Abstract
NodD1, a member of the NodD family of LysR-type transcriptional regulators (LTTRs), mediates nodulation (nod) gene expression in the soil bacterium Sinorhizobium meliloti in response to the plant-secreted flavonoid luteolin. We used genetic screens and targeted approaches to identify NodD1 residues that show altered responses to luteolin during the activation of nod gene transcription. Here we report four types of NodD1 mutants. Type I (NodD1 L69F, S104L, D134N, and M193I mutants) displays reduced or no activation of nod gene expression. Type II (NodD1 K205N) is constitutively active but repressed by luteolin. Type III (NodD1 L280F) demonstrates enhanced activity with luteolin compared to that of wild-type NodD1. Type IV (NodD1 D284N) shows moderate constitutive activity yet can still be induced by luteolin. In the absence of luteolin, many mutants display a low binding affinity for nod gene promoter DNA in vitro. Several mutants also show, as does wild-type NodD1, increased affinity for nod gene promoters with added luteolin. All of the NodD1 mutant proteins can homodimerize and heterodimerize with wild-type NodD1. Based on these data and the crystal structures of several LTTRs, we present a structural model of wild-type NodD1, identifying residues important for inducer binding, protein multimerization, and interaction with RNA polymerase at nod gene promoters.
INTRODUCTION
Sinorhizobium meliloti, a Gram-negative soil bacterium, interacts with legumes to establish an intracellular symbiosis in which it fixes molecular dinitrogen into ammonia for use by the host plant. In exchange, the host plant supplies S. meliloti with carbon compounds and other nutrients (1). S. meliloti communicates with its host plant, alfalfa (Medicago sativa), via chemical signaling. First, alfalfa secretes compounds, generally flavonoids, from its seed coats and roots. In response to flavonoids, members of the NodD family of transcriptional activators induce expression of the nod genes (2, 3). NodD1 and NodD2 require plant-derived inducers for activity (such as luteolin, dihydroxymethoxychalcone [MCh], and chrysoeriol for NodD1 and trigonelline, stachydrine, and MCh for NodD2), while NodD3 is constitutively active when overexpressed (3–7). The nod genes encode enzymes that synthesize lipochitooligosaccharide host-signaling compounds known as Nod factors (NFs) (8, 9). The host plant responds to NF by initiating root nodule morphogenesis and differentiation. S. meliloti invades the emerging nodule and fixes nitrogen as an endosymbiont of infected host cells (10, 11).
NodD thus figures prominently as a key component required for host-bacterium recognition and signaling. While the NodD proteins encoded by diverse rhizobia and allied bacteria display a high degree of homology and functional similarity, each is activated by a unique set of flavonoid inducers reflective of its symbiotic plant partner (12–16). For example, in an earlier study, we probed responsiveness of single open reading frame (ORF) constructs of NodD, using a limited set of inducers: S. meliloti nod gene transcription initiated only in response to the alfalfa-derived inducer luteolin, while Rhizobium leguminosarum bv. viciae NodD-mediated transcription (1) was activated by the flavonoids eriodictyol, 7-hydroxyflavone, and naringenin in addition to luteolin (14). NodD is a member of the LysR family of transcriptional activators (LTTRs), the largest family of prokaryotic DNA-binding proteins (>800 members) (17), and better understanding of NodD may yield insight about LTTR mechanisms. In particular, the relationship of allelic forms of NodD to its inducing ligands provides an opportunity for fine-structure understanding of NodD function and of coevolution.
NodD1 binds to a 55-bp highly conserved sequence (nod box) (18–21)made up of two half-sites that lie on the same face of the DNA helix (22). Gel filtration and in vitro cross-linking studies demonstrated that NodD1 dimerizes in solution (R. F. Fisher and S. R. Long, unpublished data), although it may bind to the nod box as a higher-order oligomer; NodD of R. leguminosarum bv. viciae has been reported to bind the nod box as an octamer (23). Binding of S. meliloti NodD1 to the nod box requires the chaperonin GroEL (24) and luteolin (14, 25) for maximal activity. The functions of individual residues in NodD remain unclear for each of these processes.
Central questions about NodD1 mechanisms remain. How is NodD1 altered upon binding luteolin? What domains of NodD1 bind to flavonoids? What domains of NodD1 are involved in transcription activation? To address these questions, here we describe the use of random and site-directed mutagenesis to generate NodD1 mutants that show altered responses to luteolin. We characterize 20 mutants whose residues are distributed along the length of the NodD1 protein. Using DNA-binding and protein oligomerization assays, we compared the behavior of NodD1 mutants to that of wild-type NodD1 during activation of nod gene transcription. With crystal structures of homologous LTTRs as a guide, we generated a molecular model of NodD1 and mapped the mutated residues to this model, allowing us to make predictions about residues important for DNA binding, inducer binding, multimerization, and interaction with RNA polymerase (RNAP), which will be useful in interpretation of future genetic and structural studies.
MATERIALS AND METHODS
Strains, plasmids, and growth conditions.
Bacterial strains and plasmids used in this work are listed in Table S1 in the supplemental material. Tetracycline (Tc)-sensitive variants of XL1-Red (Stratagene) were isolated using methods developed by Bochner et al. (26) and modified by Maloy and Nunn (27). Bacterial cultures were grown in LB medium supplemented with appropriate antibiotics (ampicillin, 50 μg ml−1; chloramphenicol, 50 μg ml−1; spectinomycin, 50 μg ml−1; tetracycline, 10 μg ml−1; streptomycin, 500 μg ml−1; and kanamycin, 25 μg ml−1). For bacterial two-hybrid analysis, cotransformants were introduced into cya Escherichia coli BTH101 and grown on M63 minimal medium with maltose as the sole carbon source at 30°C; for routine cloning and other manipulations, E. coli was grown at 37°C and S. meliloti at 30°C. Plasmids were introduced into S. meliloti by triparental mating using the helper plasmid pRK2013 (28).
Chemicals.
Luteolin was obtained from Atomergic Chemicals (Farmingdale, NY) and dissolved in N,N,-dimethylformamide (DMF).
DNA manipulation and sequencing.
Plasmid DNA isolation and purification were carried out using commercial kits, following the manufacturers' directions. In vitro mutagenesis was performed using the QuikChange site-directed mutagenesis kit (Stratagene). DNA sequencing was carried out on an Applied Biosystems Prism 310 machine (Perkin-Elmer).
Plasmid construction.
pMP60 to pMP70 and pMP86 were created by in vitro mutagenesis of pMP50. Primer sequences are available upon request. EcoRV/SpeI inserts from pMP60 to pMP70 and pMP86 were subcloned into XbaI/ScaI-digested pRF771 to create pMP150 to pMP160 and pMP173, respectively. Primers NodD1:S1-XbaI and NodD1:AS1-SalI were used to amplify nodD1 by PCR from pMP60, pMP61, pMP62, pAB100, pAB102, pAB103, and pMP86 to remove the nodD1 C-terminal stop codon and to create an in-frame fusion of NodD1 to the Strep tag (25) (Biometra). XbaI/SalI-digested PCR fragments were ligated to XbaI/SalI-digested pASK75B to yield pMP40 to pMP46, respectively. The NodD1 coding sequence was also cloned into pUT18 and pKT25 to fuse it to the T18 and T25 subdomains of Bordetella pertussis cya. Point mutations in NodD1 were introduced by site-directed mutagenesis. All mutations and vector constructions were confirmed by DNA sequencing.
