Abstract
Toxigenic Corynebacterium diphtheriae strains cause diphtheria in humans. The toxigenic C. diphtheriae isolate NCTC13129 produces three distinct heterotrimeric pili that contain SpaA, SpaD, and SpaH, making up the shaft structure. The SpaA pili are known to mediate bacterial adherence to pharyngeal epithelial cells. However, to date little is known about the expression of different pili in various clinical isolates and their importance in bacterial pathogenesis. Here, we characterized a large collection of C. diphtheriae clinical isolates for their pilin gene pool by PCR and for the expression of the respective pilins by immunoblotting with antibodies against Spa pilins. Consistent with the role of a virulence factor, the SpaA-type pili were found to be prevalent among the isolates, and most significantly, corynebacterial adherence to pharyngeal epithelial cells was strictly correlated with isolates that were positive for the SpaA pili. By comparison, the isolates were heterogeneous for the presence of SpaD- and SpaH-type pili. Importantly, using Caenorhabditis elegans as a model host for infection, we show here that strain NCTC13129 rapidly killed the nematodes, the phenotype similar to isolates that were positive for toxin and all pilus types. In contrast, isogenic mutants of NCTC13129 lacking SpaA-type pili or devoid of toxin and SpaA pili exhibited delayed killing of nematodes with similar kinetics. Consistently, nontoxigenic or toxigenic isolates that lack one, two, or all three pilus types were also attenuated in virulence. This work signifies the important role of pili in corynebacterial pathogenesis and provides a simple host model to identify additional virulence factors.
INRODUCTION
Corynebacterium diphtheriae is the etiologic agent of cutaneous and pharyngeal diphtheria in humans. This disease is characterized by the formation of a mucous pseudomembrane at the site of infection, either in the nasopharyngeal region or on skin lesions resulting from the combined effects of bacterial growth, production of diphtheria toxin (DT)—which blocks host cell protein synthesis—necrosis of underlying tissues, and the host immune response (1). Toxigenic strains of C. diphtheriae harbor the tox gene encoding DT, which is carried by the corynephage β and a family of corynebacteriophages (2). Nontoxigenic strains of C. diphtheriae infected by a corynebacteriophage that carries the tox gene undergo lysogenic conversion, thereby becoming toxigenic strains (3, 4). A key aspect of diphtheria pathogenesis that has yet to be investigated is the ability of C. diphtheriae to colonize and successfully compete in the nasopharyngeal niche (1). Of the many possible C. diphtheriae colonization factors, pili are likely to play important roles in host colonization by virtue of their ability to mediate bacterial adherence to specific host tissues (5).
Pili of C. diphtheriae were observed by electron microscopy in the late 1970s (6); however, the molecular nature of these pili and the basic mechanism of pilus assembly in Gram-positive bacteria were revealed only during the turn of the century (7). The first genome sequence of corynebacteria, a clinical isolate from the United Kingdom (strain NCTC13129) (8), led to the realization that it harbors three separate pilus gene clusters. Each of these clusters is comprised of tightly linked genes coding for one or two pilin-specific sortases and three distinct pilin substrates that produce a distinct heterotrimeric pilus, designated the SpaA-, SpaD-, and SpaH-type pili (Spa for sortase-mediated pilus assembly) (7). All three pilus types have similar architectures, having a shaft made of a major pilin joined to a specific tip pilin and a base pilin. For example, the well-studied SpaA-type pilus has a shaft made of the major pilin subunit SpaA, with SpaC located at the tip and SpaB found dispersed along the pilus shaft and at the base (7, 9). Genetic, molecular, and cytological studies revealed that the assembly of SpaA pili is a two-step process, whereby a dedicated sortase, SrtA, catalyzes the cross-linking of pilin subunits into a pilus structure, which is terminated by cell wall anchoring of the resulting polymer mediated by the housekeeping sortase SrtF via the base pilin SpaB (9). This biphasic mode of pilus assembly is found commonly in other Gram-positive pilus systems, including those of Bacillus cereus, Streptococcus agalactiae, and Streptococcus pyogenes (10–12).
Significantly, distinct corynebacterial pili mediate adhesion to specific host tissues: Using tissue cultures, Mandlik and colleagues demonstrated that the SpaA-type pili mediate corynebacterial adherence to pharyngeal epithelial cells, whereas SpaD and SpaH pili display specificity for binding to lung and laryngeal epithelial cells (13). The specific binding of corynebacteria to pharyngeal epithelial cells by SpaA pili is attributed to the two minor pilins SpaB and SpaC (13), which cannot only exist in the pilus, but most intriguingly, are linked to the bacterial cell wall in monomeric and heterodimeric forms as well (14). This display of the adhesins in the form of a fiber as well as small entities on bacterial surface may be used to mediate distant as well as tight contacts during the stages of initial encounter and the subsequent colonization. To date, however, very little is known about the distribution of the pilus gene clusters and their surface expression among clinical isolates, as well as their relative and specific contributions to corynebacterial pathogenesis. Information on the former has begun to emerge. By comparative genomic hybridization (15), Iwaki and coworkers revealed that genes similar to spaA, spaB, spaC, and srtA to -E of strain NCTC13129 were present in the genome of the widely used vaccine strain PW8 (16) but not that of the “standard” laboratory strain C7(−) (17). More recently, genome sequencing was extended to 12 strains collected from Brazilian patients with classical diphtheria, endocarditis, and pneumonia, as well as strains C7(β)—C7(−) containing β phage (18)—and PW8 (19). Noticeably, all sequenced strains harbor at least two pilus gene clusters that are located in pathogenicity islands. Amino acid sequence homology and BLASTP analyses revealed that the shaft pilin SpaA and minor pilins SpaB and SpaC were well conserved, while pilins of the SpaD- and SpaH-type pili were highly heterogenous among these isolates.
To obtain a better insight into the roles of corynebacterial pili in pathogenesis, we conducted a comparative functional genomic analysis using 42 C. diphtheriae clinical isolates gathered mostly from Russia, Canada, and the United States (20–23). The pilus gene pool was assessed by PCR amplification using primers for spa genes of NCTC13129, and pilus expression was monitored by immunoblotting as well as immunoelectron microscopy (IEM) using antibodies against respective Spa pilins. First, the majority of these clinical isolates were found to express the heterotrimeric SpaA pili, and this is correlated with bacterial adherence to pharyngeal epithelial cells. Second, we found that SpaD- and SpaH-type pili are more heterogeneous than those of the SpaA type. Finally, toward a functional analysis of bacterial virulence, we have explored Caenorhabditis elegans as a model host for C. diphtheriae because of the already proven versatility, simplicity, and efficacy of the nematode as an experimental model (7, 24). Strikingly, our results reveal that the nontoxigenic strain as well as a toxigenic strain of C. diphtheriae that lack pili are both attenuated in nematode killing compared to the toxigenic, piliated control strain NCTC13129. Our study supports the role of pili in corynebacterial colonization and virulence and provides a simple host model to further investigate the molecular mechanisms of C. diphtheriae pathogenesis.
