Abstract
Chemotaxis allows bacteria to more efficiently colonize optimal microhabitats within their larger environment. Chemotaxis in Escherichia coli is the best-studied model system, and a large number of E. coli strains have been sequenced. The Escherichia/Shigella genus encompasses a great variety of commensal and pathogenic strains, but the role of chemotaxis in their association with the host remains poorly understood. Here we show that the core chemotaxis genes are lost in many, but not all, nonmotile strains but are well preserved in all motile strains. The genes encoding the Tar, Tsr, and Aer chemoreceptors, which mediate chemotaxis to a broad spectrum of chemical and physical cues, are also nearly uniformly conserved in motile strains. In contrast, the clade of extraintestinal pathogenic E. coli strains apparently underwent an ancestral loss of Trg and Tap chemoreceptors, which sense sugars, dipeptides, and pyrimidines. The broad range of time estimated for the loss of these genes (1 to 3 million years ago) corresponds to the appearance of the genus Homo.
INTRODUCTION
Escherichia coli strains are ubiquitous colonizers of the intestines of mammals and birds (1). There are several highly adapted E. coli clones that have acquired virulence traits and cause a broad spectrum of disease, including enteric/diarrheal disease, urinary tract infections (UTIs), and sepsis/meningitis (2). Depending on the site of infection, pathogenic strains are classified as intestinal E. coli (IPEC) or extraintestinal pathogenic E. coli (ExPEC), and distinct pathotypes (based on clinical manifestation) are recognized within both categories. The most common ExPEC pathotypes include uropathogenic E. coli (UPEC), meningitis-associated E. coli (MNEC), and avian-pathogenic E. coli (APEC) (2, 3). Motility was shown to be important for the colonization of both commensal and pathogenic E. coli strains, as well as for the pathogenesis of the latter (4, 5): however, the exact role of motility and the underlying chemotaxis system in these processes remain poorly understood. The molecular machinery that controls chemotaxis in E. coli has been the subject of intensive investigation (6, 7). Its components include chemoreceptors, also known as methyl-accepting chemotaxis proteins (MCPs), a histidine kinase (CheA), an adaptor protein (CheW), a methyltransferase (CheR), and a methylesterase (CheB), as well as a response regulator (CheY) and its phosphatase (CheZ). E. coli has five chemoreceptors. Tsr mediates attractant responses to serine and quorum autoinducer AI-2 (8, 9), as well as responses to oxygen, redox, and oxidizable substrates (10, 11). It was also recently shown to mediate taxis to 3,4-dihydroxymandelic acid, a metabolite of norepinephrine that is produced by human cells (Mike Manson, personal communication). Tar mediates attractant responses to aspartate and maltose (9, 12) and negative responses to metal ions (13). Trg mediates attractant responses to ribose and galactose (14), and Tap mediates attractant responses to dipeptides and pyrimidines (15, 16). Aer mediates responses to oxygen and energy taxis (11, 17). The majority of the chemotaxis proteins are encoded in two adjacent operons, mocha (motA, motB, cheA, and cheW) and meche (tar, tap, cheR, cheB, cheY, and cheZ), whereas the remaining three chemoreceptors (Tsr, Trg, and Aer) are encoded elsewhere on the chromosome. On a large evolutionary scale, the chemotaxis system, which appeared in a common ancestor of Bacteria, underwent drastic changes displaying a wide array of variations in component design (18). Even the closest relatives of E. coli show substantial differences in the chemotaxis machinery. In Salmonella enterica, the majority of chemotaxis components are orthologous to those of E. coli, but it lacks Tap and contains additional chemoreceptors and the second adaptor protein, CheV (19). However, the driving forces that shape the chemotaxis system on a small evolutionary scale remain unknown.
E. coli is the most sequenced bacterium to date, and phylogenetic studies provided important insights into the processes of its genome evolution (20–22). E. coli strains are too closely related to each other to be resolved by classical 16S rRNA- and ribosomal protein-based phylogeny. Based on several other independent methods, including multilocus enzyme electrophoresis, multilocus sequence typing, intergenic sequence comparison, feature frequency profiles, and whole-genome phylogeny, E. coli strains are classified into several phylogenetic groups: A, B1, B2, D, E, and F (20, 22–25). The phylogenetically defined E. coli clade (1, 26, 27) also includes Shigella clones that have been previously considered a separate genus due to distinct phenotypic features, such as loss of motility, metabolic profile, and clinical manifestation (28). Chemotaxis has been studied extensively using derivatives of a single E. coli strain, K-12 (the A group), and the functionality and conservation of the chemotaxis system have not been specifically studied in members of other E. coli groups. Several studies suggested the dispensability of both core and accessory chemotaxis components in E. coli. The core genome of E. coli contains nearly 2,000 genes (21). Interestingly, only a subset of the chemotaxis genes belongs to the core genome according to this study. Key components of the chemotaxis system, i.e., CheW and CheB as well as two major chemoreceptors, Tar and Tsr, are missing from this core set, suggesting that chemotaxis might be a dispensable function in E. coli. Furthermore, several uropathogenic E. coli strains were shown to lack Trg and Tap receptors, and it was postulated that the gene loss was a result of a lack of selective pressure on sugar- and peptide-sensing receptors in the urinary tract, which is void of these substrates (29). Here, we analyzed the chemotaxis system of E. coli by comparing genomes of more than 200 strains, which included commensals and pathogens from all known phylotypes. We show that the chemotaxis system is well preserved in E coli, even among some strains that have lost motility, and that the major evolutionary event was the loss of Trg and Tap receptors that occurred not only in some uropathogenic strains but in the common ancestor of a large clade corresponding to the loosely defined B2 phylotype. We propose that among other factors, losing the ability to sense sugars, peptides, and nucleotides might have contributed to the emergence of extraintestinal clones, including pathogens.