Isolation of nodD1 mutants.
pRmE43 was treated with 100 mM potassium phosphate (pH 6.0)–5 mM EDTA–1.0 M hydroxylamine for 90 and 120 min at 70°C (29). DNA samples were desalted on G25 spin columns (Amersham Pharmacia), ethanol precipitated, and transformed into electrocompetent DH5α. Alternatively, pRmE43 was transformed into a Tc-sensitive derivative of DNA repair-defective E. coli XL1-Red. Tc-resistant transformants were pooled and cultured in LB for 65 h at 37°C. Mutagenized pRmE43 was conjugated by mass mating into S. meliloti A2105 (4); transformants were screened on TY (30) or M9-sucrose containing the appropriate antibiotics and 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal) with or without 20 μM luteolin. About 5% of the 34,000 colonies screened in the hydroxylamine-treated population appeared white on plates containing luteolin; 1,000 colonies were randomly selected and rescreened as putative activation-deficient mutants. Thirty colonies were selected for further analysis, yielding five activation-deficient mutants. Another activation-deficient mutant was isolated from screening 19,000 XL1-Red mutagenized colonies. One constitutively active mutant was isolated from screening >200,000 colonies of hydroxylamine-treated cells. To ensure that the mutant phenotype segregated with pRmE43, the mutagenized plasmid was conjugated into DH5α and then back into S. meliloti A2105. To identify the location of each mutation, the entire nodD1 ORF was sequenced. To confirm that the identified mutation caused the mutant phenotype, we recreated the point mutations for L69F, S104L, D134N, M193I, K205N, L280F, and D284N. NodD1 expression was confirmed in crude extracts of S. meliloti harboring the nodD1 mutants (data not shown). In Fig. 1B, data for the NodD1 R65C, A66T, L103F/S104L, D134N/D135N, and M193I mutants represent analysis of the mutants after reconjugation into A2105. All other data in Fig. 1B and in subsequent figures represent assays of mutants created by in vitro mutagenesis of nodD1.
Fig 1.
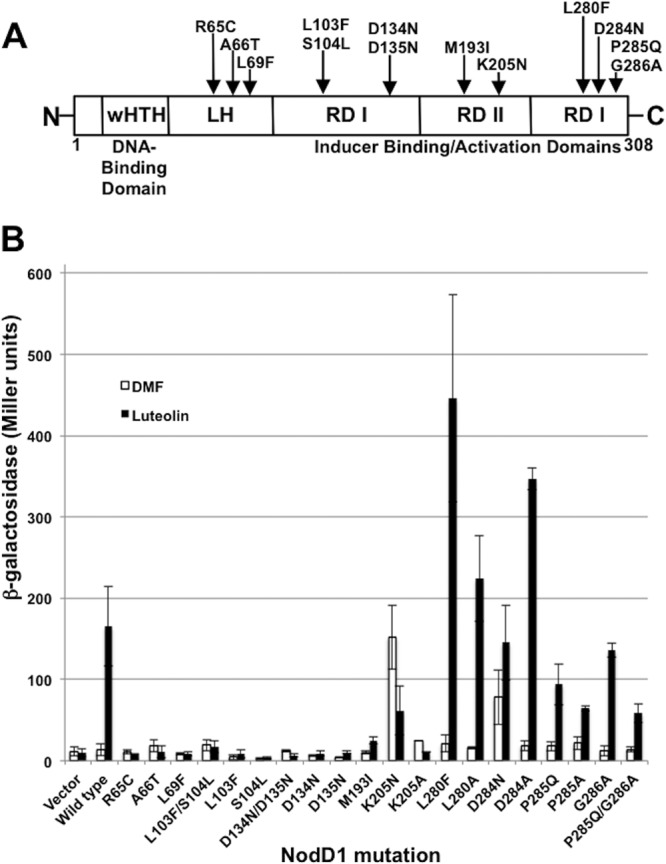
Location of mutations in NodD1. (A) Domain structure of NodD1 containing an N-terminal wHTH DBD (residues 23 to 42), LH (residues 59 to 87), RD I (residues 88 to 165 and 271 to 308), and RD II (residues 171 to 265). Domain borders are approximate and are based on domains in known LTTRs (40, 42, 57, 61). (B) β-Galactosidase activity (Miller units) of a nodC′-′lacZ reporter in S. meliloti A2105 expressing the indicated NodD1 wild-type or point mutation in the absence (white) or presence (black) of 3 μM luteolin. Data are from at least four independent assays except for mutations R65C, K205A, and D284A, for which the data are from two independent assays.
Purification of mutant NodD1 proteins.
Strep-tagged NodD1, referred to here as affinity-purified NodD1, was isolated from E. coli carrying wild-type or mutant nodD1 (pNodD1-ST or pMP40 to pMP46) grown 3 to 4 h in the presence of DMF or 3 μM luteolin as previously described (25). Total protein concentration was determined using a modified Bradford assay (Bio-Rad). Coomassie blue-stained SDS-polyacrylamide gels were scanned, and the amounts of NodD1 and GroEL, the only two bands visible, were determined to be 4.5 NodD1 monomers per monomer of GroEL. The Strep tag did not interfere with NodD1 function: mutant alleles showed similar activation of nod gene expression in S. meliloti (data not shown).
Electrophoretic mobility shift assay (EMSA).
End-labeled nodF nod box DNA (6 fmol) was mixed with increasing amounts of wild-type or mutant NodD1 (0 to 5.9 μM), and NodD1-DNA complexes and free DNA were separated on 5% Tris-Borate-EDTA (TBE) polyacrylamide gels as previously described (19, 22). Wild-type and mutant NodD1-nod box complexes migrated to the same position on the gel (data not shown). Bands representing unbound DNA and the NodD1-DNA complex were quantified on a phosphorimager (GS-363; Bio-Rad); the fraction bound represents the amount of NodD1-DNA complex/total input DNA.
Data analysis.
Because individual preparations varied in overall activity, NodD1 samples that showed consistent DNA-binding activity in the absence versus presence of luteolin were used to compare effects of different mutations in NodD1. Data were analyzed as previously described (14). Briefly, outliers were identified and discarded using a Dixon test (α < 0.05) (31, 32). Two-tailed probabilities of differences in DNA binding by NodD1 isolated from cells grown in the presence or absence of luteolin were calculated by a Wilcoxon two-sample test (31, 32). Box-plot boxes were divided at the median, and the tops and bottoms were drawn at the upper and lower quartiles (33). Top and bottom whiskers represent 1.5 interquartile ranges of the top and bottom, respectively. Observations beyond these limits were plotted as individual points.
β-Galactosidase assays.
We used a nodC-lacZ reporter to assess nod gene expression in S. meliloti cultures grown for 2 to 5 h with 3 μM luteolin unless otherwise indicated (34). For bacterial two-hybrid analyses, assays were adapted to a 96-well format as previously described (35).
NodD1 oligomerization assays.
We used DNA sequences encoding wild-type and mutant NodD1s to create fusions to the T18 and T25 catalytic subdomains of Bordetella pertussis cya (36). When appropriate NodD1 monomers interact, the catalytic subdomains restore adenylate cyclase activity and cAMP synthesis, which we monitored by assaying a cAMP-regulated β-galactosidase reporter and confirmed by growth on minimal maltose medium (36).
Molecular modeling.
We used the psiblast software program (37) and the NodD1 sequence to search the Protein Data Bank (38) to identify structural homologs and the Probcons program (39) to align NodD1 with Burkholderia sp. DntR (1UTB) (40), Acinetobacter baylyi BenM (3K1N) (41), Ralstonia eutropha CbnR (1IZ1) (42), Neisseria meningitidis CrgA (3HHG) (43), and N. meningitidis OxyR (3JV9) (44). Sequences were corrected to match their respective pdb files. NodD1 was modeled as a dimer with two chains in the program MODELLER 9v8 (45) (46) using the structural data of the aligned sequences. Graphical representations of each model were generated with the Pymol program (v1.3). The B chain of the dimer resolved with increased refinement of secondary structure and was used for the monomer graphics. The complete file for the dimer model is available at http://cmgm.stanford.edu/biology/long/NodD1_model.html.
RESULTS
Screen for nodD1 alleles showing altered responses to luteolin.
