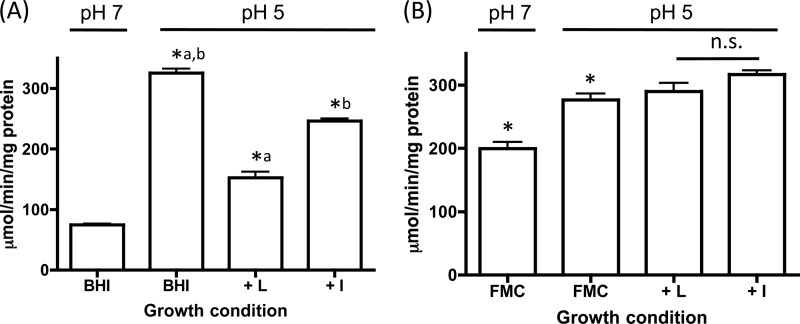
Fig 6.
ilvE promoter activity is downregulated by bcAAs and induced under nutrient-limiting conditions. CAT activity was determined for S. mutans UR213 (ilvE promoter-cat fusion in the UA159 parent strain) grown in BHI medium (A) or FMC minimal medium (B) supplemented with bcAAs. Transcriptional activity was determined as described in Materials and Methods. The growth medium was supplemented with the branched-chain amino acid isoleucine (I) or leucine (L), to a final concentration of 1 mM. Statistical significance was determined by Student's t test. *, P ≤ 0.00001 (n = 3). Pairwise comparisons are represented by lowercase letters. n.s., not statistically significant.
Transcription of ilvE is downregulated in the absence of ilvE.
bcAAs, the end products of branched-chain amino acid synthesis, are the signaling molecules that allow CodY to repress transcription of target genes, which themselves encode enzymes that synthesize bcAAs. Since the ΔilvE strain displayed a requirement for bcAAs during growth in minimal medium and a reduction in bcAA catabolism, we thus showed the contribution of IlvE to both processes of amino acid metabolism: synthesis and degradation (28). Aminotransferases (ATases) are enzymes of dual function, whose enzymatic activity can participate in both amino acid biosynthesis and degradation (43). Therefore, since ilvE encodes an ATase, the possibility existed that the gene product could contribute, via a feedback mechanism, to the expression of the cognate gene. To determine the effects that the loss of IlvE would have on its own transcription, we explored the transcriptional activity of the ilvE promoter in an ilvE deletion strain (UR217). In accordance with its role in branched-chain amino acid metabolism, the ilvE transcript level, as measured by a reporter fusion assay, was reduced in the absence of IlvE, indicating that the absence of IlvE affects transcription of the cognate gene (Fig. 7). Similar to data presented above, the most pronounced effects on the transcriptional activity occurred under low-pH growth conditions. The decreased ilvE transcript level might have been due to changes in bcAA turnover because of the loss of both enzymatic functions that IlvE provides. These data support the important role of this gene and its product in its own regulation, as well as in transcriptional regulation required for acid adaptation.
Fig 7.
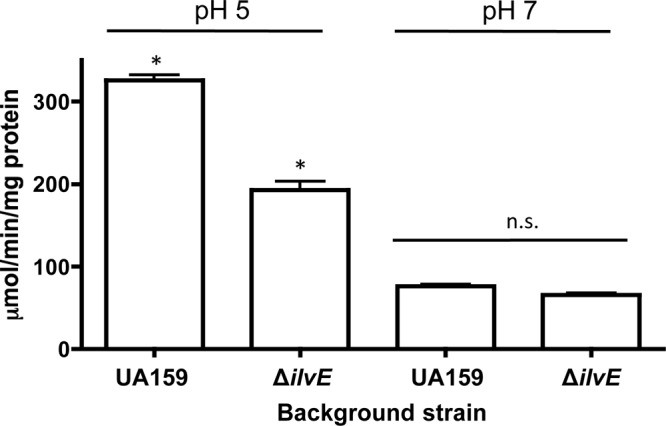
The ilvE mutant displays decreased transcriptional activity from the ilvE promoter. CAT activities are shown for ilvE promoter-cat fusions in the UA159 parent strain and ΔilvE backgrounds, grown in BHI medium. Transcriptional activity was determined for three independent cultures in duplicate for each growth condition, as described in Materials and Methods. Statistical significance was determined by Student's t test. *, P ≤ 0.00001 (n = 3). n.s., not statistically significant.
Growth under nutrient-limiting conditions induces ilvE transcription.