MATERIALS AND METHODS
Data sources and bioinformatics software.
The following software packages were used in this study: HMMER v3.0 (30), Jalview (31), MAFFT v6.847b (32), MEGA v4.0 (33), PhyML v3.0 (34), and BLAST+ v2.2.4+ (35). All multiple-sequence alignments were built in MAFFT with its l-INS-i algorithm. All maximum-likelihood phylogenetic trees were built in PhyML with standard parameters and with subtree pruning and regrafting topology search. Genomes, proteomes, and genome annotations of all distinct Escherichia and Shigella strains available in the NCBI nr database as of 12 January 2012 were collected (219 genomes). All strains and relevant information are listed in Data Set S1 in the supplemental material. The pathotype information was retrieved from primary literature and public databases.
Construction of a phylogenetic tree for Escherichia.
An Escherichia phylogenetic tree was constructed using the arcA, aroE, icd, mdh, mtlD, pgi, and rpoS genes (36). The nucleotide sequence sets for each gene were aligned individually in MAFFT. The alignments were concatenated, and the resulting alignment was used to build a maximum-likelihood tree in PhyML.
Identification of chemotaxis and accessory proteins from genomic data sets.
Chemotaxis and accessory genes and proteins were retrieved from the genome of E. coli W3110 (model wild type for chemotaxis) and used as BLAST queries against the genome set. Protein and nucleotide searches were performed to ensure retrieval of missing and partial genes. Gene neighborhoods were extracted from NCBI genome feature files.
Multiple-sequence alignment and phylogenetic analyses.
The nucleotide and protein chemotaxis sequence sets (MotA, MotB, CheA, CheW, Tar, Tap, CheR, CheB, CheY, CheZ, Tsr, Trg, and Aer) were individually aligned by MAFFT. The alignments of the chemotaxis operons, mocha and meche, were concatenated and used to build a maximum-likelihood tree in PhyML.
sSNP molecular clock calculation.
All of the chemotaxis genes (except trg and tap) and recA from clades B2 and A were individually aligned and concatenated to produce a gapless alignment. After removing sequences with errors, the final set consisted of 58 sequences (see Table S1 in the supplemental material). The alignment spanned 4,360 codons. The equation used to calculate time of divergence is as follows: (number of synonymous single-nucleotide polymorphism [sSNP] sites)/(number of potential sSNP sites × mutation rate × generations per year × 2). Potential sSNP sites were determined using the parsimonious assumption that each codon has only one potential sSNP site. Generations per year were estimated at a range from 100 to 300 to allow for a broad estimation (37–40). The experimentally determined synonymous mutation rate of 1.4 × 10−10 (41) was used.
RESULTS
Phylogenetic tree of Escherichia.
We analyzed 219 (55 complete and 164 draft) genomes of Escherichia and Shigella. This set included genomes of Escherichia fergusonii and Escherichia albertii, to serve as outgroups in the phylogenetic analysis. In order to assign newly sequenced strains to the established phylogenetic groups, we have constructed a phylogenetic tree of all 219 strains in our data set. Because relationships between such closely related strains cannot be resolved using traditional ribosomal trees, we built a maximum-likelihood tree from concatenated alignments of the arcA, aroE, icd, mdh, mtlD, pgi, and rpoS genes, as previously suggested (36). The tree (see Fig. S1 in the supplemental material) is in good agreement with previously published data, including whole-genome-based phylogeny (21). A detailed classification of all Escherichia genomes based on pathotype and phylogenetic groups is shown in Data Set S1 in the supplemental material.
Core chemotaxis genes.