To identify NodD1 residues involved in transcription activation and obtain tools for use in biochemical studies, we used random chemical and in vivo mutagenesis to isolate alleles of nodD1 that are either inactive in the presence of inducers (activation deficient) or active even in the absence of inducers (constitutively active). We used S. meliloti A2105 for mutant screening; A2105 contains insertions in all three nodD genes in addition to a nodC-lacZ fusion. This reporter fusion allows us to assess nod gene activation that is dependent on a sole source of NodD1: the introduced products of the chemical and in vivo mutageneses. We screened 34,000 colonies from cells treated with hydroxylamine and isolated five independent mutations (R65C, A66T, L69F, and the double point mutations L103F/S104L and D134N/D135N) that span the amino-terminal two-thirds of NodD1 (Fig. 1A). Each of these five mutants showed nod gene induction levels of <10% of that mediated by wild-type NodD1 with luteolin (Fig. 1B). Mutants carrying the individual point mutations L103F, S104L, D134N, and D135N from the two double point mutations above failed to activate nod gene expression in response to luteolin (Fig. 1B). We also screened 19,000 colonies containing nodD1 mutagenized in the E. coli mutator strain XL1-Red and found one mutant (M193I) that induced the nodC-lacZ fusion at ∼15% of the level seen with wild-type NodD1 (Fig. 1B). Based on homology to known LTTR structures (40, 42), we predict that the R65C, A66T, and L69F mutations cluster in the linker connecting the N-terminal helix-turn-helix (HTH) domain to the remainder of the protein, while L103F/S104L, D134N/D135N, and M193I map to the central region of the protein (Fig. 1A).
We found one constitutively active mutant (K205N) after screening >200,000 colonies treated with hydroxylamine (Fig. 1A). NodD1 K205N activates nod gene expression without luteolin to levels similar to those seen with wild-type NodD1 in the presence of luteolin (Fig. 1B). Added luteolin, however, represses NodD1 K205N-mediated nod-gene activity by 50% (Fig. 1B). We see the same effect in response to noninducing eriodictyol and 7-hydroxyflavone, flavonoids that cannot induce NodD1-mediated nod gene expression (data not shown), similar to what has been seen in other LTTR constitutively active mutants (47–49). That the K205 residue plays an important role in NodD1 function is shown by the engineered NodD1 K205A mutation, which shows < 2-fold nod gene induction with or without luteolin (Fig. 1B).
In vitro mutagenesis of NodD1.
Genetic screens for NodD mutant proteins previously carried out in closely related rhizobia that encode only a single NodD protein (50, 51) discovered residues that affected the response to their corresponding flavonoid; these were not recovered in our screens. For example, R. leguminosarum bv. trifolii NodD L280F and R. leguminosarum bv. viciae NodD D284N are both constitutively active and induce nod gene expression in response to a broader spectrum of inducers than their wild-type counterparts (29, 49). Because these residues are conserved in NodD1, we investigated their importance by constructing the corresponding point mutations in NodD1. While S. meliloti NodD1 L280F is not constitutively active, it is hyperinducible: luteolin activated nod gene expression levels 2- to 3-fold higher than those activated by wild-type NodD1 (Fig. 1B). It also partially induced nod gene expression with the noninducers eriodictyol and 7-hydroxyflavone (data not shown), making it a more promiscuous sensor than wild-type NodD1. In contrast, NodD1 D284N is constitutively active and also inducible uniquely with luteolin (Fig. 1B and data not shown). NodD1 L280A and NodD1 D284A are both hyperinducible with luteolin, highlighting the importance of these two residues in luteolin-mediated nod gene activation (Fig. 1B).
In S. meliloti, NodD3 needs no inducer for activity when ectopically expressed (3). While residues L280 and D284 are conserved between NodD proteins, the adjacent residues at positions 285 and 286 are not. To test if these residues are responsible for NodD3's constitutively active phenotype, we created single and double point mutations in NodD1 to reproduce the NodD3 sequence at these positions (P285Q, G286A, and the double mutation P285Q/G286A). Altering these residues failed to produce a constitutively active NodD protein, and such proteins, as well as NodD1 P285A, showed lower nod gene induction with luteolin than that for wild-type NodD1 (Fig. 1B).
We chose seven nodD1 mutations for further study based on their conservation between NodD alleles and on their unique phenotypes: type I, uninducible (L69F, S104L, and D134N) or poorly inducible (M193I); type II, constitutive yet repressible (K205N); type III, hyperinducible (L280F); and type IV, constitutive yet inducible (D284N) (Table 1).
Table 1.
Types of NodD1 mutantsa
| Class | NodD1 mutation | Location |
nod gene expression |
DNA binding |
Dimerization | Proposed function | ||
|---|---|---|---|---|---|---|---|---|
| − Luteolin | + Luteolin | − Luteolin | + Luteolin | |||||
| WT | None | Not active | Inducible | Yes | Increased | Yes | ||
| I | L69F | LH | Not active | Not active | No | No | Yes | DNA binding |
| S104L | RD I | Not active | Not active | No | No | Yes | Inducer binding | |
| D134N | RD I | Not active | Not active | Yes | Increased | Yes | Oligomerization/inducer binding | |
| M193I | RD II | Not active | Not active | No | Weak | Yes | Contact RNAP | |
| II | K205N | RD II | Constitutive | Repressed | Very weak | Weak | Yes | Inducer binding |
| III | L280F | RD I | Not active | Enhanced sensitivity | No | Increased | Yes | Contact RNAP |
| IV | D284N | RD I | Constitutive | Inducible | No | Increased | Yes | Contact RNAP |
WT, wild-type; LH, linker helix; RD I, regulatory domain I; RD II, regulatory domain II.
Response to luteolin.
The NodD1 mutants in this study may exhibit differences from wild-type NodD1 in one or more steps required for transcription initiation, including interaction with target DNA or with inducer, oligomerization, or the ability to load RNAP. For each of the NodD mutants, we tested the first three of these properties for altered behavior. We first tested directly for effects of luteolin on target gene expression. The M193I mutant activated nod gene expression to levels ∼ 25% of those seen with wild-type NodD1, but none of the other type I mutants responded to high luteolin (Fig. 2A). Wild-type NodD1 and the type III (L280F) and IV (D284N) mutants responded to 0.25 μM luteolin and gave maximal expression at 5 μM, while the repressible type II (K205N) mutant was sensitive to very low levels of inducer (Fig. 2B and C).
Fig 2.
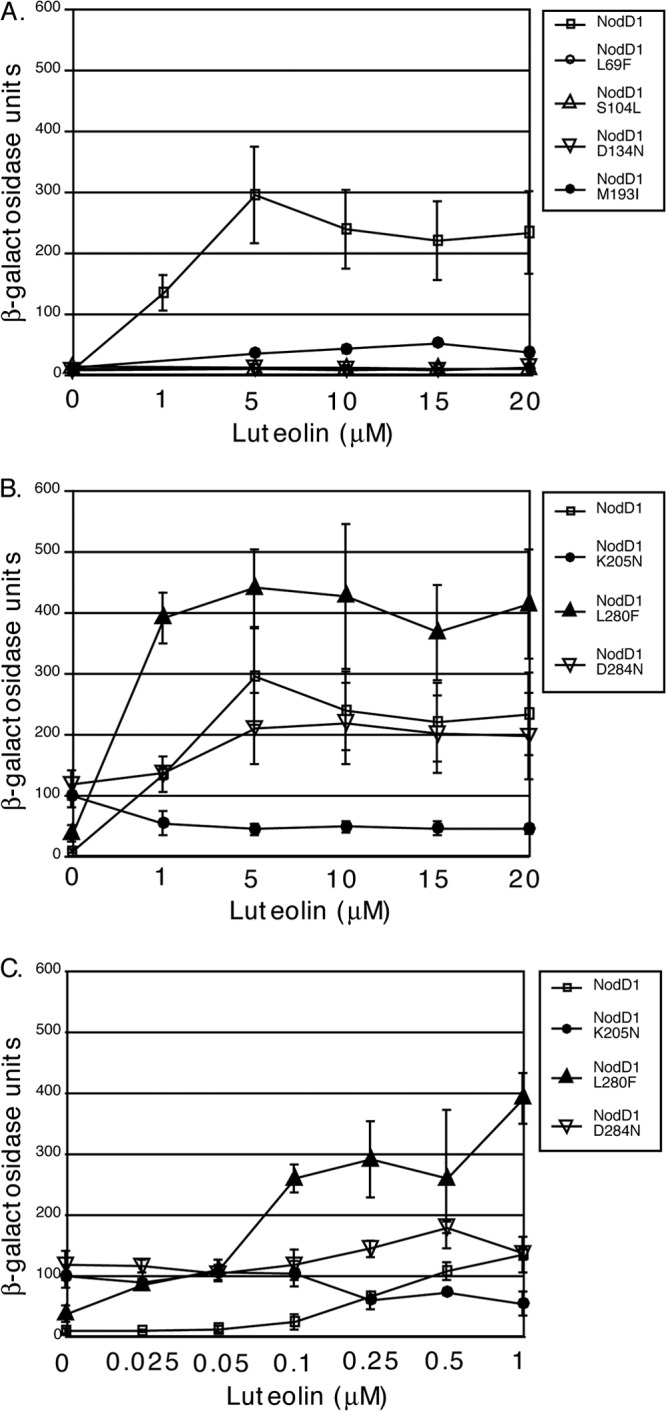
nodC′-′lacZ activity of S. meliloti expressing wild-type NodD1, NodD1 L69F, NodD1 S104L, NodD1 D134N, and NodD1 M193I (A) or wild-type NodD1, NodD1 K205N, NodD1 L280F, and NodD1 D284N (B) in response to 0 to 20 μM luteolin. Data are plotted as the means ± SD and represent ≥4 independent assays except for NodD1 S104L (n = 2). S. meliloti-expressing vector alone gave <16.5 Miller units of β-galactosidase activity under all conditions.