Following exponential-phase growth, the availability of nutrients becomes reduced as cells enter stationary phase. During stationary phase, bcAAs become limited, and as a result, genes that are under the control of CodY can be derepressed (14, 21). To examine the effect that nutritional limitation would have on ilvE transcription, UR213 was grown in FMC minimal medium (44) in the presence or absence of bcAAs. It was expected that under nutrient-limiting conditions, CodY repression would be partially alleviated, allowing for increased transcriptional activity of ilvE. When UR213 was grown in FMC minimal medium at low pH, ilvE transcriptional activity was not enhanced (Fig. 6B) compared to the results observed for cultures grown in BHI medium (Fig. 6A). However, the pH-mediated response differed between the two growth conditions (Fig. 6B). In rich medium, the ilvE transcript was responsive to environmental pH, such that an increase in transcriptional activity was observed in cultures grown at pH 5 (28). Under nutrient-limiting conditions, although the pH-dependent increase in transcription was still detectable, ilvE transcription was elevated 2-fold in cultures grown at a pH of 7 compared to cultures grown under nutrient-rich conditions (Fig. 6B). These results suggest that ilvE transcription can be affected by the availability of basic metabolites in growth medium, in addition to growth at acidic pH values, although the transcriptional response to acidic conditions is still maintained in both growth environments.
CodY represses ilvE transcription.
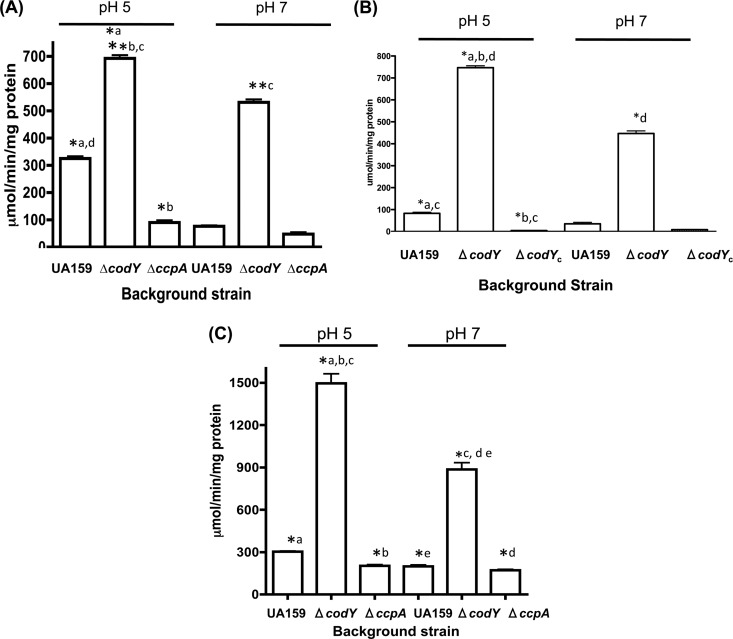
In an S. mutans codY deletion strain, a substantial majority of the genome was upregulated, supporting the role of CodY as a global repressor of gene expression (20). In previous work, we used an ilvE promoter-chloramphenicol acetyltransferase gene (cat) reporter fusion to study ilvE transcription, demonstrating that ilvE transcription was affected by extracellular pH (28). The presence of a CodY-binding domain, as well as the physical interaction between CodY and the ilvE promoter (Fig. 2 and 3), strongly implicated CodY as a regulator of ilvE transcription. To determine if CodY is a repressor of ilvE, a ΔcodY strain carrying the ilvE-cat reporter fusion (UR220) was used in transcriptional studies. In the absence of CodY, ilvE transcription was upregulated approximately 2-fold at pH 5 and approximately 7-fold at pH 7 (Fig. 8A). Although a pH-dependent response was still observed, the ilvE message was induced to a greater extent at neutral pH in the ΔcodY strain, indicating that in the absence of CodY, derepression had occurred. In order to confirm the role of CodY as a repressor of ilvE transcription, the pJL1099 promoter fusion was integrated into the codY-complemented strain (UR317). CAT assays performed on UR317 showed that introduction of a functional copy of CodY reestablished repression of ilvE transcription (Fig. 8B). Collectively, these data demonstrate that CodY acts as a repressor of ilvE transcription and that, in the absence of CodY, the effect of environmental pH on transcription is enhanced.
Fig 8.