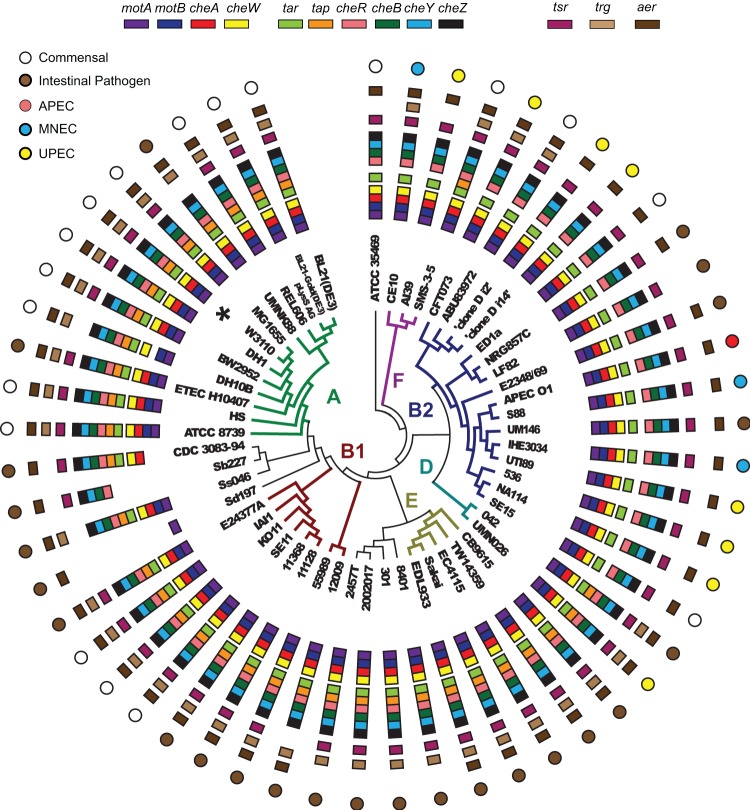
The presence or absence of 11 chemotaxis genes (cheA, cheW, cheY, cheB, cheR, cheZ, tsr, tar, trg, tap, and aer) in all 219 genomes is shown as a bird's-eye view in Fig. S2 in the supplemental material. The picture looks like a mildly used shooting target: while concentric rings representing the presence of each of the chemotaxis proteins are well preserved, there are visible holes of different sizes, showing the absence of particular genes. Many of the missing proteins can be found as pseudogenes resulting from single-nucleotide frameshifts. Sequencing errors (which occur at a rate of 1% for some next-generation sequencing methodologies) appear to be the main source of missing proteins (e.g., cheB is split as ECH7EC4401_1543 and ECH7EC4401_1544 in E. coli O157:H7 strain EC4401). Another common cause of missing genes in draft genomes is a split between different contigs (e.g., cheA is split between ZP_04536326 and ZP_04536327 in Escherichia sp. strain 3_2_53FAA). An additional cause is erroneous gene calling (e.g., a complete cheA gene in E. coli strain K-12 substrain DH10 is missing). We have analyzed each and every potential mutation in all chemotaxis genes, assigning them to obvious sequencing, assembly, and annotation errors or potentially true mutations (see Data Set S1 in the supplemental material). Completely sequenced, closed genomes served as the main internal control. The distribution of chemotaxis genes in closed genomes only is shown in Fig. 1.
Fig 1.
Presence of chemotaxis genes in completely sequenced Escherichia/Shigella genomes. Full strain names and properties are listed in Data Set S1 in the supplemental material. Phylogenetic relationships are shown in the center; a complete phylogenetic tree is available in Fig. S1 in the supplemental material. Branches are colored according to previously established phylotypes. E. coli K-12 strain W3110 (model for chemotaxis) is marked with an asterisk.
To better discriminate between potential sequencing/assembly errors and true mutations, we analyzed the nature of mutations in Shigella genomes. Shigella strains are nonmotile due to inactivation of their flagellar genes (42, 43); therefore, accumulation of mutations in their chemotaxis genes was expected. Indeed, 30% of Shigella strains had significant deletions and insertions in the mocha/meche operons (see Data Set S1 in the supplemental material). Deletions were present not only in draft genomes but also in complete genomes of Shigella, reducing the chance of these results being attributable to sequencing errors. Only 33% of Shigella strains contained complete sets of intact chemotaxis genes. In a striking contrast, none of the E. coli strains has accumulated insertions or deletions in their core chemotaxis genes (cheA, cheW, cheY, cheB, cheR, and cheZ). Single frameshift mutations in these genes were identified only in nine E. coli genomes; all of these sequences were in draft status, and the results could be due to sequencing errors. All completely finished E. coli genomes had their core chemotaxis genes intact. No events of gene duplication or horizontal gene transfer have been found among core chemotaxis genes.
Chemoreceptor loss.
In contrast to the case for core chemotaxis genes, chemoreceptor loss was observed not only in Shigella but also in some E. coli strains. In Shigella, all five chemoreceptors (Tar, Tsr, Trg, Tap, and Aer) have a nearly equal chance to be eliminated, whereas in E. coli, chemoreceptor loss was strongly biased toward Trg and Tap (Table 1). Most strikingly, this loss was observed in specific phylotypes. All B2 group strains and the majority of F group strains underwent a deletion in the tap gene. The identical nature of the deletions (Fig. 2; see Data Set S1 in the supplemental material) suggests that the event occurred prior to the B2 clade divergence. The majority (33 of 38) of B2 strains have also undergone a deletion in the trg gene. Similarly to the deletion of tap, the symmetrical nature of the trg deletion (Fig. 2; see Data Set S1 in the supplemental material) suggests that the loss was an ancestral event. Another four B2 group strains possess an identical frameshift mutation within the trg gene. The symmetrical nature of this frameshift and its presence in a completely sequenced genome of the E. coli 536 strain (Fig. 2) indicate that it is not a sequencing artifact. Thus, it appears that trg and tap deletions occurred in a common ancestor of a clade, which approximately corresponds to the B2 phylogroup. Using molecular clock calculations, we estimated a time period during which the ancestral chemoreceptor loss event occurred. We compared the number of synonymous mutations in the B2 clade, in which the loss took place, with that in the A clade, which contains the chemotaxis wild-type K-12 strains. The B2 clade has overall and on average more sSNPs than the A clade, indicating a longer time period of divergence from respective common ancestors. Our estimates indicate that B2 diverged from ∼1 to 3 million years ago (Ma), whereas the A clade did so from ∼0.4 to 1.2 Ma (assuming 300 to 100 generations per year).