DNA binding activity.
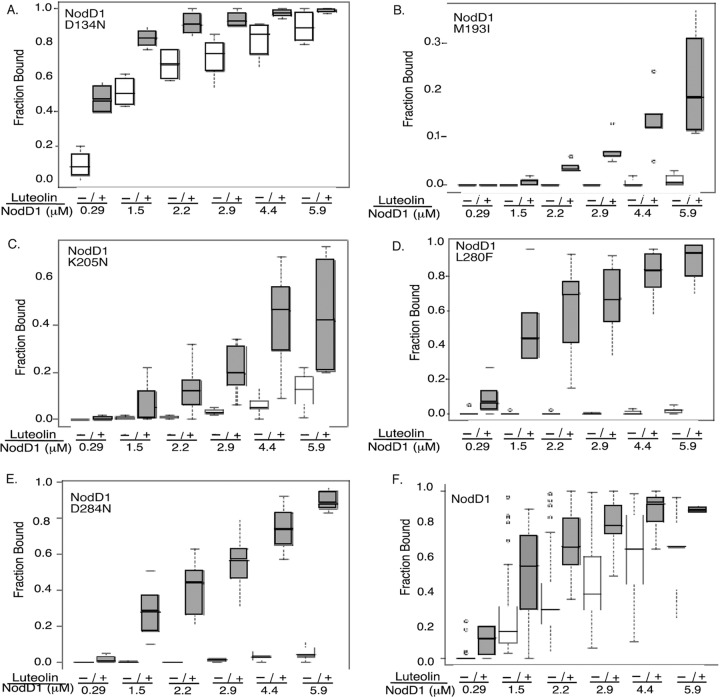
In previous work, we demonstrated that NodD1 shows increased binding to nodF nod box DNA in the presence of luteolin (14, 25). To test for DNA binding activity and luteolin recognition, we affinity purified wild-type and mutant NodD1 proteins from E. coli grown with or without luteolin and titrated binding to the nodF nod box (Fig. 3). The L69F and S104L type I NodD1 mutants, which map near the winged helix-turn-helix DNA-binding domain (wHTH DBD) (52–54), failed to bind the nod box with or without luteolin (data not shown). All of the other mutants tested showed increased nod box binding in the presence of luteolin, indicating their ability to bind luteolin (Fig. 3). Consistent with its weak luteolin-dependent nod gene induction in vivo, the M193I type I mutant weakly bound the nodF promoter in the presence of inducer (Fig. 3B). Much to our surprise, the remaining mutants showed no correlation between in vitro nod box binding and in vivo nod gene expression. For example, the D134N mutant, which failed to activate nod gene expression in vivo (Fig. 1B), unexpectedly showed the most robust nod box binding with and without luteolin (Fig. 3A). In contrast, the type II (K205N) and IV (D284N) mutants, which are constitutively active in vivo (Fig. 1B), showed very little binding to the nodF promoter in the absence of luteolin (Fig. 3C and E). Moreover, while luteolin decreased NodD1 K205N-mediated nod gene transcription in vivo (Fig. 1B), it weakly stimulated its binding to the nod box in vitro (Fig. 3C). Finally, the type III (L280F) mutant and wild-type NodD1 showed comparable DNA binding in the presence of luteolin (Fig. 3D and F) despite the mutant's much higher levels of nod gene induction (Fig. 1B).
Fig 3.
EMSA of wild-type and mutant NodD1 proteins isolated from cells grown in the absence or presence of luteolin. (A to E) NodD1 mutants. (F) Wild-type NodD1. NodD1 L69F and NodD1 S104L demonstrated no binding to the nod gene promoter in the absence or presence of luteolin (data not shown). Increasing amounts of affinity-purified NodD1 containing GroEL isolated from cells grown in the absence (−) versus presence (+) of 3 μM luteolin were incubated with end-labeled nodF nod box DNA as described in Materials and Methods. Data are presented as box plots (see Materials and Methods). Note the difference in scale in panels B and C. Each point represents at least three independent protein purifications and DNA binding assays. P < 0.05 at each protein concentration except for the M1931 (0.29 μM, 1.5 μM), K205N (0.29 μM), and D284N (0.29 μM) mutants, where P < 0.20.
NodD1 oligomerization.
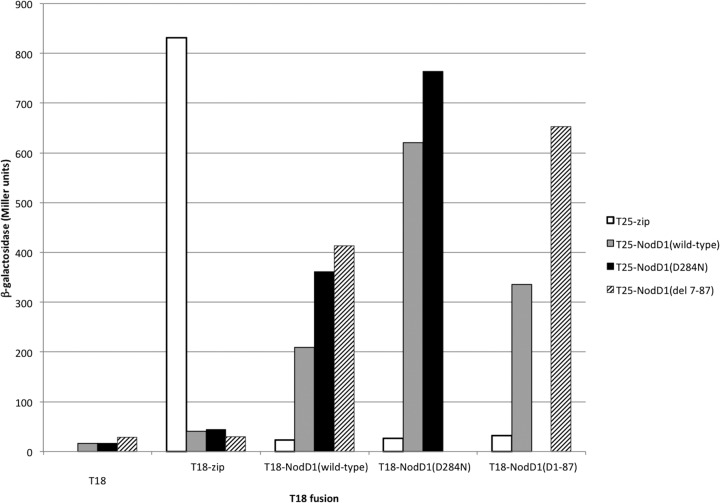
To determine if the NodD1 mutants were still able to dimerize, a property likely critical for proper transcription activation, we used a bacterial two-hybrid system that depends on interaction-mediated reconstitution of adenylate cyclase activity in cya mutant E. coli (36). Restoration of activity, as seen in the positive control with the leucine zipper fused to the two domains of adenylate cyclase, permits synthesis of cAMP, which we monitored qualitatively by growth on minimal maltose medium (see Table S2 in the supplemental material) and quantitatively via a cAMP-regulated β-galactosidase reporter (Fig. 4; see also Table S2). Conversely, low negative-control background levels are seen when the T25-NodD1 fusions are presented with either the T18 catalytic subdomain (T18) alone or T18 fused to the yeast GCN4 leucine zipper (T18-zip) (Fig. 4). NodD1 forms homodimers (Fig. 4), and each of the NodD1 mutants tested retained the ability to form heterodimers with wild-type NodD1 (see Table S2). A representative example of one of the mutant NodD1s, NodD1 D284N, shows that it can form dimers with itself and with wild-type NodD1 (Fig. 4). As with wild-type NodD1, the mutant fails to reconstitute adenylate cyclase activity with the negative controls T18 and T18-zip (Fig. 4). We also created an 81-amino-acid deletion (residues 7 to 87) in the N-terminal domain (NTD) of NodD1, which includes the wHTH DBD (Fig. 1A), and found that it could still interact with both wild-type NodD1 and itself but not with the negative controls (Fig. 4). We thus conclude that the NTD of NodD1 is not absolutely required for NodD1 dimerization.