(A) Transcription of ilvE is regulated by CodY and CcpA. CAT activities are shown for a full-length ilvE promoter-cat fusion in the following background strains: UA159, ΔcodY strain, and ΔccpA strain. Cultures were grown in BHI medium as described in Materials and Methods. Statistical significance was determined by Student's t test. *, P ≤ 0.00001; **, P ≤ 0.0000001 (n = 3). Pairwise comparisons are represented by lowercase letters. (B) Genetic complementation of CodY restores transcriptional repression of ilvE. CAT activities are shown for a full-length ilvE promoter-cat fusion in the following background strains: UA159, ΔcodY strain, and complemented ΔcodY strain (ΔcodYc). Cultures were grown in BHI medium as described in Materials and Methods. Statistical significance was determined by Student's t test. *, P ≤ 0.00001 (n = 3). Pairwise comparisons are represented by lowercase letters. (C) Nutritional limitation and the absence of CodY enhance transcription of ilvE. CAT activities are shown for a full-length ilvE promoter-cat fusion in the following background strains: UA159, ΔcodY strain, and ΔccpA strain. Cultures were grown in FMC minimal medium as described in Materials and Methods. Statistical significance was determined by Student's t test. *, P ≤ 0.0000001 (n = 3). Pairwise comparisons are represented by lowercase letters.
CcpA activates transcription of ilvE.
The ilv-leu operon in B. subtilis is regulated not only by CodY but also by CcpA (26). The operon contains the prototypical cre site in its promoter region, which is characteristic of genes regulated by CcpA. Unlike the ilv-leu genes in other bacteria, which are arranged in operons, ilvE in S. mutans is located independently from other branched-chain amino acid biosynthetic genes in the genome, suggesting that regulation of ilvE could be distinct from that of the other genes involved in bcAA metabolism.
The ilvE promoter region used in transcriptional assays in this study did not contain a strong cre site; however, given data from studies performed on other organisms showing a link between carbon catabolite repression and bcAA synthesis (26), we explored this relationship in S. mutans. Transcription of ilvE was determined via a cat reporter fusion construct in a ccpA deletion strain (UR268). In the absence of CcpA, transcription of ilvE was reduced compared to the expression observed in the parent strain or in the ΔcodY strain (Fig. 8A). The data support the role of CcpA as an activator of branched-chain amino acid biosynthetic genes. Despite the absence of a well-defined cre site, the loss of CcpA had an effect on ilvE expression, suggesting that CcpA indirectly plays a role in regulation of ilvE or that a less-conserved binding site or binding site location is present within the intergenic region preceding the gene (Fig. 1). Similar to results seen for expression of ilvE in a ΔcodY strain, the effect of pH on transcription was not obvious, as ilvE transcription in UR268 was diminished in an equivalent manner in both strains, independent of the pH value of the growth media.
Transcription of ilvE under nutrient-limiting conditions is independent of CcpA but enhanced in the absence of CodY.
The data presented thus far show that transcription of ilvE is affected by the loss of either the CodY or CcpA regulator. Since both of these regulators of gene expression are, in turn, influenced by nutrient levels in the growth medium, we explored the effect on ilvE transcription under nutrient-limiting conditions in the absence of either regulator. In FMC minimal medium, similar levels of transcription were observed in both the parent strain background and the ΔccpA background when cultures were grown under identical conditions (Fig. 8C). The data show that under conditions of nutritional limitation, ilvE transcription was induced independently of CcpA. In the absence of CodY and under nutrient-limited growth conditions, ilvE transcription was upregulated approximately 5-fold at pH 5 and pH 7 compared to levels seen in the parent strain background (Fig. 8B). The results support the role of CodY in regulation of ilvE transcription in response to both nutritional availability and environmental pH.