MATERIALS AND METHODS
Bacterial strains and media.
Corynebacterium diphtheriae NCTC13129 was obtained from the American Type Culture Collection (ATCC), and its isogenic deletion mutants were generated according to a published protocol (7). Forty-two C. diphtheriae clinical isolates were collected between 1997 and 2000. All C. diphtheriae strains (see Table S1 in the supplemental material) were cultured at 37°C in heart infusion broth (HIB), on heart infusion agar (HIA), or on Trypticase soy agar supplemented with 5% sheep blood (Hemostat) (TSASB). Reagents were purchased from Fisher Scientific or Sigma-Aldrich unless otherwise indicated.
Chromosomal DNA extraction and PCR.
Chromosomal DNA was extracted from corynebacteria using the Promega Wizard genomic DNA purification kit according to the manufacturer's protocol. Single primer pairs were designed for PCR-amplification of tox and 16S, based on the genome sequence of the sequenced strain NCTC13129 (8) and conservation of those genes (19); with the potential heterogeneity in the spa pilin genes among the isolates, multiple primer pairs for PCR amplification of spa genes were used (see Table S2 in the supplemental material). PCR amplification was performed as previously described (25).
Extraction of C. diphtheriae pili.
Pili were extracted from the corynebacterial cell wall as previously described (7). Briefly, cells from an overnight culture were washed twice in SMM buffer (0.5 M sucrose, 10 mM MgCl2, and 10 mM maleate [pH 6.8]) and treated with muramidase (300 U/ml) at 37°C for 4 h. Soluble pili were precipitated with 7.5% trichloroacetic acid and washed with acetone. Protein samples were boiled in sodium dodecyl sulfate (SDS) containing sample buffer, separated by 4 to 12% Tris-glycine gradient gels (Invitrogen), and blotted with specific antisera at the following dilutions: anti-SpaA and anti-SpaB, 1:20,000; anti-SpaG, 1:10,000; anti-SpaC, anti-SpaD, anti-SpaH, and anti-SpaI 1:5,000; and anti-SpaE and anti-SpaF, 1:2,000. Polyclonal antibodies raised against recombinant pilins were obtained as previously described (7, 26).
ELISA.
Corynebacteria were grown overnight at 37°C in HIB with shaking. Cells were pelleted (3,000 × g, 2 min), washed, and resuspended in carbonate buffer (15 mM sodium carbonate, 35 mM sodium bicarbonate [pH 9.6]). Enzyme-linked immunosorbent assay (ELISA) wells were coated with 100 μl of the cell suspension. Blocking agent (5% nonfat dry milk in phosphate-buffered saline [PBS] with 0.2% Tween 20) was added to wells to prevent nonspecific binding. For detection, sample wells were incubated with pilin-specific rabbit-raised antibodies diluted in blocking agent (anti-SpaA and anti-SpaD, 1:10,000; anti-SpaH, 1:2,000) for 1 h at room temperature with shaking, followed by addition of horseradish peroxidase (HRP)-conjugated anti-rabbit IgG (1:10,000) and the 3,3′,5,5′-tetramethylbenzidine (TMB) substrate reagent set (BD OptEIA). The reaction was stopped by addition of 1 M sulfuric acid, and the A420 was read on an ELISA plate reader (Tecan Infinite M1000).
Immunoelectron microscopy.
Corynebacteria grown on blood agar plates were scraped, washed in 100 mM NaCl, and resuspended in phosphate-buffered saline (PBS) before immunolabeling according to a previous protocol (26). Briefly, a drop of bacterial suspension was placed onto carbon-coated nickel grids, washed three times with PBS containing 2% bovine serum albumin (BSA), and blocked for 1 h in PBS with 0.1% gelatin. Samples were stained with a specific antiserum (1:100 dilution) for 1 h, followed by washing and blocking. Subsequently, samples were treated with 12- or 18-nm-diameter gold particle-conjugated goat anti-rabbit IgG (Jackson ImmunoResearch) diluted 1:20 in PBS with 2% BSA for 1 h. Samples were washed five times with water before being stained with 1% uranyl acetate and viewed in a JEOL JEM-1400 transmission electron microscope.
Cell cultures and adhesion assays.
Detroit 562 cells (human pharynx carcinoma CCL-138; ATCC) were maintained in Eagle's minimum essential medium (EMEM; ATCC) supplemented with 100 U penicillin, 100 μg/ml streptomycin (penicillin-streptomycin solution; ATCC), and 10% fetal bovine serum (FBS; Gibco) at 37°C with 5% CO2. For adherence assays, epithelial cells at 90% confluence in 12-well tissue culture plates were washed with PBS, and fresh medium (without antibiotics and FBS) was added to the wells. Epithelial cells were infected with corynebacteria grown to the mid-exponential phase at a multiplicity of infection (MOI) of 10. After 1 h of incubation, epithelial cells were washed to remove unbound bacteria and detached with trypsin and 0.025% Triton X-100. Appropriate dilutions of detached cells were plated on TSASB to enumerate adherent bacteria. The percentage of adherent bacteria was determined using the titer of adherent bacteria for each strain compared to the input titer, which was obtained by counting colonies on plates as described above. Each assay was performed in triplicates and repeated three times. Statistical analysis was performed by using Student's t test.
Caenorhabditis elegans killing assays.
C. elegans strain N2 was maintained on nematode growth (NG) agar plates containing a bacterial lawn of OP50 Escherichia coli. Killing assays were performed as previously described (27), with some modifications. Briefly, 15 μl of overnight cultures of C. diphtheriae grown in brain heart infusion (BHI) broth was spread onto BHI agar plates, supplemented with 25 μg ml−1 nalidixic acid and 50 μg/ml 5-fluoro-2-deoxyuridine (FuDR), and incubated at 37°C for 24 h. L4-stage nematodes were transferred to BHI plates containing C. diphtheriae strains and incubated at 25°C for the remainder of the experiment. Dead nematodes were counted and removed every 24 h. For each corynebacterial strain, ∼90 nematodes were used and the assays were performed twice. Kaplan-Meier survival analysis was used, and all statistical analysis was performed with Prism 5.0 (GraphPad, CA), with P values of <0.05 considered significant. LT50, the time required to kill 50% of nematodes, was determined for each bacterial strain.
RESULTS
Heterogeneity of major pilin genes and surface expression of pili in C. diphtheriae clinical isolates.