Fig 4.
Wild-type NodD1 forms homodimers with wild-type NodD1 and heterodimers with mutant NodD1. β-Galactosidase activity (Miller units) of a lacZ reporter in cya mutant E. coli BTH101 expressing the indicated fusions to the T18 and T25 cya subdomains is shown. Interaction between NodD1 monomers reconstitutes active Cya and results in increased β-galactosidase activity. Each of the T25-NodD1 fusions interacts with each of the T18-NodD1 fusions. None of the T25 fusions interact with the T18 catalytic subdomain alone. Only the T25-zip fusion interacts with T18-zip.
Structural model of NodD1.
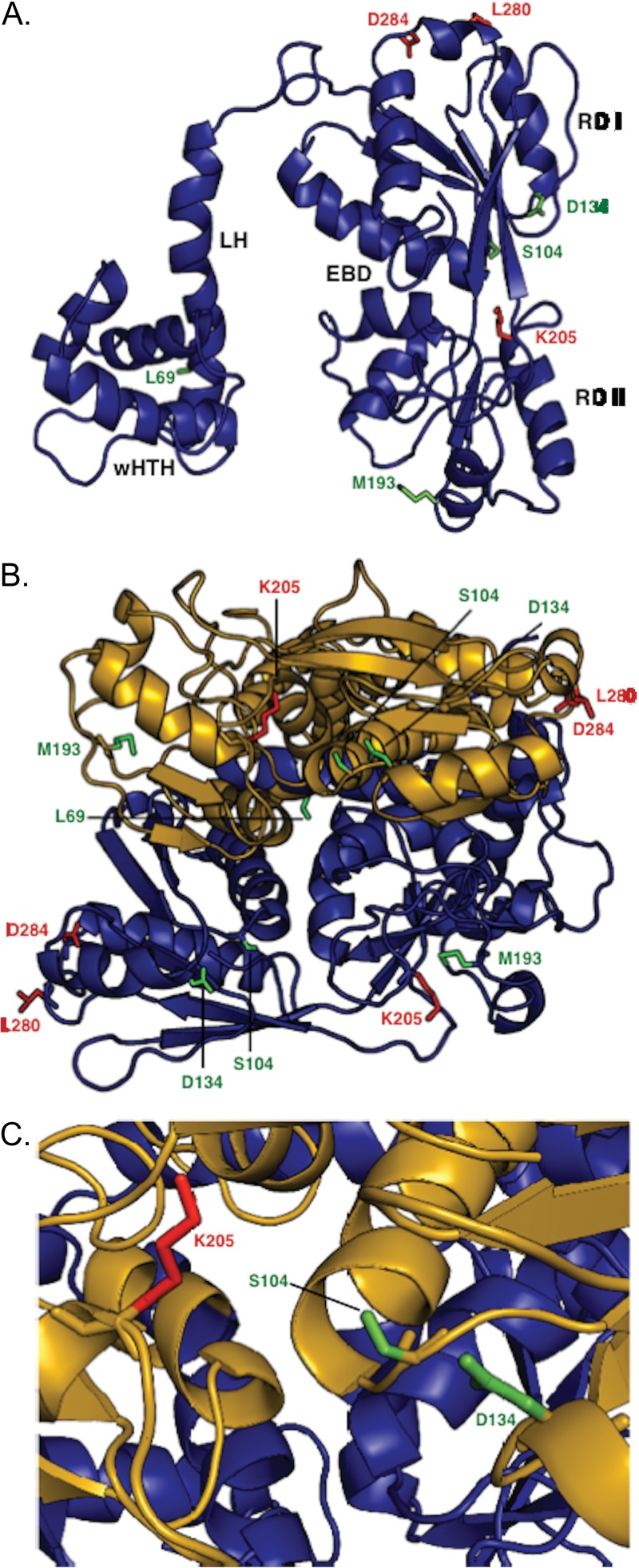
We used the MODELLER computer program (55) to generate a preliminary model of the structure of NodD1 based on the crystal structures of LTTRs with conserved sequence identity: DntR (31.6%) (40), BenM (19.6%) (41), CbnR (19.5%) (42), CrgA (19.1%) (56), and OxyR (18.8%) (44). We took advantage of the fact that crystal structures are highly conserved among LTTRs despite <20% overall sequence identity. Each monomer consists of a wHTH DBD, linker helix (LH), and two α/β domains, termed regulatory domain I (RD I) and regulatory domain II (RD II), enclosing a cavity postulated to be the effector binding domain (EBD) based on cocrystallization with effector molecules and on LTTR mutant studies (40, 42, 43, 57–62). LTTRs can make either symmetric or asymmetric dimers. The best-fitting model of NodD1 required an asymmetric dimer: each monomer (Fig. 5A) is related by 2-fold symmetry, so that the N and C termini of each monomer lie on opposite sides of the dimer (Fig. 5B). We mapped the locations of the NodD1 mutations onto the monomer model (Fig. 5A): L69F is in the linker region, mapping near the site of the wHTH DBD. S104L and K205N both map to the entrance of the EBD (Fig. 5C), while the spatially adjacent D134 maps to the outer face of RD I. M193 is located on the outer face of RD II, while L280 and D284 map to the outer face of RD I near the carboxy terminus.
Fig 5.
Structural model of NodD1 based on crystal structures of Burkholderia sp. DntR (1UTB) (40), Acinetobacter baylyi BenM (3K1N) (41), Ralstonia eutropha CbnR (1IZ1) (42), Neisseria meningitidis CrgA (3HHG) (43), and N. meningitidis OxyR (3JV9) (44). All structures shown in backbone ribbon denote α-helices and β-sheets. Mutant residues are shown in stick figure. Residues L69, S104, D134, and M193, representing type I NodD1 mutants, are shown in green. Residues K205, L280F, and D284, representing type II to IV NodD1 mutants, are shown in red. (A) The modeled NodD1 monomer is shown with the various domains labeled: wHTH, winged helix-turn-helix; LH, linker helix; RD I, regulatory domain I; RD II, regulatory domain II; EBD, effector binding domain. (B) Model of NodD1 as a dimer. The EBDs are formed by clefts in each monomer. (C) Closeup view of the modeled NodD1 EBD.
DISCUSSION
NodD1 plays a key role in establishment of nitrogen-fixing symbiosis by sensing the initial flavonoid signal from the host plant and activating expression of the bacterial genes required to make the responding NF signal compound. Our goal is to advance knowledge of NodD1 structure and function so as to better understand the mechanism of NodD1 action. Here we sought to deduce which parts of the protein are involved in perception and transduction of the flavonoid signal. We used random and direct mutageneses to construct, isolate, and characterize NodD1 mutations (Table 1). Type I mutants (L69F, S104L, D134N, and M193I) fail to activate or weakly activate nod gene expression with or without luteolin. The single type II mutant (K205N) is constitutively active for nod gene expression yet represses with luteolin. The type III mutant (L280F), inactive without inducer, is 3-fold more sensitive to luteolin than is wild-type NodD1. The type IV mutant (D284N) is constitutively active yet still induced by luteolin to absolute levels similar to that of wild-type NodD1 with luteolin.
With the exception of one type I mutant (D134N), all variants show lower promoter binding affinities in the absence of luteolin than wild-type NodD1 (Fig. 3). Other LTTRs have also shown no correlation between in vitro DNA binding and in vivo activation: constitutively active mutants isolated in OxyR show very weak binding to target promoters in vitro (52); constitutively active mutant NodD S95P in R. leguminosarum bv. viciae cannot autoregulate nodD expression (49), a likely consequence of weakened binding to nod gene promoters. For the S. meliloti type II and IV mutants described here, there is no correlation of in vitro and in vivo activity. This might be explained if the mutant NodD1 proteins are less stable than wild-type NodD1 during purification and assay of DNA binding properties, thus leading to less DNA binding in vitro than what occurs in vivo. We note that we did not detect differences in expression when we examined crude extracts from cells harboring the mutant NodD1s (data not shown). Alternatively, mutant NodD1-nod box binding may be stabilized by RNAP in vivo, allowing more rapid promoter escape and constitutive activation, which is not reflected in our purified gel shift system.