Table 1.
Loss of chemoreceptor genes in E. coli and Shigella genomes
| Lost genea | No. of genomes |
|||
|---|---|---|---|---|
|
E. coli |
Shigella |
|||
| All (n = 183) | Finished (n = 46) | All (n = 28) | Finished (n = 8) | |
| tar | 0 | 0 | 12 | 2 |
| tsr | 4 | 2 | 4 | 1 |
| aer | 1 | 1 | 12 | 4 |
| trg | 34 | 16 | 7 | 3 |
| tap | 41 | 18 | 10 | 2 |
Excluding detected sequencing/assembly/annotation errors (see Data Set S1 in the supplemental material for details).
Fig 2.
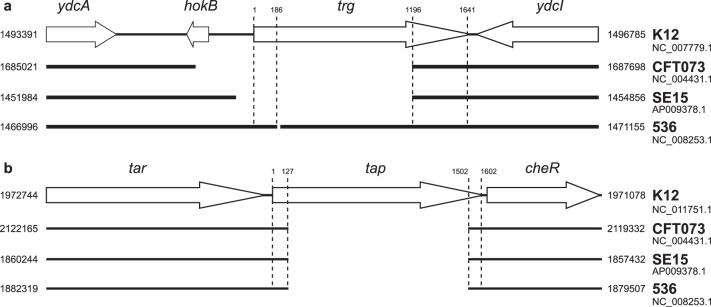
Deletions in tap and trg genes in B2 group strains. Gene neighborhoods in representative genomes are shown. Full strain names and genomic locations of deletions are listed in Data Set S1 in the supplemental material.
Chemoreceptor acquisition.
While no chemoreceptor gene duplication was observed in any analyzed genome, we detected several receptor acquisition events (Table 2). All acquired chemoreceptor genes were plasmid borne. In E. fergusonii ECD227, an acquired chemoreceptor is 99% identical to the MCP from Salmonella enterica subsp. enterica serovar Kentucky strain CVM29188, the gene for which is also located on a plasmid. These plasmids are similar and were implicated in antimicrobial resistance in Salmonella and virulence in E. fergusonii (44). This chemoreceptor is significantly different from canonical E. coli MCPs in sequence, although it belongs to the same class 36H (45) and has the same predicted membrane topology. E. coli O157:H7 strain EC4024 acquired a chemoreceptor that was identified from its N-terminal portion (residues 1 to 350) located at a contig end. This fragment was 99% identical to an MCP from an Enterobacter hormaechei strain (GI 334124148) and showed limited similarity to Trg (Table 2). The MCP gene is found neighboring a sucrose metabolism gene cluster both on the plasmid and in the Enterobacter genomes, suggesting a possible role of the MCP as a sucrose sensor. Finally, seven E. coli genomes were found to possess an aer-like MCP gene likely acquired from Aeromonas caviae, which is also known to cause gastroenteritis (46). In six genomes, these MCP genes are identical, suggesting a single recent acquisition event.
Table 2.
Horizontally transferred chemoreceptor genes in Escherichia genomes
| Genome | Acquired gene |
Closest BLAST hit |
||
|---|---|---|---|---|
| Name/GI no. | % Sequence identity with E. coli K-12 homolog | Organism/GI no. | % Sequence identity | |
| E. fergusonii ECD227 | Tsr (MCPI)/424819104 | 37 | S. enterica/194447140 | 99 |
| E. coli | ||||
| O157:H7 strain EC4024 | Trg (MCPIII)/195941089 | 29 | E. hormaechei/334124148 | 99 |
| 101-1 | Aer (MCPV)/19443928 | 33 | A. caviae/51470604 | 99 |
| E1520 | Aer (MCPV)/323937477 | 33 | A. caviae/51470604 | 100 |
| G58-1 | Aer (MCPV)/345368913 | 33 | A. caviae/51470604 | 100 |
| MS 84-1 | Aer (MCPV)/300904008 | 33 | A. caviae/51470604 | 100 |
| MS 85-1 | Aer (MCPV)/315252457 | 33 | A. caviae/51470604 | 100 |
| MS 124-1 | Aer (MCPV)/301305681 | 33 | A. caviae/51470604 | 100 |
| TA007 | Aer (MCPV)/323969140 | 33 | A. caviae/51470604 | 100 |
DISCUSSION
Despite a relatively short timeline of divergence, the chemotaxis system in the genus Escherichia has undergone substantial changes. First, the loss of the entire chemotaxis function, manifested as severe mutations in core chemotaxis genes, was observed. This event was unambiguously detected only in the nonmotile, intracellular Shigella. All E. coli genomes contain intact core chemotaxis genes, indicating that chemotaxis is critical for motile strains. On the other hand, not all Shigella strains lost their chemotaxis genes. For example, in S. flexneri K-671, the entire chemotaxis system appears to be intact, whereas flagella are absent due to mutations in the flhDC flagellar master operon (47). Several Shigella strains retain intact mocha and meche operons. Thus, the chemosensory apparatus in these strains might be used for other functions. This is a common trend in the evolution of the chemotaxis system on a larger evolutionary scale; it was coopted to control such processes as gene expression in many bacterial species (18, 48). On the other hand, severe defects in Shigella metabolism were linked to mutations in the promoter region, in the absence of nonsense mutations in corresponding genes (49, 50), and therefore it is possible that the mocha and/or meche operon is not fully functional in Shigella, while