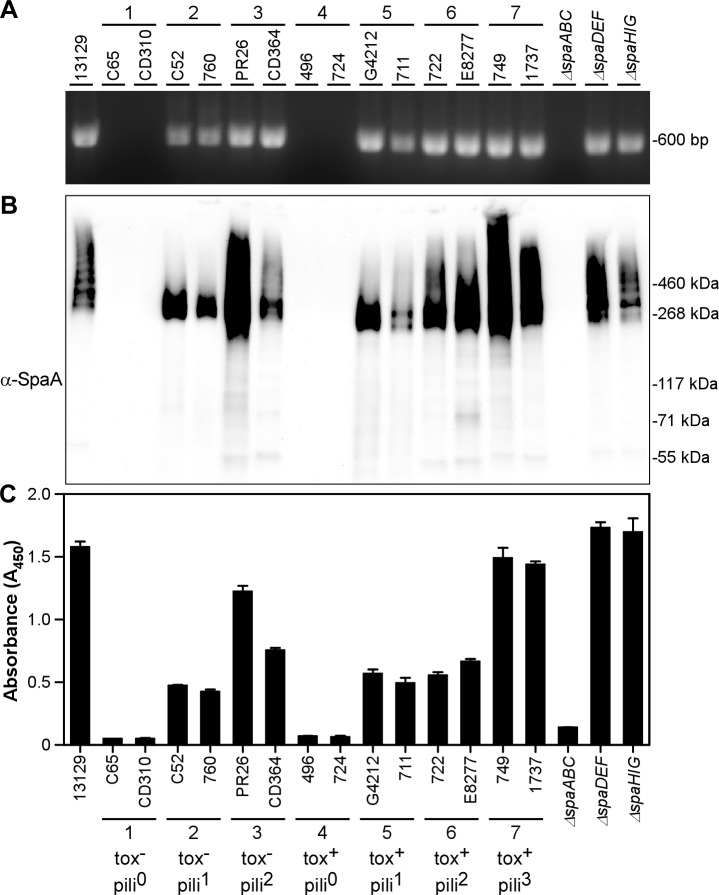
We performed PCR and Western blotting (WB) to examine toxigenicity as well as the presence of major pilin genes and their expression in 42 clinical isolates collected between 1979 and 2000 from different parts of the world (4, 16, 20–23). For PCR analysis, primers targeting the toxin gene (tox) and the major pilin subunits of each pilus type (spaA, spaD, and spaH) were designed based on the sequence of the type strain NCTC13129 (8). Due to potential sequence divergence among the clinical isolates, multiple sets of primers (see Table S2 in the supplemental material) were used for PCR amplification of each pilus type. As a control, primers that target the 16S ribosomal subunit gene were used. Additional control strains in these experiments included the type strain NCTC13129 as well as isogenic strains that lack all pilin genes corresponding to specific pilus gene clusters, i.e., ΔspaABC, ΔspaDEF, and ΔspaHIG strains or all pilin genes (i.e., the ΔspaA–I strain). Western blot analysis was done using a published protocol (14) with preparations of pili extracted from the cell wall by digestion with muramidase; solubilized proteins were trichloroacetic acid (TCA) precipitated and dissolved in hot SDS-sample buffer for gel electrophoresis and immunoblotting with specific antisera. Selected PCR and Western blot results for the spaA locus and the SpaA pili, respectively, are presented in Fig. 1A and B, while the complete set of results for all 42 clinical isolates are summarized in Table S1 in the supplemental material.
Fig 1.
Characterization of C. diphtheriae clinical isolates. Representative clinical isolates from each subgroup were examined for the presence of spaA by PCR amplification (A) and for the expression of SpaA by Western blotting (B) and ELISA (C) with antibodies to SpaA (α-SpaA). ELISA analyses were performed in triplicate, and error bars represent standard deviations. Strain names, subgroups 1 to 7 without toxin (tox−) or with toxin (tox+) and having 0, 1, 2, or 3 types of pili (pili0, pili1, etc.), and markers are indicated.
The 42 clinical isolates that we analyzed can be categorized into seven groups based on the presence or absence of toxin and specific pilus genes (i.e., tox, spaA, spaD, and spaH), of which two representatives from each group are shown in Table 1 and Fig. 1. Groups 1 to 3 are isolates that have no toxin gene and zero, one, or two major pilin subunit genes, respectively. Conversely, groups 4 to 6 contain the tox gene and zero, one, or two major pilin subunit genes. Isolates in group 7 all exhibited a genotype similar to that of the type strain (i.e., they have tox and all three major pilin subunit genes).
Table 1.
Heterogeneity of tox and major pilin genes and pilus expression in corynebacterial strains by PCR and Western blot analysesa
| Group | Strain | Origin | Toxigenicity |
SpaA |
SpaD |
SpaH |
Source or reference | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Reported | PCR | PCR | WB | PCR | WB | PCR | WB | ||||
| 1 | C65 | Canada | − | − | − | − | − | − | − | − | 20 |
| CD310 | Ohio | NDb | − | − | − | − | − | − | − | This study | |
| 2 | C52 | Kazakhstan | − | − | + | + | − | − | − | − | 21 |
| 760 | Russia | − | − | + | + | − | − | − | − | 22 | |
| 3 | PR26 | South Dakota | − | − | + | + | − | − | + | + | 20 |
| CD364 | New York | ND | − | + | + | − | − | + | + | This study | |
| 4 | 496 | Russia | + | + | − | − | − | − | − | − | 22 |
| 724 | Russia | + | + | − | − | − | − | − | − | 22 | |
| 5 | G4212 | Russia | + | + | + | + | − | − | − | − | 22 |
| 711 | Russia | + | + | + | + | − | − | − | − | 22 | |
| 6 | 722 | Russia | + | + | + | + | − | − | + | + | 22 |
| E8277 | South Dakota | + | + | + | + | + | + | − | − | 20 | |
| 7 | 1737 | Russia | + | + | + | + | + | + | + | + | 22 |
| 749 | Russia | + | + | + | + | + | + | + | + | 22 | |
| Sequence strain | 13129c | U.K. | + | + | + | + | + | + | + | + | 8 |
A full list of corynebacterial strains is shown in Table S1 in the supplemental material. Plus and minus signs indicate the presence and absence, respectively, of PCR products or Western immunoblotting (WB) signal.
ND, not determined.
Sequence strain NTCT13129.
To determine whether the strains that contain the various pilin genes encode and express the respective pili, we analyzed cell-wall-linked pili by Western blotting (WB) with antibodies against individual major pilin subunits SpaA, SpaD, or SpaH (Fig. 1B; see Table S1 in the supplemental material). Isolates of groups 2, 3, 5, 6, and 7, which contain the spaA gene, produced respective high-molecular-mass SpaA polymers with various degrees of cross-linking; in contrast, no SpaA polymers are observed in group 1 and 4 isolates, which displayed no PCR product for SpaA (Fig. 1B). Similarly, our analyses of the SpaD and SpaH major pilin subunits (see Table S1) showed that the presence of spaD and/or spaH genes in the categorized strains corresponds to expression of SpaD and/or SpaH polymers as well. Of note, only SpaA monomers were observed in isolates 1899 (group 5) and 765 (group 6), and their surface display was confirmed by immunoelectron microscopy (IEM) (see Table S1 and Fig. S1 in the supplemental material).