DISCUSSION
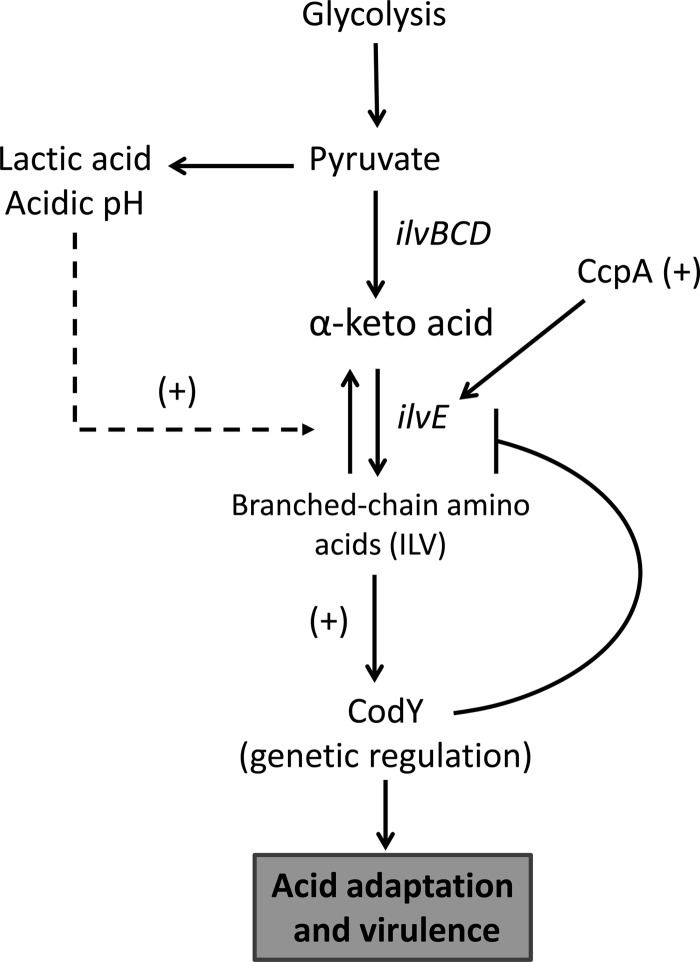
Previous reports from our group and many others have focused on the nature and extent of the acid response pathways in oral streptococci (7). One mechanism of protection from acidification is the rerouting of carbon away from lactic acid. Rerouting carbon away from pyruvate would have the effect of reducing organic acid production from fermentation and would generate molecules important for bacterial homeostasis. An overflow pathway directly linked to acidic end product rerouting is provided by branched-chain amino acid biosynthesis. Proteins responsible for branched-chain amino acid biosynthesis are upregulated during acid stress and, due to their link in rerouting pyruvate, are important for acid tolerance and amino acid metabolism (Fig. 9) (7). Aminotransferases, which are key metabolic enzymes for bcAA biosynthesis, are central players in the modulation of acidic end products. The importance of IlvE, a branched-chain amino acid aminotransferase, was demonstrated when a strain carrying a deletion in the gene exhibited reduced acid tolerance (28). These findings indicated that altering the metabolic profile of S. mutans toward production of bcAAs affected the acid-adaptive capabilities of the organism and cemented the link between amino acid metabolism and acid resistance. Expression of ilvE was responsive to acidification of the growth medium, which also implied that transcription of the gene was either required or, at minimum, important during acid adaptation.
Fig 9.
Pyruvate overflow pathway through branched-chain amino acid metabolism. The metabolism of carbohydrates leads to the generation of pyruvate, which results in the generation of lactic acid, the major acidic end product responsible for tooth demineralization and caries formation. Pyruvate can be rerouted to the synthesis of branched-chain amino acids by the action of the ilv biosynthetic pathway. CcpA aids in the activation of ilv when cells are exposed to an acidic environment. Branched-chain amino acids, in turn, are important molecules for genetic regulation via their role as coeffectors for CodY transcriptional regulation. As branched-chain amino acid synthesis increases, CodY-dependent repression of the ilv genes also increases, resulting in repression of these gene transcripts when bcAAs are in excess.
As a branched-chain amino acid biosynthetic gene, it was expected that ilvE could also be controlled by a combination of regulatory signals. Chief among these is regulation by CodY, a global regulator of genes in S. mutans known to be important for acid tolerance as well as virulence. The function of CodY as a repressor is dependent upon branched-chain amino acid levels, to which IlvE could contribute via its role as an aminotransferase. The importance of ilvE in acid tolerance and the identification of the gene as a target for CodY regulation led us to explore transcriptional regulation of ilvE by CodY in combination with the role of acidification of the extracellular environment.
The presence of a CodY consensus binding sequence and previous studies (11, 17, 20, 22) identifying ilvE as a target for CodY regulation prompted an investigation to determine if CodY was able to interact physically with the ilvE promoter region in S. mutans. EMSAs showed that CodY bound the ilvE promoter region in a specific manner, since the removal of the CodY consensus sequence caused an abrogation of the interaction (Fig. 5). The interaction was also shown to be similar to CodY binding in L. lactis, where isoleucine enhanced the binding of CodY to ilvE without a further requirement for GTP. These data show that in S. mutans, branched-chain amino acids are the predominant coeffectors in mediating an interaction between CodY and ilvE and that CodY repression responds to changes in amino acid levels, not to changes in the levels of GTP (Fig. 3 and 4). More importantly, when transcriptional reporter assays were performed, branched-chain amino acids were shown to repress ilvE transcription. The role of these metabolites as effector molecules would be expected, since biosynthetic genes usually exhibit a response to their cognate biosynthetic end products. In this case, the bcAA products of ilvE have been shown to enhance binding of CodY to the ATase encoded by ilvE (Fig. 6). Although ilvE transcription was not dramatically elevated in the parent strain under nutrient-limiting conditions, bcAAs were unable to repress transcription as they did in rich medium, thus demonstrating the requirement of these amino acids for growth in minimal medium and, in part, explaining the derepression of ilvE transcription (Fig. 6B). A characteristic of ilvE transcription in minimal medium was the 2-fold increase in transcriptional activity at pH 7 compared to expression in rich medium. This may have been caused by nutrient limitation conditions, where bcAA-mediated repression of ilvE transcription would not occur, potentially due to the fact that bcAA levels cannot accumulate and therefore CodY is unable to exert its repressive action on ilvE. In support of the increased transcriptional activity observed under nutrient-limiting conditions, experiments performed using FMC minimal medium showed that ATase activity for all three branched-chain amino acids was also increased under these conditions (data not shown). These findings suggest that there is a greater requirement for bcAA synthesis under nutrient-limiting conditions that outpaces the increased expression resulting from the low pH of the growth environment (Fig. 6B).