remaining apparently intact. Second, we detected changes in the chemoreceptor repertoire caused by gene loss and, to a lesser extent, by horizontal gene transfer, but not gene duplication. The major chemoreceptors Tar and Tsr are well preserved in E. coli. This is consistent with their roles as modulators of important behaviors, which, in addition to sensing of various attractants and repellents, include energy taxis (11), thermotaxis (51), and pH taxis (52). Tar and Tsr are equally important for commensal and pathogenic strains. These chemoreceptors are also necessary and sufficient for chemotaxis toward urine in the pathogenic E. coli strain CFT073 (53). Although the aerotaxis receptor Aer has been categorized as a minor receptor according to its low abundance in the cell (54), it is also well preserved in E. coli, likely due to its role in energy taxis and thermotaxis. Consequently, we propose to refer to Aer as a major chemoreceptor, in addition to Tar and Tsr.
We have found evidence for at least three independent events of new chemoreceptor acquisitions by E. coli strains. A Trg-like chemoreceptor was found to be encoded in a sucrose metabolism gene cluster. Both gene order conservation for this receptor (together with fructokinase) in Enterobacteriaceae plasmids and the known role for Trg of mediating chemotaxis to ribose and galactose suggest that it might sense sucrose. Sucrose and fructose metabolism gene clusters have been reported in several E. coli extraintestinal strains (55, 56). Another interesting case is an additional Aer-like chemoreceptor, which is present in several E. coli strains but appears to be a result of a single acquisition event. Multiple copies of Aer are not uncommon among gammaproteobacteria. For example, they are present in such pathogens as Vibrio cholerae (57) and Pseudomonas aeruginosa (58).
Unambiguously, loss can be established only for Trg and Tap, where large deletions were identified in corresponding genes in many E. coli genomes. The overwhelming majority of these strains belong to the B2 clade, which contains major extraintestinal pathogens. The deletions occurred in the same chromosomal position in all B2 strains, strongly suggesting a single ancestral event. This loss does not appear to be a result of relaxed selective pressure on sensors to sugars and dipeptides that are exceedingly rare in urine from individuals with healthy kidneys. Genomes that lost trg and tap contain intact genes coding for ribose, galactose/glucose, and dipeptide periplasmic-binding proteins that mediate the sensing of these compounds through Trg and Tap. This suggests continuing exposure to these molecules, which is not in line with a selection-driven loss due to minimal exposure or nonexposure. Furthermore, some B2 strains are persistent in the intestine, expressing enhanced features for colonization (59), and function as commensals until they are outside the intestinal tract. Thus, they are not exclusively under selection pressure from the urinary environment. Finally, some extraintestinal B2 strains are not found in the urinary tract but preferentially migrate elsewhere (for example, MNEC strains). Taken together, these observations imply that it is possible that the ancestral loss of trg and tap predisposed gut-inhabiting strains to seek other niches to occupy or to develop new adaptive strategies to remain fully competitive in the gut.
The molecular clock analysis of the chemotaxis system of the B2 strains suggests that they branched off fairly early, which is in agreement with the previously published data (25). Even with as broad an estimation as ∼1 to 3 Ma, this places the divergence of the B2 clade in the ballpark of the estimated appearance of the genus Homo (2.3 to 2.4 Ma) (60) and provides yet another intriguing temporal link between human specialization and E. coli pathogenicity.
Supplementary Material
ACKNOWLEDGMENTS
We thank Harry L. T. Mobley for discussion and helpful suggestions and Michael D. Manson for communicating results prior to publication and helpful suggestions.
This work was supported by National Institutes of Health grant GM072295 (to I.B.Z.). K.B. and A.D.F. received support from the Graduate Program in Genome Science and Technology, University of Tennessee—Oak Ridge National Laboratory.
Footnotes
Published ahead of print 7 June 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JB.00421-13.