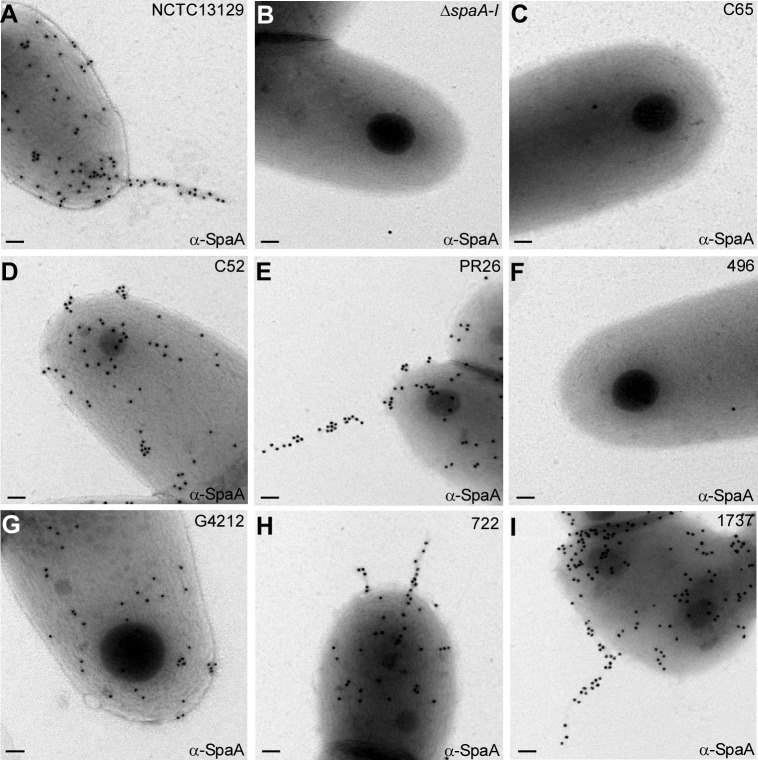
The Western blot analysis described above shows the expression and polymerization of pilins, but it does not reveal whether the assembled pilus is incorporated in the cell wall and surface displayed. To confirm that the SpaA pilus polymers are surface displayed by strains that express them, we performed ELISA and IEM (see Materials and Methods). Strains that harbor the spaA gene and assembled SpaA polymers exhibited a SpaA-positive signal in ELISA, whereas isolates that were negative by PCR and immunoblotting analyses showed no SpaA signal, similar to the isogenic mutant that lacks SpaA pili (Fig. 1C). Representative isolates in each group were then examined by IEM, whereby a corynebacterial cell suspension was spotted on nickel grids and washed cells were labeled with anti-SpaA, followed by IgG-conjugated gold particles. In agreement with the results above, gold particle-labeled SpaA pili were observed in isolates C52 (group 2), PR26 (group 3), G4212 (group 5), 722 (group 6), and 1737 (group 7) (Fig. 2D, E, G, H, and I, respectively), similar to the type strain NCTC13129 (Fig. 2A). In contrast, no gold particles were detected in isolates C65 (group 1) (Fig. 2C) and 496 (group 4) (Fig. 2F), which lack the spaA gene, similar to the ΔspaA–I negative-control strain (Fig. 2B). Altogether, these data indicate (i) that the distributions of pilus genes of different types are varied, (ii) that the presence of pili is not limited to toxigenic strains of C. diphtheriae, and (iii) that the SpaA-type pilus is most widely expressed in the clinical isolates.
Fig 2.
Corynebacteria were immobilized on carbon-coated nickel grids and stained with specific antibodies against SpaA (α-SpaA) and goat anti-rabbit IgG conjugated to 18-nm-diameter gold particles. Samples were viewed by transmission electron microscopy. Scale bars indicate a length of 0.2 μm.
Heterogeneity of minor pilin subunits in C. diphtheriae clinical isolates.
As mentioned above, strain NCTC13129 produces three distinct pilus types, each comprised of a major shaft pilin and two minor pilins—forming the pilus tip and the pilus base. To examine whether the presence of the shaft pilin in the clinical isolates as shown in Fig. 1 correlates with the presence of minor pilin genes and their corresponding expression, we next performed PCR amplification for analysis of the presence of spaB and spaC (minor pilins of SpaA-type pilus), spaE and spaF (minor pilins of SpaD-type pilus), and spaI and spaG (minor pilins of SpaH-type pilus), using the clinical isolates presented in Table 1. Western blot analysis with specific antisera for the respective cell-wall-linked minor pilins of these isolates was also carried out (Table 2). Interestingly, neither minor pilin genes nor minor pilus proteins were observed in the group 1 isolates, which do not contain any shaft pilin genes. Except for the group 4 isolates, which are toxigenic but nonpiliated, isolates of each of the remaining groups all contained spaB and spaC genes as well as the SpaB and SpaC pilins expressed (Fig. 1 and Table 1).
Table 2.
Heterogeneity of minor pilins in corynebacterial strains by PCR and Western blot analyses
| Groupa | Strain | SpaBb |
SpaCb |
SpaEc |
SpaFc |
SpaId |
SpaGd |
||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| PCR | WB | PCR | WB | PCR | WB | PCR | WB | PCR | WB | PCR | WB | ||
| C65 | − | − | − | − | − | − | − | − | − | − | − | − | |
| CD310 | − | − | − | − | − | − | − | − | − | − | − | − | |
| 2 | C52 | + | + | + | + | − | − | − | − | − | − | − | − |
| 760 | + | + | + | + | − | + | + | − | + | − | + | − | |
| 3 | PR26 | + | + | + | + | − | − | − | − | + | + | + | + |
| CD364 | + | + | + | + | + | + | + | + | + | + | + | + | |
| 4 | 496 | − | − | − | − | − | − | − | − | + | − | + | − |
| 724 | + | − | + | − | + | − | + | − | + | − | + | − | |
| 5 | G4212 | + | + | + | + | + | − | − | − | − | − | − | − |
| 711 | + | + | + | + | − | − | − | − | − | − | − | − | |
| 6 | 722 | + | + | + | + | + | − | − | − | + | − | + | − |
| E8277 | + | + | + | + | + | + | + | + | + | + | − | − | |
| 7 | 1737 | + | + | + | + | + | + | + | − | + | + | + | + |
| 749 | + | + | + | + | + | + | + | + | + | + | + | + | |
| Sequence strain | 13129e | + | + | + | + | + | + | + | + | + | + | + | + |
Arbitrary groups based on the presence of different types of pili and toxin.
Minor pilins of the SpaA-type pili.
Minor pilins of the SpaD-type pili.
Minor pilins of the SpaH-type pili.
Sequence strain NTCT13129.
While the above analyses revealed that clinical isolates expressing shaft pilin SpaA also expressed the respective minor pilins SpaB and SpaC, similar analyses for the minor pilins of the SpaD and SpaH types produced mixed results. PCR and Western blot analyses of the nontoxigenic strains PR26 and CD364 (group 3) and the toxigenic strains 1737 and 749 (group 7) gave positive results for the presence of minor pilins SpaI and SpaG, in agreement with the positive results for the shaft pilin SpaH (Table 1). Interestingly, strain CD364 also contained spaE and spaF and expressed the corresponding pilin; however, in this case, the PCR and immunoblotting analyses for the shaft SpaD pilin were negative (Tables 1 and 2). As mentioned above for the group 4 isolates, while the PCR and Western blot analyses for the shaft pilins SpaA, SpaD, and SpaH showed negative results, the PCR analysis of strain 724 showed the presence of all minor pilin genes for three pilus types (Table 2). Notably, the corresponding pilins were not observed by immunoblotting with anti-SpaB, anti-SpaC, anti-SpaE, anti-SpaF, anti-SpaI, and anti-SpaG. However, similar analyses of strain 496 revealed the presence of only the spaI and spaG genes.