Enzymatic characterization of the ΔilvE strain demonstrated that IlvE degrades branched-chain amino acids, as evidenced by the reduced catabolic properties of the deletion strain compared to the parent strain. It was also shown by our group that deletion of the gene causes a nutritional requirement for bcAAs, thereby establishing a contribution to bcAA synthesis (28). In the ΔilvE strain, ilvE transcription was reduced approximately 2-fold (Fig. 7). In our previous work, we demonstrated that isoleucine catabolism in a ΔilvE strain was reduced approximately 75% compared to that in the parent strain. Therefore, larger intracellular amounts of isoleucine in the absence of IlvE could enhance CodY repression and account, in part, for the reduced levels of ilvE transcriptional activity in this background.
As suggested by transcriptional and binding assays, CodY was shown to be a regulator of ilvE transcription. In the absence of CodY, ilvE expression is derepressed, as illustrated by the 2-fold increase in ilvE-cat specific activity compared to the levels seen in the parent strain background (Fig. 8B). The absence of CodY also had an effect on the transcriptional activity assayed at neutral pH, where a 5-fold increase in transcription of ilvE was detected in the codY deletion background compared to that in the parent strain. A similar, nearly 2-fold increase in ilvE transcription was also observed when activity was assayed in minimal medium, a condition that could mimic the absence of CodY, since bcAAs are limited. The data indicate that CodY is also important for pH-dependent regulation of ilvE transcription and that, in its absence, derepression occurs at nonacidic pH values.
Strong repression by CodY led us to explore the roles of other regulatory proteins implicated in regulation of biosynthetic genes, such as CcpA, in regulation of ilvE. In a ΔccpA strain, ilvE transcription was decreased compared to that in the parent strain, demonstrating that CcpA activates ilvE transcription (Fig. 8A), despite the presence of a consensus cre sequence within the intergenic region preceding ilvE. This could indicate that a lower-affinity binding site may exist within the ilvE promoter that functions as a binding site for CcpA or that CcpA is indirectly involved with the activation of ilvE transcription. In the absence of an activator, CodY repression is the predominant regulatory effector and may explain why there is no pH-dependent effect on ilvE transcriptional activity in the ccpA deletion strain. Under nutrient-limiting conditions, the ΔccpA strain behaved similarly to the parent strain, displaying derepression independent of the pH of the growth medium.
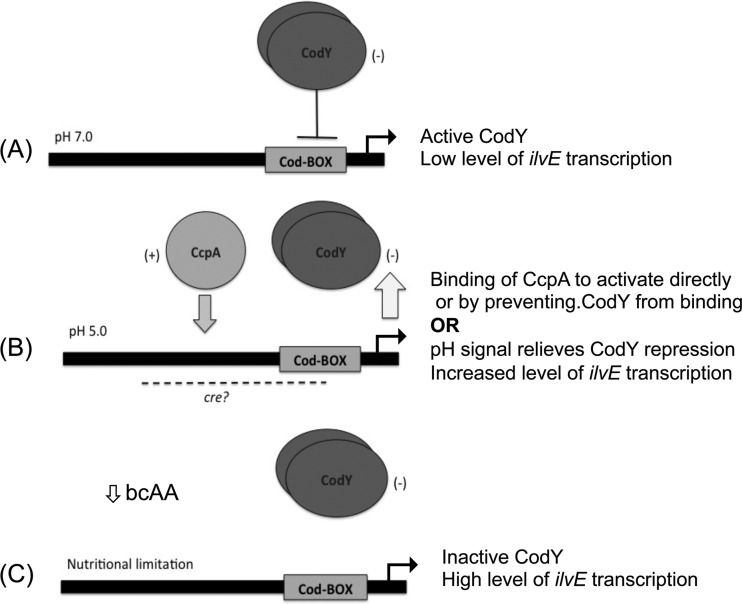
The mechanism of coordinated regulation by these two proteins is based on our current understanding of ilvE regulation. Under rich, nonacidic growth conditions, the “basal” level of ilvE transcription (Fig. 10A) is maintained by CodY, which plays a major role in repression of the gene. When conditions change, due to rapid sugar metabolism and consequent acidification of the environment, CodY repression is partially alleviated, possibly by pH sensing in combination with CcpA activation, which results in higher transcript levels of ilvE at pH 5 (Fig. 10B). Activation seems to be self-limiting, as in many other organisms, since transcript levels of ilvE in the UA159 parent strain background at pH 5, where increased transcription of ilvE occurs, never reach those obtained in a codY deletion background. Positive regulation cannot overcome the negative regulation of ilvE unless CodY is either absent or induced by nutrient limitation, allowing for higher levels of transcription in response to the nutritional needs of the organism (Fig. 10C). This regulatory control allows for expression of ilvE under acid and nutritional stress.