REFERENCES
- 1. Chaudhuri RR, Henderson IR. 2012. The evolution of the Escherichia coli phylogeny. Infect. Genet. Evol. 12: 214– 226 [DOI] [PubMed] [Google Scholar]
- 2. Kaper JB, Nataro JP, Mobley HLT. 2004. Pathogenic Escherichia coli. Nat. Rev. Microbiol. 2: 123– 140 [DOI] [PubMed] [Google Scholar]
- 3. Croxen MA, Finlay BB. 2010. Molecular mechanisms of Escherichia coli pathogenicity. Nat. Rev. Microbiol. 8: 26– 38 [DOI] [PubMed] [Google Scholar]
- 4. Giron JA, Torres AG, Freer E, Kaper JB. 2002. The flagella of enteropathogenic Escherichia coli mediate adherence to epithelial cells. Mol. Microbiol. 44: 361– 379 [DOI] [PubMed] [Google Scholar]
- 5. Lane MC, Lockatell V, Monterosso G, Lamphier D, Weinert J, Hebel JR, Johnson DE, Mobley HLT. 2005. Role of motility in the colonization of uropathogenic Escherichia coli in the urinary tract. Infect. Immun. 73: 7644– 7656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Hazelbauer GL, Falke JJ, Parkinson JS. 2008. Bacterial chemoreceptors: high-performance signaling in networked arrays. Trends Biochem. Sci. 33: 9– 19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Wadhams GH, Armitage JP. 2004. Making sense of it all: bacterial chemotaxis. Nat. Rev. Mol. Cell Biol. 5: 1024– 1037 [DOI] [PubMed] [Google Scholar]
- 8. Hegde M, Englert DL, Schrock S, Cohn WB, Vogt C, Wood TK, Manson MD, Jayaraman A. 2011. Chemotaxis to the quorum-sensing signal AI-2 requires the Tsr chemoreceptor and the periplasmic LsrB AI-2-binding protein. J. Bacteriol. 193: 768– 773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Springer MS, Goy MF, Adler J. 1977. Sensory transduction in Escherichia coli: two complementary pathways of information processing that involve methylated proteins. Proc. Natl. Acad. Sci. U. S. A. 74: 3312– 3316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Greer-Phillips SE, Alexandre G, Taylor BL, Zhulin IB. 2003. Aer and Tsr guide Escherichia coli in spatial gradients of oxidizable substrates. Microbiology 149: 2661– 2667 [DOI] [PubMed] [Google Scholar]
- 11. Rebbapragada A, Johnson MS, Harding GP, Zuccarelli AJ, Fletcher HM, Zhulin IB, Taylor BL. 1997. The Aer protein and the serine chemoreceptor Tsr independently sense intracellular energy levels and transduce oxygen, redox, and energy signals for Escherichia coli behavior. Proc. Natl. Acad. Sci. U. S. A. 94: 10541– 10546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Hazelbauer GL. 1975. Maltose chemoreceptor of Escherichia coli. J. Bacteriol. 122: 206– 214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Tso WW, Adler J. 1974. Negative chemotaxis in Escherichia coli. J. Bacteriol. 118: 560– 576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Harayama S, Palva ET, Hazelbauer GL. 1979. Transposon-insertion mutants of Escherichia coli K12 defective in a component common to galactose and ribose chemotaxis. Mol. Gen. Genet. 171: 193– 203 [DOI] [PubMed] [Google Scholar]
- 15. Liu X, Parales RE. 2008. Chemotaxis of Escherichia coli to pyrimidines: a new role for the signal transducer Tap. J. Bacteriol. 190: 972– 979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Manson MD, Blank V, Brade G, Higgins CF. 1986. Peptide chemotaxis in E. coli involves the Tap signal transducer and the dipeptide permease. Nature 321: 253– 256 [DOI] [PubMed] [Google Scholar]
- 17. Bibikov SI, Biran R, Rudd KE, Parkinson JS. 1997. A signal transducer for aerotaxis in Escherichia coli. J. Bacteriol. 179: 4075– 4079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Wuichet K, Zhulin IB. 2010. Origins and diversification of a complex signal transduction system in prokaryotes. Sci. Signal. 3: ra50. 10.1126/scisignal.2000724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Frye J, Karlinsey JE, Felise HR, Marzolf B, Dowidar N, McClelland M, Hughes KT. 2006. Identification of new flagellar genes of Salmonella enterica serovar Typhimurium. J. Bacteriol. 188:2233– 2243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Jaureguy F, Landraud L, Passet V, Diancourt L, Frapy E, Guigon G, Carbonnelle E, Lortholary O, Clermont O, Denamur E, Picard B, Nassif X, Brisse S. 2008. Phylogenetic and genomic diversity of human bacteremic Escherichia coli strains. BMC Genomics 9: 560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Touchon M, Hoede C, Tenaillon O, Barbe V, Baeriswyl S, Bidet P, Bingen E, Bonacorsi S, Bouchier C, Bouvet O, Calteau A, Chiapello H, Clermont O, Cruveiller S, Danchin A, Diard M, Dossat C, Karoui ME, Frapy E, Garry L, Ghigo JM, Gilles AM, Johnson J, Le Bouguenec C, Lescat M, Mangenot S, Martinez-Jehanne V, Matic I, Nassif X, Oztas S, Petit MA, Pichon C, Rouy Z, Ruf CS, Schneider D, Tourret J, Vacherie B, Vallenet D, Medigue C, Rocha EP, Denamur E. 2009. Organised genome dynamics in the Escherichia coli species results in highly diverse adaptive paths. PLoS Genet. 