The lack of PCR products for spaE/F and spaI/G with the above isolates using primers based on strain NCTC13129 and immunoblotting signal with antibodies against the SpaE/F and SpaI/G proteins of NCTC13129 implicates sequence variation in the nucleotide and protein levels found in various clinical isolates. This is consistent with the recent genomic study based on whole-genome sequencing showing heterogeneity of SpaD and SpaH pilus gene clusters in many C. diphtheriae clinical isolates (19).
Variations in pilus gene pools and pilus expression in the fully sequenced strain C. diphtheriae PW8.
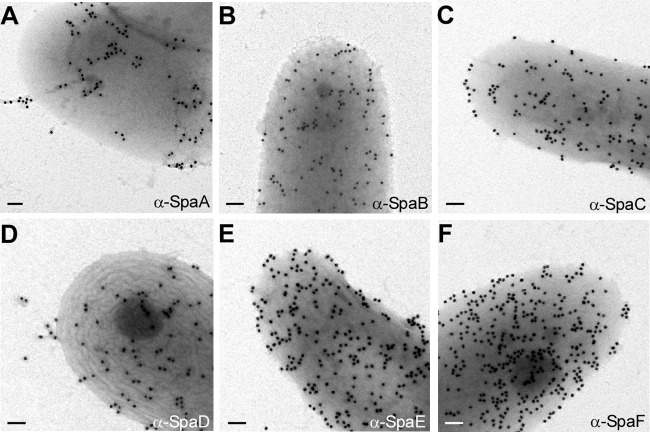
A bioinformatics analysis of the genome of the strain PW8 mentioned above revealed the presence of both SpaA and SpaD pilus gene clusters (19). The SpaA gene cluster in this strain, however, harbors genes encoding intact SpaA and SpaB pilin and the SrtA homologs. A sequence comparison between the spaC gene of PW8 and that of NCTC13129 revealed that the former lacked a segment between nucleotides 2474 and 2943. Of note, Western blotting with anti-SpaA and anti-SpaC revealed SpaA- and SpaC-reactive species, respectively, in the cell wall fractions of strain PW8 (see Fig. S2 in the supplemental material). On the other hand, the SpaD pilus gene cluster contains several copies of spaD, spaF, and sortase srtC, and the majority of these genes are disrupted by mobile elements (19). To determine surface expression of these pilins in strain PW8, we employed immunoelectron microscopy using antibodies against the SpaA- and SpaD-type pilins of strain 13129. As shown in Fig. 3A and D, short pili were detected with antibodies against the shaft pilins SpaA and SpaD, whereas abundant signal of minor pilins SpaB, SpaC, SpaE, and SpaF was observed on the cell surface of strain PW8 (Fig. 3B, C, E, and F, respectively). Future experiments will examine how these Spa pilins are assembled on the bacterial surface.
Fig 3.
Cells of strain PW8 were immobilized on carbon-coated nickel grids and subjected to immunoelectron microscopy with specific antibodies against SpaA- and SpaD-type pilins similarly described in the legend to Fig. 2. A mutant lacking all Spa pilin was used as a control (i.e., the ΔspaA–I mutant). Scale bars indicate a length of 0.2 μm.
Correlation between SpaA pilus expression and the adherence of C. diphtheriae clinical isolates to human pharyngeal epithelial cells.
As noted in the introduction, previous work by Mandlik et al. (13) has established that the SpaA-type pili are the major adhesin that mediates C. diphtheriae adherence to human pharyngeal epithelial cells, the relevant tissue affected by corynebacterial infection underlying diphtheria. To determine whether individual corynebacterial clinical isolates analyzed in the seven subgroups above have the ability to adhere to nasopharyngeal tissue, we performed adhesion assays with human pharyngeal epithelial cells (D562) as reported previously (13). The epithelial cells were cultured in multiwell plates to near confluence and infected with individual isolates of corynebacteria. The infected host cells were then washed to remove nonadherent corynebacteria, detached from wells, and then plated on agar plates to enumerate bacteria adherent to the epithelial cells. The binding of the individual clinical isolates to pharyngeal cells was compared to that of strain NCTC13129, which was set at 100% (Fig. 4). As a negative control, we used an isogenic mutant of strain NCTC13129 devoid of all six sortase genes (the ΔsrtA–F mutant) that exhibits a severe defect in adherence to epithelial cells due to the absence of surface pili as reported previously (13). When comparing the C. diphtheriae NCTC13129 strain with corynebacterial clinical isolates in groups 1 (nontoxigenic) and 4 (toxigenic), each of which does not express any pilin subunits, a defect in adherence similar to that of the negative control was observed (Fig. 4). This indicates that adherence to pharyngeal cells is independent of diphtheria toxin, as the group 4 isolates contain tox and the group 1 isolates do not. Significantly, isolates of groups 2, 3, 5, 6, and 7, which all expressed SpaA-type pili (Fig. 1), exhibited significant binding to pharyngeal epithelial cells, with an adherence level of the isolates of groups 2, 3, and 7 comparable to that of strain NCTC13129 (Fig. 4). Of note, strain PW8 also exhibited adherence to D562 epithelial cells, albeit at a reduced level compared to strain NCTC13129 (15).
Fig 4.
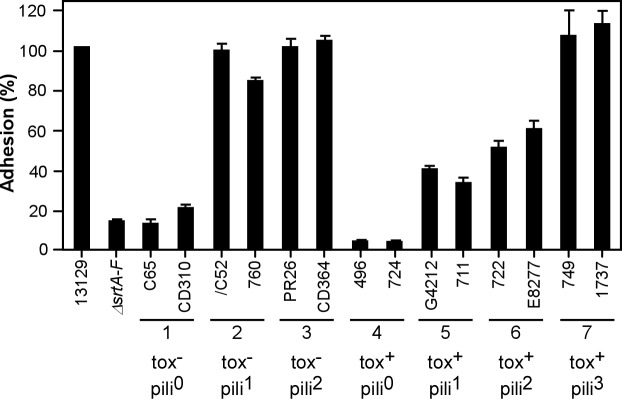
Adherence of C. diphtheriae to pharyngeal epithelial cells (D562). Epithelial cells were infected with corynebacteria at an MOI of 10, and adherent bacteria were then enumerated. Data are presented as percentages of adhesion relative to that of NCTC13129. The mean adhesion percentage of NCTC13129 to D562 cells was ∼18%. The results are presented as averages from at least three independent experiments performed in triplicate. Strain names and subgroups 1 to 7 without toxin (tox−) or with toxin (tox+) and having 0, 1, 2, or 3 types of pili (pili0, pili1, etc.) are indicated.