Fig 10.
Proposed model for coordinated regulation of ilvE by CcpA and CodY. (A) The regulation of ilvE in S. mutans combines multiple signals, depending on the growth conditions and extracellular environment. Under nonacidic, nutrient-rich environmental conditions (pH 7.0), ilvE transcription is repressed by the action of CodY. (B) Under acidic conditions (pH 5.0), resulting from the metabolism of carbohydrates and subsequent depletion of nutrients, CodY derepression in combination with CcpA activation allows for increased transcription of ilvE. The CodY and CcpA proteins regulate ilvE by binding to domains within the ilvE intergenic region. The CodY-binding domain is required for interaction. The CcpA-binding domain has yet to be determined. (C) Nutritional regulation is solely dependent on CodY. CodY senses the levels of intracellular amino acids, which drop under nutrient-limiting conditions. Binding of CodY to its target genes is enhanced in the presence of isoleucine; hence, as the levels of bcAAs decrease, CodY repression is alleviated.
A recent publication demonstrating an interaction between CcpA and CodY in Bacillus subtilis (45) encourages us to investigate the possibility of a direct interaction between these regulatory proteins as a mechanism for regulation of ilvE. Other genes involved in the acid tolerance response of S. mutans are currently being examined as potential targets for CodY and CcpA control. Determining whether regulation is due to complex formation between the two regulatory partners is of interest and may be important for the linkage of nutrition, acid adaptation, and virulence in S. mutans.
ACKNOWLEDGMENTS
This study was supported by NIH/NIDCR grants DE017425 and DE017157 and by the Training Program in Oral Infectious Diseases (grant T90DE021985-2).
We thank Cory Hubbard for his assistance with molecular methods and with preparation of the manuscript. We thank José Lemos for purified DnaK and Jacqueline Abranches for helpful discussions.
Footnotes
Published ahead of print 7 June 2013
REFERENCES
- 1. Fozo EM, Quivey RG., Jr 2004. Shifts in the membrane fatty acid profile of Streptococcus mutans enhance survival in acidic environments. Appl. Environ. Microbiol. 70:929–936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Griswold AR, Jameson-Lee M, Burne RA. 2006. Regulation and physiologic significance of the agmatine deiminase system of Streptococcus mutans UA159. J. Bacteriol. 188:834–841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Kuhnert WL, Zheng G, Faustoferri RC, Quivey RG., Jr 2004. The F-ATPase operon promoter of Streptococcus mutans is transcriptionally regulated in response to external pH. J. Bacteriol. 186:8524–8528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Quivey RG, Jr, Kuhnert WL, Hahn K. 2000. Adaptation of oral streptococci to low pH. Adv. Microb. Physiol. 42:239–274 [DOI] [PubMed] [Google Scholar]
- 5. Matsui R, Cvitkovitch D. 2010. Acid tolerance mechanisms utilized by Streptococcus mutans. Future Microbiol. 5:403–417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Chia JS, Lee YY, Huang PT, Chen JY. 2001. Identification of stress-responsive genes in Streptococcus mutans by differential display reverse transcription-PCR. Infect. Immun. 69:2493–2501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Len AC, Harty DW, Jacques NA. 2004. Proteome analysis of Streptococcus mutans metabolic phenotype during acid tolerance. Microbiology 150:1353–1366 [DOI] [PubMed] [Google Scholar]
- 8. Sonenshein AL. 2007. Control of key metabolic intersections in Bacillus subtilis. Nat. Rev. Microbiol. 5:917–927 [DOI] [PubMed] [Google Scholar]
- 9. Bennett HJ, Pearce DM, Glenn S, Taylor CM, Kuhn M, Sonenshein AL, Andrew PW, Roberts IS. 2007. Characterization of relA and codY mutants of Listeria monocytogenes: identification of the CodY regulon and its role in virulence. Mol. Microbiol. 63:1453–1467 [DOI] [PubMed] [Google Scholar]