5: e1000344. 10.1371/journal.pgen.1000344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Wirth T, Falush D, Lan R, Colles F, Mensa P, Wieler LH, Karch H, Reeves PR, Maiden MC, Ochman H, Achtman M. 2006. Sex and virulence in Escherichia coli: an evolutionary perspective. Mol. Microbiol. 60:1136– 1151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Ochman H, Selander RK. 1984. Standard reference strains of Escherichia coli from natural populations. J. Bacteriol. 157: 690– 693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Escobar-Paramo P, Clermont O, Blanc-Potard AB, Bui H, Le Bouguenec C, Denamur E. 2004. A specific genetic background is required for acquisition and expression of virulence factors in Escherichia coli. Mol. Biol. Evol. 21: 1085– 1094 [DOI] [PubMed] [Google Scholar]
- 25. White AP, Sibley KA, Sibley CD, Wasmuth JD, Schaefer R, Surette MG, Edge TA, Neumann NF. 2011. Intergenic sequence comparison of Escherichia coli isolates reveals lifestyle adaptations but not host specificity. Appl. Environ. Microbiol. 77: 7620– 7632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Zhang Y, Lin K. 2012. A phylogenomic analysis of Escherichia coli/Shigella group: implications of genomic features associated with pathogenicity and ecological adaptation. BMC Evol. Biol. 12: 174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Skippington E, Ragan MA. 2012. Phylogeny rather than ecology or lifestyle biases the construction of Escherichia coli-Shigella genetic exchange communities. Open Biol. 2: 120112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Yang F, Yang J, Zhang X, Chen L, Jiang Y, Yan Y, Tang X, Wang J, Xiong Z, Dong J, Xue Y, Zhu Y, Xu X, Sun L, Chen S, Nie H, Peng J, Xu J, Wang Y, Yuan Z, Wen Y, Yao Z, Shen Y, Qiang B, Hou Y, Yu J, Jin Q. 2005. Genome dynamics and diversity of Shigella species, the etiologic agents of bacillary dysentery. Nucleic Acids Res. 33:6445– 6458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Lane MC, Lloyd AL, Markyvech TA, Hagan EC, Mobley HLT. 2006. Uropathogenic Escherichia coli strains generally lack functional Trg and Tap chemoreceptors found in the majority of E. coli strains strictly residing in the gut. J. Bacteriol. 188: 5618– 5625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Eddy SR. 1998. Profile hidden Markov models. Bioinformatics 14: 755– 763 [DOI] [PubMed] [Google Scholar]
- 31. Waterhouse AM, Procter JB, Martin DMA, Clamp M, Barton GJ. 2009. Jalview Version 2-a multiple sequence alignment editor and analysis workbench. Bioinformatics 25: 1189– 1191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Katoh K, Toh H. 2008. Recent developments in the MAFFT multiple sequence alignment program. Brief Bioinform. 9: 286– 298 [DOI] [PubMed] [Google Scholar]
- 33. Tamura K, Dudley J, Nei M, Kumar S. 2007. MEGA4: molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol. Biol. Evol. 24: 1596– 1599 [DOI] [PubMed] [Google Scholar]
- 34. Guindon S, Delsuc JFF, Dufayard W, Gascuel O. 2009. Estimating maximum likelihood phylogenies with PhyML. Methods Mol. Biol. 538: 113– 137 [DOI] [PubMed] [Google Scholar]
- 35. Camacho C, Coulouris G, Avagyan V, Ma N, Papadopoulos J, Bealer K, Madden TL. 2009. BLAST plus: architecture and applications. BMC Bioinformatics 10: 421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Miquel S, Peyretaillade E, Claret L, de Vallee A, Dossat C, Vacherie B, Zineb el H, Segurens HB, Barbe V, Sauvanet P, Neut C, Colombel JF, Medigue C, Mojica FJ, Peyret P, Bonnet R, Darfeuille-Michaud A. 2010. Complete genome sequence of Crohn's disease-associated adherent-invasive E. coli strain LF82. PLoS One 5:e12714. 10.1371/journal.pone.0012714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Achtman M, Morelli G, Zhu P, Wirth T, Diehl I, Kusecek B, Vogler AJ, Wagner DM, Allender CJ, Easterday WR, Chenal-Francisque V, Worsham P, Thomson NR, Parkhill J, Lindler LE, Carniel E, Keim P. 2004. Microevolution and history of the plague bacillus, Yersenia pestis. Proc. Natl. Acad. Sci. U. S. A. 10:17837– 17842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Foster JT, Beckstrom-Sterberg SM, Pearson T, Beckstrom-Sternberg JS, Chain PSG, Roberto FF, Hinath J, Brettin T, Keim P. 2009. Whole-genome-based phylogeny and divergence of the genus Brucella. J. Bacteriol. 191: 2864– 2870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Galloway-Pena P, Roh JH, Latorre M, Qin X, Murray BE. 2012. Genomic and SNP analyses demonstrate a distant separation of the hospital and community-associated clades of Enterococcus faecium. PLoS One 7: e30187. 10.1371/journal.pone.0030187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Pearson T, Giffard P, Beckstrom-Sternberg S, Auerbach R, Hornstra H, Tuanyok A, Price EP, Glass MB, Leadem B, Beckstrom-Sternberg JS, Allan GJ, Foster JT, Wagner DM, Okinaka RT, Sim SH, Pearson O, Wu Z, Chang J, Kaul R, Hoffmaster AR, Brettin TS, Robison RA, Mayo M, Gee JE, Tan P, Currie BJ, Keim P. 2009. Phylogeographic reconstruction of a bacterial species with high levels of lateral gene transfer. BMC Biol. 7:78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Lenski RE, Winkworth CL, Riley MA. 2003. Rates of DNA sequence evolution in experimental populations of Escherichia coli during 20,000 generations. J. Mol. Evol. 56:498– 508 [DOI] [PubMed] [Google Scholar]