All mutant NodD1 proteins except the L69F and S104L type I mutants exhibited enhanced binding to nod gene promoters when isolated from cells exposed to luteolin (Fig. 3A to E), implying that they can still recognize and bind luteolin. Because NodD1 L69F and NodD1 S104L failed to bind to nod gene promoters in either the absence or presence of luteolin, we cannot infer anything about their interaction with luteolin. Type II mutant NodD1 K205N doesn't bind nod box DNA in the presence of an inducer as well as does wild-type NodD1 (compare Fig. 3C and F), implying that it has a decreased affinity for luteolin. That idea is consistent with findings of studies of the LTTR Cbl, where a similar mutation (Cbl T202A) yielded a constitutively active mutant with significantly reduced inducer binding affinity (60). We previously demonstrated that noninducing flavonoids can still stimulate NodD1 binding to target promoters (14), quantitatively uncoupling the processes of DNA binding and transcriptional activation. An example of this is seen in the D134N type I mutant, which shows increased nod gene promoter binding in the presence of luteolin (Fig. 3A) but fails to activate nod gene expression (Fig. 1B). Our findings are consistent with a model where the ligand-binding pocket remains intact but the mutant NodD1 proteins cannot undergo the downstream interactions necessary for transcription activation.
The NodD1 point mutants characterized in this study each retained the ability to form homo- and heterodimers in vivo (Fig. 4; see also Table S2 in the supplemental material). We could not assess with the bacterial two-hybrid assay if higher-order oligomerization occurs. While most LTTRs are thought to function as tetramers (54), only the LTTR CbnR crystallized as a tetramer (42); the other LTTR crystal structures all formed asymmetric dimers (40, 41, 57, 60–63). Differences in oligomerization states may be due to differences in crystallization conditions or the absence of DNA-binding domains in some of the crystallized LTTRs (58). Sequence similarity between proposed LTTR oligomerization domains is low (58), suggesting that multiple weak interactions at relatively small interfaces contribute to oligomerization, rather than specific amino acid interactions (57). For example, in OxyR, seven amino acids mapping to the dimer interface identified as necessary for oligomerization by site-directed mutagenesis are not conserved among OxyR orthologs (64).
An ideal goal is to obtain crystal structures of NodD1 with and without its ligands (both inducing[i.e., luteolin] and noninducing [e.g., eriodictyol and 7-hydroxyflavone]) and of the NodD1 mutant proteins isolated in this study. To date, repeated attempts at obtaining purified NodD1 for crystallization have been confounded by the presence of the molecular chaperonin GroEL, which copurifies with NodD1 through multiple purification schemes (25, 65) (Fisher and Long, unpublished). We speculate that NodD1 is intrinsically a somewhat unstable protein that requires GroEL to maintain its fully active form and to fold around inducing ligands. As an alternative approach to gain insight into NodD1 structure/function, we generated a structural model (Fig. 5) based on the highly conserved crystal structures of related LTTRs. This approach is speculative, but it provides an opportunity to make functional predictions based on the positions of the mutant NodD1 residues.
Each modeled NodD1 monomer displays the expected conserved features of a wHTH DBD, LH, RDI and RD II, and EBD (Fig. 5A) (66). The NodD1 mutants cluster to four locations on the NodD1 monomer (Fig. 5A). The site represented by the type I mutation L69F, located in the LH, helps form a hydrophobic cluster with other LH residues and the NTD (66), an interaction likely important for maintaining relative orientation of the wHTHs with respect to each other (in the dimer) and to their DNA binding sites. The site represented by the type I mutation D134N lies on the inner face of RD I (Fig. 5A), a region that in other LTTRs maps to a conserved dimer-dimer interface (41, 42, 57, 59–62) (see Fig. S1 in the supplemental material). While the D134N mutant protein forms homodimers, we cannot rule out the possibility that it may be defective in forming biologically relevant dimers involved in tetramer formation, which may be required for nod gene activation. The site represented by the type I mutation S104L and type II mutation K205N lies at the interface of RD I and RD II, i.e., the EBD, which in this case is the putative flavonoid binding pocket (17) (Fig. 5). Both S104 and K205 are located in mutational hotspots clustered around the ligand-binding cavity (61) (see Fig. S1), where point mutations in other LTTRs have constitutively active or activation-deficient phenotypes thought to result from altered effector interactions (29, 47, 48, 52, 60, 67–71). Finally, the site defined by the type I, II, and IV mutations M193I, L280F, and D284N, respectively, maps to the outer face of NodD1 (Fig. 5A), perhaps constituting a domain that interacts with RNAP, as seen in CysB and OxyR (72–74).
In our pursuit of mechanistic understanding of NodD activity, the mutant studies we present here will serve as a foundation for further characterization of this important family of transcriptional activators. One of the strengths of this study is that our model of NodD1 can guide targeted future studies aimed at defining the overall structure and function of NodD1. One enticing prospect is to extend both mutant studies and structural analysis to examine how NodD proteins from different symbiotic bacteria interact with the distinct flavonoid signals produced by their respective host plants, an early step that is absolutely critical for the specificity seen in N-fixing symbioses.
Supplementary Material
ACKNOWLEDGMENTS
We thank Anh Bui and David Keating for assistance with isolation of the activation-deficient NodD1 mutants, Jerry Tsai (University of the Pacific) for assistance with generating the NodD1 model, Daniel Ledant (Institut Pasteur) for providing plasmids, strains, and instruction in using the bacterial two-hybrid system, Sid Shaw (Indiana University) for discussions of two-hybrid potential, Dan Gage (University of Connecticut) for the reference to carrying out β-galactosidase assays in a 96-well format, and Dong Wang and other members of the Long laboratory for helpful comments and scientific discussions. We also thank anonymous reviewers for their constructive comments on presentation of this work.
This work was supported by NIH grants RO1 GM30962 and RO1 GM093628 to S.R.L.
Footnotes
Published ahead of print 14 June 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JB.00309-13.