- 10. Dineen SS, McBride SM, Sonenshein AL. 2010. Integration of metabolism and virulence by Clostridium difficile CodY. J. Bacteriol. 192:5350–5362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Hendriksen WT, Bootsma HJ, Estevao S, Hoogenboezem T, de Jong A, de Groot R, Kuipers OP, Hermans PW. 2008. CodY of Streptococcus pneumoniae: link between nutritional gene regulation and colonization. J. Bacteriol. 190:590–601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Serror P, Sonenshein AL. 1996. Interaction of CodY, a novel Bacillus subtilis DNA-binding protein, with the dpp promoter region. Mol. Microbiol. 20:843–852 [DOI] [PubMed] [Google Scholar]
- 13. Slack FJ, Serror P, Joyce E, Sonenshein AL. 1995. A gene required for nutritional repression of the Bacillus subtilis dipeptide permease operon. Mol. Microbiol. 15:689–702 [DOI] [PubMed] [Google Scholar]
- 14. Sonenshein AL. 2005. CodY, a global regulator of stationary phase and virulence in Gram-positive bacteria. Curr. Opin. Microbiol. 8:203–207 [DOI] [PubMed] [Google Scholar]
- 15. Molle V, Nakaura Y, Shivers RP, Yamaguchi H, Losick R, Fujita Y, Sonenshein AL. 2003. Additional targets of the Bacillus subtilis global regulator CodY identified by chromatin immunoprecipitation and genome-wide transcript analysis. J. Bacteriol. 185:1911–1922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Shivers RP, Sonenshein AL. 2005. Bacillus subtilis ilvB operon: an intersection of global regulons. Mol. Microbiol. 56:1549–1559 [DOI] [PubMed] [Google Scholar]
- 17. Majerczyk CD, Dunman PM, Luong TT, Lee CY, Sadykov MR, Somerville GA, Bodi K, Sonenshein AL. 2010. Direct targets of CodY in Staphylococcus aureus. J. Bacteriol. 192:2861–2877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Villapakkam AC, Handke LD, Belitsky BR, Levdikov VM, Wilkinson AJ, Sonenshein AL. 2009. Genetic and biochemical analysis of the interaction of Bacillus subtilis CodY with branched-chain amino acids. J. Bacteriol. 191:6865–6876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. den Hengst CD, van Hijum SA, Geurts JM, Nauta A, Kok J, Kuipers OP. 2005. The Lactococcus lactis CodY regulon: identification of a conserved cis-regulatory element. J. Biol. Chem. 280:34332–34342 [DOI] [PubMed] [Google Scholar]
- 20. Lemos JA, Nascimento MM, Lin VK, Abranches J, Burne RA. 2008. Global regulation by (p)ppGpp and CodY in Streptococcus mutans. J. Bacteriol. 190:5291–5299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Brinsmade SR, Kleijn RJ, Sauer U, Sonenshein AL. 2010. Regulation of CodY activity through modulation of intracellular branched-chain amino acid pools. J. Bacteriol. 192:6357–6368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Chambellon E, Yvon M. 2003. CodY-regulated aminotransferases AraT and BcaT play a major role in the growth of Lactococcus lactis in milk by regulating the intracellular pool of amino acids. Appl. Environ. Microbiol. 69:3061–3068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Tojo S, Satomura T, Morisaki K, Deutscher J, Hirooka K, Fujita Y. 2005. Elaborate transcription regulation of the Bacillus subtilis ilv-leu operon involved in the biosynthesis of branched-chain amino acids through global regulators of CcpA, CodY and TnrA. Mol. Microbiol. 56:1560–1573 [DOI] [PubMed] [Google Scholar]
- 24. Henkin TM. 1996. The role of CcpA transcriptional regulator in carbon metabolism in Bacillus subtilis. FEMS Microbiol. Lett. 135:9–15 [DOI] [PubMed] [Google Scholar]
- 25. Abranches J, Nascimento MM, Zeng L, Browngardt CM, Wen ZT, Rivera MF, Burne RA. 2008. CcpA regulates central metabolism and virulence gene expression in Streptococcus mutans. J. Bacteriol. 190:2340–2349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Ludwig H, Meinken C, Matin A, Stulke J. 2002. Insufficient expression of the ilv-leu operon encoding enzymes of branched-chain amino acid biosynthesis limits growth of a Bacillus subtilis ccpA mutant. J. Bacteriol. 184:5174–5178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Commichau FM, Forchhammer K, Stulke J. 2006. Regulatory links between carbon and nitrogen metabolism. Curr. Opin. Microbiol. 9:167–172 [DOI] [PubMed] [Google Scholar]