- 42. Giron JA. 1995. Expression of flagella and motility by Shigella. Mol. Microbiol. 18: 63– 75 [DOI] [PubMed] [Google Scholar]
- 43. Pupo GM, Lan RT, Reeves PR. 2000. Multiple independent origins of Shigella clones of Escherichia coli and convergent evolution of many of their characteristics. Proc. Natl. Acad. Sci. U. S. A. 97: 10567– 10572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Fricke WF, McDermott PF, Mammel MK, Zhao S, Johnson TJ, Rasko DA, Fedorka-Cray PJ, Pedroso A, Whichard JM, Leclerc JE, White DG, Cebula TA, Ravel J. 2009. Antimicrobial resistance-conferring plasmids with similarity to virulence plasmids from avian pathogenic Escherichia coli strains in Salmonella enterica serovar Kentucky isolates from poultry. Appl. Environ. Microbiol. 75: 5963– 5971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Alexander RP, Zhulin IB. 2007. Evolutionary genomics reveals conserved structural determinants of signaling and adaptation in microbial chemoreceptors. Proc. Natl. Acad. Sci. U. S. A. 104: 2885– 2890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Deodhar LP, Saraswathi K, Varudkar A. 1991. Aeromonas spp. and their association with human diarrheal disease. J. Clin. Microbiol. 29: 853– 856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Tominaga A, Lan R, Reeves PR. 2005. Evolutionary changes of the flhDC flagellar master operon in Shigella strains. J. Bacteriol. 187: 4295– 4302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Kirby JR. 2009. Chemotaxis-like regulatory systems: unique roles in diverse bacteria. Annu. Rev. Microbiol. 63: 45– 59 [DOI] [PubMed] [Google Scholar]
- 49. Manson MD, Yanofsky C. 1976. Naturally occurring sites within the Shigella dysenteriae tryptophan operon severely limit tryptophan biosynthesis. J. Bacteriol. 126: 668– 678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Miozzari G, Yanofsky C. 1978. Naturally occurring promoter down mutation: nucleotide sequence of the trp promoter/operator/leader region of Shigella dysenteriae 16. Proc. Natl. Acad. Sci. U. S. A. 75: 5580– 5584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Nishiyama S, Ohno S, Ohta N, Inoue Y, Fukuoka H, Ishijima A, Kawagishi I. 2010. Thermosensing function of the Escherichia coli redox sensor Aer. J. Bacteriol. 192: 1740– 1743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Yang Y, Sourjik V. 2012. Opposite responses by different chemoreceptors set a tunable preference point in Escherichia coli pH taxis. Mol. Microbiol. 86: 1482– 1489 [DOI] [PubMed] [Google Scholar]
- 53. Raterman EL, Welch RA. 2013. Chemoreceptors of Escherichia coli CFT073 play redundant roles in chemotaxis toward urine. PLoS One 8: e54133. 10.1371/journal.pone.0054133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Gosink KK, Buron-Barral MC, Parkinson JS. 2006. Signaling interactions between the aerotaxis transducer Aer and heterologous chemoreceptors in Escherichia coli. J. Bacteriol. 188:3487– 3493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Alteri CJ, Mobley HLT. 2012. Escherichia coli physiology and metabolism dictates adaptation to diverse host microenvironments. Curr. Opin. Microbiol. 15: 3– 9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Porcheron G, Kut E, Canepa S, Maurel MC, Schouler C. 2011. Regulation of fructooligosaccharide metabolism in an extra-intestinal pathogenic Escherichia coli strain. Mol. Microbiol. 81: 717– 733 [DOI] [PubMed] [Google Scholar]
- 57. Boin MA, Austin MJ, Hase CC. 2004. Chemotaxis in Vibrio cholerae. FEMS Microbiol. Lett. 239: 1– 8 [DOI] [PubMed] [Google Scholar]
- 58. Watts KJ, Taylor BL, Johnson MS. 2011. PAS/poly-HAMP signalling in Aer-2, a soluble haem-based sensor. Mol. Microbiol. 79: 686– 699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Nowrouzian FL, Adlerberth I, Wold AE. 2006. Enhanced persistence in the colonic microbiota of Escherichia coli strains belonging to phylogenetic group B2: role of virulence factors and adherence to colonic cells. Microbes Infect. 8: 834– 840 [DOI] [PubMed] [Google Scholar]
- 60. Pickering R, Dirks PHGM, Jinnah Z, de Ruiter DJ, Churchill SE, Herries AIR, Woodhead JD, Hellstrom JC, Berger LR. 2011. Australopithecus sediba at 1.977 Ma and implications for the origins of the genus Homo. Science 333: 1421– 1423 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.