REFERENCES
- 1. Young JP, Johnston AW. 1989. The evolution of specificity in the legume-Rhizobium symbiosis. Trends Ecol. Evol. 4: 341– 349 [DOI] [PubMed] [Google Scholar]
- 2. Egelhoff TT, Fisher RF, Jacobs TW, Mulligan JT, Long SR. 1985. Nucleotide sequence of Rhizobium meliloti 1021 nodulation genes: nodD is read divergently from nodABC. DNA 4: 241– 248 [DOI] [PubMed] [Google Scholar]
- 3. Mulligan JT, Long SR. 1989. A family of activator genes regulates expression of Rhizobium meliloti nodulation genes. Genetics 122: 7– 18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Honma MA, Asomaning M, Ausubel FM. 1990. Rhizobium meliloti nodD genes mediate host-specific activation of nodABC. J. Bacteriol. 172: 901– 911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Hartwig UA, Maxwell CA, Joseph CM, Phillips DA. 1989. Interactions among flavonoid nod gene inducers released from alfalfa seeds and roots. Plant Physiol. 91: 1138– 1142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Hartwig UA, Maxwell CA, Joseph CM, Phillips DA. 1990. Chrysoeriol and luteolin released from alfalfa seeds induce nod genes in Rhizobium meliloti. Plant Physiol. 92: 116– 122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Phillips D, Joseph C, Maxwell C. 1992. Trigonelline and stachydrine released from alfalfa seeds activate NodD2 protein in Rhizobium meliloti. Plant Physiol. 99: 1526– 1531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Denarie J, Cullimore J. 1993. Lipo-oligosaccharide nodulation factors: a minireview new class of signaling molecules mediating recognition and morphogenesis. Cell 74: 951– 954 [DOI] [PubMed] [Google Scholar]
- 9. Fisher R, Long SR. 1992. Rhizobium-plant signal exchange. Nature 357: 655– 660 [DOI] [PubMed] [Google Scholar]
- 10. Jones KM, Kobayashi H, Davies BW, Taga ME, Walker GC. 2007. How rhizobial symbionts invade plants: the Sinorhizobium-Medicago model. Nat. Rev. Microbiol. 5: 619– 633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Oldroyd GE, Downie JA. 2008. Coordinating nodule morphogenesis with rhizobial infection in legumes. Annu. Rev. Plant Biol. 59: 519– 546 [DOI] [PubMed] [Google Scholar]
- 12. Bender GL, Nayudu M, Le Strange KK, Rolfe BG. 1988. The nodD1 gene from Rhizobium strain NGR234 is a key determinant in the extension of host range to the nonlegume Parasponia. Mol. Plant Microbe Interact. 1: 259– 266 [Google Scholar]
- 13. Horvath B, Bachem CW, Schell J, Kondorosi A. 1987. Host-specific regulation of nodulation genes in Rhizobium is mediated by a plant-signal, interacting with the nodD gene product. EMBO J. 6: 841– 848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Peck MC, Fisher RF, Long SR. 2006. Diverse flavonoids stimulate NodD1 binding to nod gene promoters in Sinorhizobium meliloti. J. Bacteriol. 188: 5417– 5427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Spaink HP, Wijffelman CA, Pees E, Okker RJH, Lugtenberg BJJ. 1987. Rhizobium nodulation gene nodD as a determinant of host specificity. Nature 328: 337– 340 [Google Scholar]
- 16. Zaat SAJ, Schripsema J, Wijffelman CA, Van Brussel AAN, Lugtenberg BJJ. 1989. Analysis of the major inducers of the Rhizobium nodA promoter from Vicia sativa root exudate and their activity with different nodD genes. Plant Mol. Biol. 13: 175– 188 [DOI] [PubMed] [Google Scholar]
- 17. Maddocks SE, Oyston PC. 2008. Structure and function of the LysR-type transcriptional regulator (LTTR) family proteins. Microbiology 154: 3609– 3623 [DOI] [PubMed] [Google Scholar]
- 18. Fisher R, Long SR. 1989. DNA footprint analysis of the transcriptional activator proteins NodD1 and NodD3 on inducible nod gene promoters. J. Bacteriol. 171: 5492– 5502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Fisher RF, Egelhoff TT, Mulligan JT, Yelton MM, Long SR. 1988. Rhizobium meliloti NodD binds to DNA sequences upstream of inducible nodulation genes, p 391–398 In Bothe H, de Bruijn FJ, Newton WE. (ed), Nitrogen fixation: hundred years after. Gustav Fischer, New York, NY [Google Scholar]
- 20. Rostas K, Kondorosi E, Horvath B, Simoncsits A, Kondorosi A. 1986. Conservation of extended promoter regions of nodulation genes in Rhizobium. Proc. Natl. Acad. Sci. U. S. A. 83: 1757– 1761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Hong GF, Burn JE, Johnston AW. 1987. Evidence that DNA involved in the expression of nodulation (nod) genes in Rhizobium binds to the product of the regulatory gene nodD. Nucleic Acids Res. 15: 9677– 9690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Fisher RF, Long SR. 1993. Interactions of NodD at the nod box: NodD binds to two distinct sites on the same face of the helix and induces a bend in the DNA. J. Mol. Biol. 233: 336– 348 [DOI] [PubMed] [Google Scholar]
- 23. Liu S, Lu H, Hong G. 1998. NodD binds to target DNA in isologous octamer. Sci. China C Life Sci. 41: 592– 599 [DOI] [PubMed] [Google Scholar]
- 24. Ogawa J, Long SR. 1995. The Rhizobium meliloti groELc locus is required for regulation of early nod genes by the transcription activator NodD. Genes Dev. 9: 714– 729 [DOI] [PubMed] [Google Scholar]
- 25. Yeh KC, Peck MC, Long SR. 2002. Luteolin and GroESL modulate in vitro activity of NodD. J. Bacteriol. 184: 525– 530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Bochner BR, Huang HC, Schieven GL, Ames BN. 1980. Positive selection for loss of tetracycline resistance. J. Bacteriol. 143: 926– 933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Maloy SR, Nunn WD. 1981. Selection for loss of tetracycline resistance by Escherichia coli. J. Bacteriol. 145: 1110– 1111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Figurski DH, Helinski DR. 1979. Replication of an origin-containing derivative of plasmid RK2 dependent on a plasmid function provided in trans. Proc. Natl. Acad. Sci. U. S. A. 76: 1648– 1652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Burn J, Rossen L, Johnston AWB. 1987. Four classes of mutations in the nodD gene of Rhizobium leguminosarum biovar viciae that affect its ability to autoregulate and/or activate other nod genes in the presence of flavonoid inducers. Genes Dev. 1: 456– 464 [Google Scholar]
- 30. Beringer JE. 1974. R factor transfer in Rhizobium leguminosarum. J. Gen. Microbiol. 84: 188– 198 [DOI] [PubMed] [Google Scholar]
- 31. Rohlf FJ, Sokal RR. 1995. Statistical tables, 3rd ed. W. H. Freeman and Company, New York, NY [Google Scholar]
- 32. Sokal RR, Rohlf FJ. 1995. Biometry, 3rd ed. W. H. Freeman and Company, New York, NY [Google Scholar]
- 33. Gentleman R, Ihaka R. 1997. The R language. The Interface Foundation of North America, Fairfax Station, VA [Google Scholar]
- 34. Miller BE, Kredich NM. 1987. Purification of the cysB protein from Salmonella typhimurium. J. Biol. Chem. 262: 6006– 6009 [PubMed] [Google Scholar]
- 35. Griffith KL, Wolf RE., Jr 2002. Measuring beta-galactosidase activity in bacteria: cell growth, permeabilization, and enzyme assays in 96-well arrays. Biochem. Biophys. Res. Commun. 290: 397– 402 [DOI] [PubMed] [Google Scholar]
- 36. Karimova G, Pidoux J, Ullmann A, Ladant D. 1998. A bacterial two-hybrid system based on a reconstituted signal transduction pathway. Proc. Natl. Acad. Sci. U. S. A. 95: 5752– 5756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Altschul SF, Lipman DJ. 1990. Protein database searches for multiple alignments. Proc. Natl. Acad. Sci. U. S. A. 87: 5509– 5513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H, Shindyalov IN, Bourne PE. 2000. The Protein Data Bank. Nucleic Acids Res. 28: 235– 242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Do CB, Mahabhashyam MS, Brudno M, Batzoglou S. 2005. ProbCons: probabilistic consistency-based multiple sequence alignment. Genome Res. 15: 330– 340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Smirnova IA, Dian C, Leonard GA, McSweeney S, Birse D, Brzezinski P. 2004. Development of a bacterial biosensor for nitrotoluenes: the crystal structure of the transcriptional regulator DntR. J. Mol. Biol. 340: 405– 418 [DOI] [PubMed] [Google Scholar]
- 41. Ruangprasert A, Craven SH, Neidle EL, Momany C. 2010. Full-length structures of BenM and two variants reveal different oligomerization schemes for LysR-type transcriptional regulators. J. Mol. Biol. 404: 568– 586 [DOI] [PubMed] [Google Scholar]