- 28. Santiago B, MacGilvray M, Faustoferri RC, Quivey RG., Jr 2012. The branched-chain amino acid aminotransferase encoded by ilvE is involved in acid tolerance in Streptococcus mutans. J. Bacteriol. 194:2010–2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Guedon E, Sperandio B, Pons N, Ehrlich SD, Renault P. 2005. Overall control of nitrogen metabolism in Lactococcus lactis by CodY, and possible models for CodY regulation in Firmicutes. Microbiology 151:3895–3909 [DOI] [PubMed] [Google Scholar]
- 30. Ajdic D, McShan WM, McLaughlin RE, Savic G, Chang J, Carson MB, Primeaux C, Tian R, Kenton S, Jia H, Lin S, Qian Y, Li S, Zhu H, Najar F, Lai H, White J, Roe BA, Ferretti JJ. 2002. Genome sequence of Streptococcus mutans UA159, a cariogenic dental pathogen. Proc. Natl. Acad. Sci. U. S. A. 99:14434–14439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Murchison HH, Barrett JF, Cardineau GA, Curtiss R., 3rd 1986. Transformation of Streptococcus mutans with chromosomal and shuttle plasmid (pYA629) DNAs. Infect. Immun. 54:273–282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Aslanidis C, de Jong PJ. 1990. Ligation-independent cloning of PCR products (LIC-PCR). Nucleic Acids Res. 18:6069–6074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Lau PC, Sung CK, Lee JH, Morrison DA, Cvitkovitch DG. 2002. PCR ligation mutagenesis in transformable streptococci: application and efficiency. J. Microbiol. Methods 49:193–205 [DOI] [PubMed] [Google Scholar]
- 34. Sheng J, Marquis RE. 2007. Malolactic fermentation by Streptococcus mutans. FEMS Microbiol. Lett. 272:196–201 [DOI] [PubMed] [Google Scholar]
- 35. Derr AM, Faustoferri RC, Betzenhauser MJ, Gonzalez K, Marquis RE, Quivey RG., Jr 2012. Mutation of the NADH oxidase gene (nox) reveals an overlap of the oxygen- and acid-mediated stress responses in Streptococcus mutans. Appl. Environ. Microbiol. 78:1215–1227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Terleckyj B, Willett NP, Shockman GD. 1975. Growth of several cariogenic strains of oral streptococci in a chemically defined medium. Infect. Immun. 11:649–655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Bradford MM. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248–254 [DOI] [PubMed] [Google Scholar]
- 38. Ratnayake-Lecamwasam M, Serror P, Wong KW, Sonenshein AL. 2001. Bacillus subtilis CodY represses early-stationary-phase genes by sensing GTP levels. Genes Dev. 15:1093–1103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Shaw WV, Hopwood DA. 1976. Chloramphenicol acetylation in Streptomyces. J. Gen. Microbiol. 94:159–166 [DOI] [PubMed] [Google Scholar]
- 40. Jerga A, Rock CO. 2009. Acyl-acyl carrier protein regulates transcription of fatty acid biosynthetic genes via the FabT repressor in Streptococcus pneumoniae. J. Biol. Chem. 284:15364–15368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Handke LD, Shivers RP, Sonenshein AL. 2008. Interaction of Bacillus subtilis CodY with GTP. J. Bacteriol. 190:798–806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Majerczyk CD, Sadykov MR, Luong TT, Lee C, Somerville GA, Sonenshein AL. 2008. Staphylococcus aureus CodY negatively regulates virulence gene expression. J. Bacteriol. 190:2257–2265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Madsen SM, Beck HC, Ravn P, Vrang A, Hansen AM, Israelsen H. 2002. Cloning and inactivation of a branched-chain-amino-acid aminotransferase gene from Staphylococcus carnosus and characterization of the enzyme. Appl. Environ. Microbiol. 68:4007–4014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Terleckyj B, Shockman GD. 1975. Amino acid requirements of Streptococcus mutans and other oral streptococci. Infect. Immun. 11:656–664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Wunsche A, Hammer E, Bartholomae M, Volker U, Burkovski A, Seidel G, Hillen W. 2012. CcpA forms complexes with CodY and RpoA in Bacillus subtilis. FEBS J. 279:2201–2214 [DOI] [PubMed] [Google Scholar]