C. diphtheriae clinical isolates lacking toxin and/or pili are attenuated in nematode killing.
While undertaking the characterization of corynebacterial clinical isolates, we sought to develop an animal model for C. diphtheriae infection. We chose to use Caenorhabditis elegans as a model host because of its proven versatility. It has been shown that the C. elegans infection model can be used to study a variety of bacterial pathogens (both Gram negative and Gram positive) and give insights into bacterial virulence factors and host-pathogen interactions (24, 29). Therefore, we employed a standard C. elegans killing assay with the clinical isolates, whereby L4-stage nematodes of strain N2 were acutely fed with various C. diphtheriae cultures and dead nematodes were enumerated every 24 h during the course of the experiments. To evaluate the role of toxin and pili in killing of nematodes, using the parental strain NCTC13129, we generated isogenic mutants that lack genes encoding SpaA-type pili (ΔspaABC) or spaABC and tox genes (ΔspaABC Δtox). We then examined their ability to kill nematodes overtime. As shown in Fig. 5, deletion of spaABC resulted in a significantly delayed killing of nematodes compared to that of the parental strain, the phenotype that was similar to deletion of both spaABC and tox. Of note, no significant difference in the survival curves of the two mutants was observed.
Fig 5.
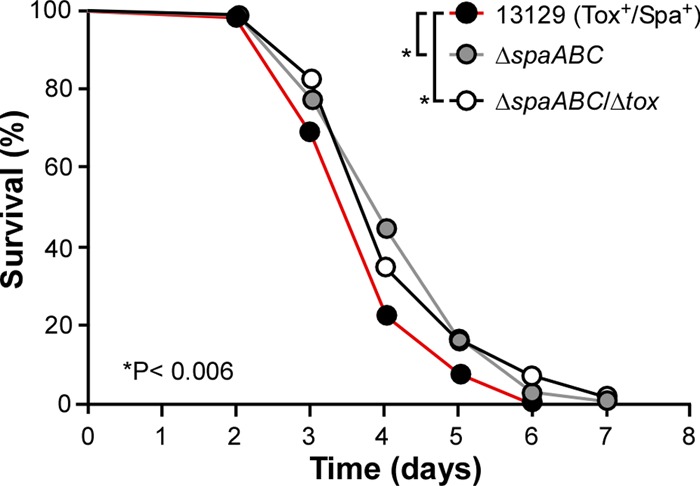
The importance of pili and toxin in C. diphtheriae virulence in the C. elegans host model. L4-stage nematodes of strain N2 were fed on corynebacteria of NCTC13129 (Tox+/Spa+; black circles), as well as its isogenic ΔspaABC mutant lacking SpaA-type pili (gray circles) or its isogenic ΔspaABC Δtox mutant lacking SpaA-type pili and toxin (white circles). Nematode survival was observed over time. The results are presented as percentages, and statistical analysis was performed using GraphPad Prism 5.0. Asterisks indicate P values of <0.006.
To examine whether the presence of various pilus types in nontoxigenic and toxigenic isolates correlates with degree of bacterial virulence, we subjected representative strains in seven groups (Table 2) to the nematode killing assay to determine the LT50 (time required to kill 50% of nematodes) of each strain. For strain NCTC13129, which produces the SpaA-, SpaD-, and SpaH-type pili as well as DT, 50% of nematodes were killed around 3 days (LT50 of 3.18) (Table 3). A very similar phenotype was observed for the group 7 isolates 1737 and 749, toxigenic strains expressing all pilus types like NCTC13129 (LT50s of 3.71 and 2.88, respectively). By comparison, representative isolates of groups 1 to 6 all exhibited longer rates of killing the nematodes than those of strain NCTC13129 as well as isolates 1737 and 749 of group 7 (Table 3). Interestingly, isolates G4212 and 711 of group 5, which were toxigenic and positive for SpaA-type pili, displayed the most delayed killing phenotype (LT50s of 8.88 and 9.06, respectively). On the other hand, group 1 isolates C65 and CD310 were devoid of toxin and all pilus types, but their rates of killing were significantly faster (LT50s of 5.65 and 5.71, respectively) than those of isolates G4212 and 711. A similar phenotype was observed for group 2 isolates C52 and 760, which are negative for toxin but positive for SpaA-type pili (LT50s of 5.65 and 5.71, respectively).
Table 3.
Lethality analysis of C. elegans infected by Corynebacterium diphtheriae isolates
| Group | Strain | Toxigenicity | Pilus type(s) present | LT50 (days)a |
|---|---|---|---|---|
| 1 | C65 | − | Spa− | 5.65 |
| CD310 | − | Spa− | 5.71 | |
| 2 | C52 | − | SpaA | 5.44 |
| 760 | − | SpaA | 5.48 | |
| 3 | PR26 | − | SpaA, SpaH | 7.02 |
| CD364 | − | SpaA SpaH | 8.01 | |
| 4 | 496 | + | Spa− | 5.70 |
| 724 | + | Spa− | 6.03 | |
| 5 | G4212 | + | SpaA | 8.88 |
| 711 | + | SpaA | 9.06 | |
| 6 | 722 | + | SpaA, SpaH | 5.46 |
| E8277 | + | SpaA, SpaD | 7.95 | |
| 7 | 1737 | + | SpaA, SpaD, SpaH | 3.71 |
| 749 | + | SpaA, SpaD, SpaH | 2.88 | |
| Sequence strain | 13129b | + | SpaA, SpaD, SpaH | 3.18 |
Time (days) required for killing 50% of the nematodes. The results are averages of two independent experiments.
Sequence strain NTCT13129.
While there was no real correlation between pilus type and bacterial virulence in the C. elegans model, full virulence of C. diphtheriae required toxin and all pilus types. Altogether, the results support the notion that both toxin and pili are the major factors for C. diphtheriae virulence in nematodes and that the C. elegans model can be used to evaluate corynebacterial virulence.
DISCUSSION
Soon after the identification of C. diphtheriae (30), classic microbiologic experiments led to the discovery of the organism's potent disease-causing exotoxin, DT, which remained the major known virulence factor of this pathogen. The phenomenal success of diphtheria vaccines generated from the inactive DT has eclipsed the need to search for additional virulence factors that may contribute to the severity and complexity of the disease that once killed humans in major epidemic proportions. Nevertheless, the genome sequence of C. diphtheriae has uncovered a plethora of factors presumed to be involved in the pathogenesis of diphtheria (8). It was previously shown (7, 13) that C. diphtheriae strain NCTC13129 harbors three pilus gene clusters encoding the SpaA-, SpaD- and SpaH-type pili. As described in the introduction, the assembly of corynebacterial pili occurs by the sortase-mediated pathway found commonly in Gram-positive bacteria (5). In spite of the active research on the C. diphtheriae genomics and the molecular mechanisms of assembly of corynebacterial pili, little information has been available about the surface expression of various pilin genes and their relationship to bacterial virulence.