- 42. Muraoka S, Okumura R, Ogawa N, Nonaka T, Miyashita K, Senda T. 2003. Crystal structure of a full-length LysR-type transcriptional regulator, CbnR: unusual combination of two subunit forms and molecular bases for causing and changing DNA bend. J. Mol. Biol. 328: 555– 566 [DOI] [PubMed] [Google Scholar]
- 43. Sainsbury S, Ren J, Saunders NJ, Stuart DI, Owens RJ. 2008. Crystallization and preliminary X-ray analysis of CrgA, a LysR-type transcriptional regulator from pathogenic Neisseria meningitidis MC58. Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 64: 797– 801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Sainsbury S, Ren J, Nettleship JE, Saunders NJ, Stuart DI, Owens RJ. 2010. The structure of a reduced form of OxyR from Neisseria meningitidis. BMC Struct. Biol. 10: 10. 10.1186/1472-6807-10-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Sali A, Blundell TL. 1993. Comparative protein modelling by satisfaction of spatial restraints. J. Mol. Biol. 234:779– 815 [DOI] [PubMed] [Google Scholar]
- 46. Martí-Renom MA, Stuart AC, Fiser A, Sánchez R, Melo F, Sali A. 2000. Comparative protein structure modeling of genes and genomes. Annu. Rev. Biophys. Biomol. Struct. 29: 291– 325 [DOI] [PubMed] [Google Scholar]
- 47. Dangel AW, Gibson JL, Janssen AP, Tabita FR. 2005. Residues that influence in vivo and in vitro CbbR function in Rhodobacter sphaeroides and identification of a specific region critical for co-inducer recognition. Mol. Microbiol. 57: 1397– 1414 [DOI] [PubMed] [Google Scholar]
- 48. Jorgensen C, Dandanell G. 1999. Isolation and characterization of mutations in the Escherichia coli regulatory protein XapR. J. Bacteriol. 181: 4397– 4403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. McIver J, Djordjevic MA, Weinman JJ, Bender GL, Rolfe BG. 1989. Extension of host range of Rhizobium leguminosarum bv. trifolii caused by point mutations in nodD that result in alterations in regulatory function and recognition of inducer molecules. Mol. Plant Microbe Interact. 2: 97– 106 [DOI] [PubMed] [Google Scholar]
- 50. Schofield PR, Djordevic MA, Rolfe B, Shine J, Watson JM. 1983. A molecular linkage map of nitrogenase and nodulation genes in Rhizobium trifolii. Mol. Gen. Genet. 192: 459– 465 [Google Scholar]
- 51. Shearman CA, Rossen L, Johnston AWB, Downie JA. 1986. The Rhizobium leguminosarum nodulation gene nodF encodes a polypeptide similar to acyl-carrier protein and is regulated by nodD plus a factor in pea root exudate. EMBO J. 5: 647– 652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Kullik I, Stevens J, Toledano MB, Storz G. 1995. Mutational analysis of the redox-sensitive transcriptional regulator OxyR: regions important for DNA binding and multimerization. J. Bacteriol. 177: 1285– 1291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Lochowska A, Iwanicka-Nowicka R, Plochocka D, Hryniewicz MM. 2001. Functional dissection of the LysR-type CysB transcriptional regulator. Regions important for DNA binding, inducer response, oligomerization, and positive control. J. Biol. Chem. 276: 2098– 2107 [DOI] [PubMed] [Google Scholar]
- 54. Schell MA. 1993. Molecular biology of the LysR family of transcriptional regulators. Annu. Rev. Microbiol. 47: 595– 626 [DOI] [PubMed] [Google Scholar]
- 55.Sali A, Webb B.2010. Modeller: a program for protein structure modeling, release 9.8.
- 56. Sainsbury S, Lane LA, Ren J, Gilbert RJ, Saunders NJ, Robinson CV, Stuart DI, Owens RJ. 2009. The structure of CrgA from Neisseria meningitidis reveals a new octameric assembly state for LysR transcriptional regulators. Nucleic Acids Res. 37: 4545– 4558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Choi H, Kim S, Mukhopadhyay P, Cho S, Woo J, Storz G, Ryu S. 2001. Structural basis of the redox switch in the OxyR transcription factor. Cell 105: 103– 113 [DOI] [PubMed] [Google Scholar]
- 58. Ezezika OC, Haddad S, Clark TJ, Neidle EL, Momany C. 2007. Distinct effector-binding sites enable synergistic transcriptional activation by BenM, a LysR-type regulator. J. Mol. Biol. 367: 616– 629 [DOI] [PubMed] [Google Scholar]
- 59. Ezezika OC, Haddad S, Neidle EL, Momany C. 2007. Oligomerization of BenM, a LysR-type transcriptional regulator: structural basis for the aggregation of proteins in this family. Acta Crystallogr. Sect. F Struct. Biol. Cryst Commun. 63: 361– 368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Stec E, Witkowska-Zimny M, Hryniewicz MM, Neumann P, Wilkinson AJ, Brzozowski AM, Verma CS, Zaim J, Wysocki S, Bujacz GD. 2006. Structural basis of the sulphate starvation response in E. coli: crystal structure and mutational analysis of the cofactor-binding domain of the Cbl transcriptional regulator. J. Mol. Biol. 364: 309– 322 [DOI] [PubMed] [Google Scholar]
- 61. Tyrrell R, Verschueren KH, Dodson EJ, Murshudov GN, Addy C, Wilkinson AJ. 1997. The structure of the cofactor-binding fragment of the LysR family member, CysB: a familiar fold with a surprising subunit arrangement. Structure 5: 1017– 1032 [DOI] [PubMed] [Google Scholar]
- 62. Monferrer D, Tralau T, Kertesz MA, Dix I, Sola M, Uson I. 2010. Structural studies on the full-length LysR-type regulator TsaR from Comamonas testosteroni T-2 reveal a novel open conformation of the tetrameric LTTR fold. Mol. Microbiol. 75: 1199– 1214 [DOI] [PubMed] [Google Scholar]
- 63. Balcewich MD, Reeve TM, Orlikow EA, Donald LJ, Vocadlo DJ, Mark BL. 2010. Crystal structure of the AmpR effector binding domain provides insight into the molecular regulation of inducible AmpC beta-lactamase. J. Mol. Biol. 400: 998– 1010 [DOI] [PubMed] [Google Scholar]
- 64. Knapp GS, Tsai JW, Hu JC. 2009. The oligomerization of OxyR in Escherichia coli. Protein Sci. 18: 101– 107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Fisher RF, Egelhoff TT, Mulligan JT, Long SR. 1988. Specific binding of proteins from Rhizobium meliloti cell-free extracts containing NodD to DNA sequences upstream of inducible nodulation genes. Genes Dev. 2: 282– 293 [DOI] [PubMed] [Google Scholar]
- 66. Zaim J, Kierzek AM. 2003. The structure of full-length LysR-type transcriptional regulators. Modeling of the full-length OxyR transcription factor dimer. Nucleic Acids Res. 31: 1444– 1454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Cebolla A, Sousa C, de Lorenzo V. 1997. Effector specificity mutants of the transcriptional activator NahR of naphthalene degrading Pseudomonas define protein sites involved in binding of aromatic inducers. J. Biol. Chem. 272: 3986– 3992 [DOI] [PubMed] [Google Scholar]
- 68. Schell MA, Brown PH, Raju S. 1990. Use of saturation mutagenesis to localize probable functional domains in the NahR protein, a LysR-type transcription activator. J. Biol. Chem. 265: 3844– 3850 [PubMed] [Google Scholar]
- 69. Burn JE, Hamilton WD, Wootton JC, Johnston AWB. 1989. Single and multiple mutations affecting properties of the regulatory gene nodD of Rhizobium. Mol. Microbiol. 3: 1567– 1577 [DOI] [PubMed] [Google Scholar]
- 70. Akakura R, Winans SC. 2002. Constitutive mutations of the OccR regulatory protein affect DNA bending in response to metabolites released from plant tumors. J. Biol. Chem. 277: 5866– 5874 [DOI] [PubMed] [Google Scholar]
- 71. Cho K, Winans SC. 1993. Altered-function mutations in the Agrobacterium tumefaciens OccR protein and in an OccR-regulated promoter. J. Bacteriol. 175: 7715– 7719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Lochowska A, Iwanicka-Nowicka R, Zaim J, Witkowska-Zimny M, Bolewska K, Hryniewicz MM. 2004. Identification of activating region (AR) of Escherichia coli LysR-type transcription factor CysB and CysB contact site on RNA polymerase alpha subunit at the cysP promoter. Mol. Microbiol. 53: 791– 806 [DOI] [PubMed] [Google Scholar]
- 73. Tao K, Fujita N, Ishihama A. 1993. Involvement of the RNA polymerase alpha subunit C-terminal region in co-operative interaction and transcriptional activation with OxyR protein. Mol. Microbiol. 7: 859– 864 [DOI] [PubMed] [Google Scholar]
- 74. Tao K, Zou C, Fujita N, Ishihama A. 1995. Mapping of the OxyR protein contact site in the C-terminal region of RNA polymerase alpha subunit. J. Bacteriol. 177: 6740– 6744 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.