In this study, making use of a large number of clinical isolates collected from different parts of the world, we asked two simple but important questions: (i) whether the expression of three distinct types of pili varies in clinical isolates or not and (ii) whether there is a relationship between pilus expression and virulence of representative clinical isolates using a nematode model host. Consistent with a genomic study published recently (19), our studies reveal that the SpaA-type pilus is the most common pilus type expressed among these isolates (Fig. 1 and 2; see Table S1 in the supplemental material). Because the SpaA-type pili serve as the major adhesins required for corynebacterial adherence to epithelial cells of the pharynx (13), the predominant infection site, it is likely that the SpaA pilins are major colonization factors for the establishment of C. diphtheriae infection. Consistent with this notion, while multiple pilus gene clusters are also detected in the genome of many toxigenic and nontoxigenic clinical isolates of C. diphtheriae, the SpaA pilus gene cluster was observed at high frequency (19). Remarkably, the SpaA pilus gene cluster was also shown to be more prevalent than the SpaD and SpaH gene clusters among different corynebacterial species (30). Interestingly, our analysis of the draft genome of C. diphtheriae bv. intermedius NCTC5011, an isolate collected prior to mass vaccination in the United Kingdom, assuming that this strain was “not subject to the evolutionary selective pressure of vaccination” (31), reveals the presence of SpaA- and SpaD-type pilus gene clusters in this strain. The SpaA pilins are extremely highly conserved structurally (data not shown), based on their primary sequences compared to that of strain NCTC13129, which is a C. diphtheriae bv. gravis isolate from a United Kingdom patient with clinical diphtheria who traveled abroad in 1997 (8). As SpaA pili are the major adhesin of C. diphtheriae for pharyngeal epithelial cells, these findings suggest that the spaA gene cluster was selectively maintained throughout the organism's evolution.
While the SpaA-type pili appear to be the most common pilus and are associated with bacterial adherence to pharyngeal epithelial cells (Fig. 4), the SpaD- and SpaH-type pili are detected less frequently by our DNA and protein analyses based on strain NCTC13129 (see Table S1 in the supplemental material). This examination is consistent with our previous finding that the amino acid sequences of the SpaD- and SpaH-type pilins are highly diverged among many toxigenic and nontoxigenic strains, including the vaccine strain PW8 (19). Intriguingly, while several genes of the PW8 spaD locus appear to be disrupted by mobile elements (19), their products are abundantly detected on the bacterial surface by IEM (Fig. 3). These immunoreactive signals are specific as none of them were observed in strains lacking all pilins (data not shown). It is possible that truncated pilins were still produced, secreted, and bound to the bacterial surface. Our previous studies suggest that these pilins may be specific for other tissues (13), since C. diphtheriae strains are also found on skin, nasal tissues, and larynx (32). A major medically relevant question that still remains unanswered is whether the strains possessing SpaD and SpaH pili represent a distinct clinical outcome.
Another major gap in our knowledge is how these pili are involved in bacterial infection. In an attempt to address this question, we employed C. elegans as a simple but versatile animal model for killing by bacterial pathogens. Like many other Gram-positive pathogens described so far (24), C. diphtheriae rapidly kills the nematodes; this killing appears to involve SpaA pili since strains lacking SpaA pili or both SpaA pili and toxin do not exhibit significant differences in their attenuated virulence in the nematode killing assay (Fig. 5). We also compared the nematode killing rates (LT50) of strain NCTC13129 and clinical isolates representative for groups 1 to 7. With an exception of group 7 isolates, which are positive for toxin and all three pilus types like NCTC13129, isolates of the other groups display a delayed killing phenotype (Table 3). However, there is no clear correlation between different types of pili and bacterial virulence. We also compared the survival rates of strain NCTC13129 and individual isogenic mutants lacking pilins of the SpaA type, SpaD type, or both types. Although all mutants exhibited attenuation in nematode killing, no significant difference was found in the LT50 values of these mutants (data not shown). Perhaps, the nematode model may not differentiate subtle defects, or each pilus type may have a comparable role in the killing of nematodes. Interestingly, while the nontoxigenic isolates C65 and CD310 (group 1) do not possess any pilus types, they have a significantly reduced rate of killing (i.e., greater LT50) than that of strain NCTC13129, suggesting other factors may contribute to corynebacterial virulence. Thus, the C. elegans host model can serve as a valuable tool to speedily identify additional virulence factors of C. diphtheriae and better understand the pathogenesis caused by this pathogen. This will be an important goal since vaccination with DT appears to lead to the selection of nontoxigenic pathogens in the affected population (33–36).
Supplementary Material
ACKNOWLEDGMENTS
We thank Pamela K. Cassiday and Maria L. Tondella (Centers for Disease Control and Prevention) for providing C. diphtheriae clinical isolates, Danielle Garsin, Ransome van der Hoeven, and Xin Ma for technical help, and Melissa Reardon-Robinson for critical review of the manuscript and discussion.
Research reported in this publication was supported by the National Institute of Allergy and Infectious Diseases of the National Institutes of Health under award no. R56AI061381 to H. T.-T.
Footnotes
Published ahead of print 14 June 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JB.00500-13.
REFERENCES
- 1. Murphy JR. 1996. Corynebacterium diphtheriae. In Baron S. (ed), Medical microbiology, 4th ed, chapter 32 The University of Texas Medical Branch, Galveston, TX: [PubMed] [Google Scholar]
- 2. Holmes RK. 2000. Biology and molecular epidemiology of diphtheria toxin and the tox gene. J. Infect. Dis. 181(Suppl 1):S156–S167 [DOI] [PubMed] [Google Scholar]
- 3. Freeman VJ, Morse IU. 1952. Further observations on the change to virulence of bacteriophage-infected avirulent strains of Corynebacterium diphtheriae. J. Bacteriol. 63:407–414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Holmes RK, Barksdale L. 1969. Genetic analysis of tox+ and tox− bacteriophages of Corynebacterium diphtheriae. J. Virol. 3:586–598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Mandlik A, Swierczynski A, Das A, Ton-That H. 2008. Pili in Gram-positive bacteria: assembly, involvement in colonization and biofilm development. Trends Microbiol. 16:33–40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Yanagawa R, Honda E. 1976. Presence of pili in species of human and animal parasites and pathogens of the genus Corynebacterium. Infect. Immun. 13:1293–1295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Ton-That H, Schneewind O. 2003. Assembly of pili on the surface of Corynebacterium diphtheriae. Mol. Microbiol. 50:1429–1438 [DOI] [PubMed] [Google Scholar]
- 8. Cerdeno-Tarraga AM, Efstratiou A, Dover LG, Holden MT, Pallen M, Bentley SD, Besra GS, Churcher C, James KD, De Zoysa A, Chillingworth T, Cronin A, Dowd L, Feltwell T, Hamlin N, Holroyd S, Jagels K, Moule S, Quail MA, Rabbinowitsch E, Rutherford KM, Thomson NR, Unwin L, Whitehead S, Barrell BG, Parkhill J. 2003. The complete genome sequence and analysis of Corynebacterium diphtheriae NCTC13129. Nucleic Acids Res. 31:6516–6523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Mandlik A, Das A, Ton-That H. 2008. The molecular switch that activates the cell wall anchoring step of pilus assembly in gram-positive bacteria. Proc. Natl. Acad. Sci. U. S. A. 105:14147–14152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Budzik JM, Marraffini LA, Schneewind O. 2007. Assembly of pili on the surface of Bacillus cereus vegetative cells. Mol. Microbiol. 66:495–510 [DOI] [PubMed] [Google Scholar]
- 11. Nobbs AH, Rosini R, Rinaudo CD, Maione D, Grandi G, Telford JL. 2008. Sortase A utilizes an ancillary protein anchor for efficient cell wall anchoring of pili in Streptococcus agalactiae. Infect. Immun. 76:3550–3560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Smith WD, Pointon JA, Abbot E, Kang HJ, Baker EN, Hirst BH, Wilson JA, Banfield MJ, Kehoe MA. 2010. Roles of minor pilin subunits Spy0125 and Spy0130 in the serotype M1 Streptococcus pyogenes strain SF370. J. Bacteriol. 192:4651–4659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Mandlik A, Swierczynski A, Das A, Ton-That H. 2007. Corynebacterium diphtheriae employs specific minor pilins to target human pharyngeal epithelial cells. Mol. Microbiol. 64:111–124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Chang C, Mandlik A, Das A, Ton-That H. 2011. Cell surface display of minor pilin adhesins in the form of a simple heterodimeric assembly in Corynebacterium diphtheriae. Mol. Microbiol. 79:1236–1247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Iwaki M, Komiya T, Yamamoto A, Ishiwa A, Nagata N, Arakawa Y, Takahashi M. 2010. Genome organization and pathogenicity of Corynebacterium diphtheriae C7(−) and PW8 strains. Infect. Immun. 78:3791–3800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Park WH, Williams AW. 1896. The production of diphtheria toxin. J. Exp. Med. 1:164–185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Freeman VJ. 1951. Studies on the virulence of bacteriophage-infected strains of Corynebacterium diphtheriae. J. Bacteriol. 61:675–688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Barksdale WL, Pappenheimer AM., Jr 1954. Phage-host relationships in nontoxigenic and toxigenic diphtheria bacilli. J. Bacteriol. 67:220–232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Trost E, Blom J, de Castro Soares S, Huang IH, Al-Dilaimi A, Schroder J, Jaenicke S, Dorella FA, Rocha FS, Miyoshi A, Azevedo V, Schneider MP, Silva A, Camello TC, Sabbadini PS, Santos CS, Santos LS, Hirata R, Jr, Mattos-Guaraldi AL, Efstratiou A, Schmitt MP, Ton-That H, Tauch A. 2012. Pangenomic study of Corynebacterium diphtheriae that provides insights into the genomic diversity of pathogenic isolates from cases of classical diphtheria, endocarditis, and pneumonia. J. Bacteriol. 194:3199–3215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Marston CK, Jamieson F, Cahoon F, Lesiak G, Golaz A, Reeves M, Popovic T. 2001. Persistence of a distinct Corynebacterium diphtheriae clonal group within two communities in the United States and Canada where diphtheria is endemic. J. Clin. Microbiol. 39:1586–1590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Mothershed EA, Cassiday PK, Pierson K, Mayer LW, Popovic T. 2002. Development of a real-time fluorescence PCR assay for rapid detection of the diphtheria toxin gene. J. Clin. Microbiol. 40:4713–4719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Popovic T, Kombarova SY, Reeves MW, Nakao H, Mazurova IK, Wharton M, Wachsmuth IK, Wenger JD. 1996. Molecular epidemiology of diphtheria in Russia, 1985–1994. J. Infect. Dis. 174:1064–1072 [DOI] [PubMed] [Google Scholar]
- 23. Henricson B, Segarra M, Garvin J, Burns J, Jenkins S, Kim C, Popovic T, Golaz A, Akey B. 2000. Toxigenic Corynebacterium diphtheriae associated with an equine wound infection. J. Vet. Diagn. Invest. 12:253–257 [DOI] [PubMed] [Google Scholar]
- 24. Schulenburg H, Ewbank JJ. 2004. Diversity and specificity in the interaction between Caenorhabditis elegans and the pathogen Serratia marcescens. BMC Evol. Biol. 4:49. 10.1186/1471-2148-4-49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Nakao H, Popovic T. 1997. Development of a direct PCR assay for detection of the diphtheria toxin gene. J. Clin. Microbiol. 35:1651–1655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Gaspar AH, Ton-That H. 2006. Assembly of distinct pilus structures on the surface of Corynebacterium diphtheriae. J. Bacteriol. 188:1526–1533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Garsin DA, Sifri CD, Mylonakis E, Qin X, Singh KV, Murray BE, Calderwood SB, Ausubel FM. 2001. A simple model host for identifying Gram-positive virulence factors. Proc. Natl. Acad. Sci. U. S. A. 98:10892–10897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Reference deleted.
- 29. Sifri CD, Begun J, Ausubel FM. 2005. The worm has turned—microbial virulence modeled in Caenorhabditis elegans. Trends Microbiol. 13:119–127 [DOI] [PubMed] [Google Scholar]
- 30. Rogers EA, Das A, Ton-That H. 2011. Adhesion by pathogenic corynebacteria. Adv. Exp. Med. Biol. 715:91–103 [DOI] [PubMed] [Google Scholar]
- 31. Sangal V, Tucker NP, Burkovski A, Hoskisson PA. 2012. Draft genome sequence of Corynebacterium diphtheriae biovar intermedius NCTC 5011. J. Bacteriol. 194:4738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Hadfield TL, McEvoy P, Polotsky Y, Tzinserling VA, Yakovlev AA. 2000. The pathology of diphtheria. J. Infect. Dis. 181(Suppl 1):S116–S120 [DOI] [PubMed] [Google Scholar]
- 33. Wilson AP. 1995. The return of Corynebacterium diphtheriae: the rise of non-toxigenic strains. J. Hosp. Infect. 30(Suppl):306–312 [DOI] [PubMed] [Google Scholar]
- 34. Romney MG, Roscoe DL, Bernard K, Lai S, Efstratiou A, Clarke AM. 2006. Emergence of an invasive clone of nontoxigenic Corynebacterium diphtheriae in the urban poor population of Vancouver, Canada. J. Clin. Microbiol. 44:1625–1629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Reacher M, Ramsay M, White J, De Zoysa A, Efstratiou A, Mann G, Mackay A, George RC. 2000. Nontoxigenic Corynebacterium diphtheriae: an emerging pathogen in England and Wales? Emerg. Infect. Dis. 6:640–645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Soubeyrand B, Plotkin SA. 2002. Microbial evolution: antitoxin vaccines and pathogen virulence. Nature 417:609–610 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.