Abstract
Type IV pili play important roles in a wide array of processes, including surface adhesion and twitching motility. Although archaeal genomes encode a diverse set of type IV pilus subunits, the functions for most remain unknown. We have now characterized six Haloferax volcanii pilins, PilA[1-6], each containing an identical 30-amino-acid N-terminal hydrophobic motif that is part of a larger highly conserved domain of unknown function (Duf1628). Deletion mutants lacking up to five of the six pilin genes display no significant adhesion defects; however, H. volcanii lacking all six pilins (ΔpilA[1-6]) does not adhere to glass or plastic. Consistent with these results, the expression of any one of these pilins in trans is sufficient to produce functional pili in the ΔpilA[1-6] strain. PilA1His and PilA2His only partially rescue this phenotype, whereas ΔpilA[1-6] strains expressing PilA3His or PilA4His adhere even more strongly than the parental strain. Most surprisingly, expressing either PilA5His or PilA6His in the ΔpilA[1-6] strain results in microcolony formation. A hybrid protein in which the conserved N terminus of the mature PilA1His is replaced with the corresponding N domain of FlgA1 is processed by the prepilin peptidase, but it does not assemble functional pili, leading us to conclude that Duf1628 can be annotated as the N terminus of archaeal PilA adhesion pilins. Finally, the pilin prediction program, FlaFind, which was trained primarily on archaeal flagellin sequences, was successfully refined to more accurately predict pilins based on the in vivo verification of PilA[1-6].
INTRODUCTION
Biofilms are considered a natural state for most microbes and allow protection from environmental stresses (1–3). Biofilm formation begins with attachment of cells to a surface. As organisms accumulate, they begin to spread along the surface and aggregate, forming microcolonies while at the same time producing a thick exopolysaccharide matrix (EPS) and eventually developing into the macrocolonies that are found in a mature biofilm (4).
Bacterial type IV pili are thin, multifunctional, filamentous protein complexes that can extend from the cell surface, attach to a foreign surface, and retract. Thus, pili facilitate adherence to abiotic surfaces, the formation of close intracellular associations, and twitching motility, all functions that are critical to biofilm formation (5–9).
Pilin precursor proteins are translocated through the cytoplasmic membrane via the Sec translocation pathway (10, 11). However, while most Sec substrates contain class I or class II signal peptides, which are processed at sites subsequent to the membrane-inserted hydrophobic portion of the signal peptide (H domain), a specific prepilin peptidase, PilD (PibD in archaea), cleaves the class III signal peptides of pilin precursors at a site preceding the H domain (12–14). Integration of the mature pilins into pili occurs after the release of the H domain from the membrane. Extracellular interactions between hydrophobic N termini of the pilins that result in the formation of a hydrophobic central core provide a scaffold for assembly of a pilus (15–17).
In addition to the prepilin peptidase, two other highly conserved proteins are crucial to the biosynthesis of a pilus filament: PilB, an ATPase that transforms the energy necessary for pilus assembly, and PilC, a multispanning membrane protein that may serve as an assembly platform (18, 19). The retraction of type IV pili, which has thus far been observed only in Gram-negative bacteria in which these structures have been studied most extensively, also requires a second ATPase, PilT (20, 21). While the assembly of functional type IV pili in Gram-negative bacteria requires several additional components, the exact functions of many of these other components are unknown (18, 19).
Typically, a type IV pilus is composed of major pilins, the primary structural component of the pilus, and several minor pilin-like proteins that play important roles in pilus assembly or function or both (22). Deletion of the gene encoding the major pilin results in loss of pilus structure and function in most organisms, while deleting genes that encode minor pilins can have a variety of effects (23–25).
A single species can display a diverse set of pili. This diversity may be required for different functions, although pili may also exhibit some functional overlap depending on growth conditions. For example, Vibrio cholerae grown in culture medium expresses the MshA pili, which the organism uses to adhere to borosilicate, while expressing distinct type IV pili, the TcpA pili, to attach to epithelial cells in vivo (26, 27).
In archaea, the first proteins shown to be processed by a prepilin peptidase were flagellins which, unlike bacterial flagellins, are not transported to the tip of a flagellum via a type III secretion system (28, 29). The abundant archaeal major and minor flagellin sequences available were used to train an archaeal prepilin prediction program, FlaFind, which predicted a large diverse set of type IV pilin-like proteins in euryarchaea and crenarchaea by detecting signal peptide signals similar to those of the flagellins (30). In vivo studies on Methanococcus maripaludis have confirmed that a subset of pilins containing a unique EppA prepilin peptidase processing site is involved in surface adhesion (31). Two additional sets of adhesion pilins have been identified in the Sulfolobales, one of which was also involved in Sulfolobus acidocaldarius autoaggregation (32, 33). Microcolony formation has been observed in many archaeal species, including S. acidocaldarius and Sulfolobus solfataricus (34, 35).
As in other archaeal species, haloarchaea in biofilms display cell surface structures (36). Moreover, Haloferax volcanii, which has a genome that encodes 38 potential pilin-like proteins as predicted by FlaFind, can adhere to abiotic surfaces (30, 37). This surface adhesion requires the presence of a functional PibD, implicating type IV pili in this process. However, unlike many other prokaryotes, H. volcanii surface adhesion does not require the presence of flagella (37).
In this study, we have shown that six H. volcanii pilins play an important role in the surface adhesion of this haloarchaeon. Surprisingly, each of these pilins can, by itself, promote pilus formation and rescue, to some degree, the adhesion defect of an H. volcanii strain that lacks all six pilins. However, the rescued adhesion phenotypes vary depending upon the pilin expressed. Each of these proteins contains a domain of unknown function, Duf1628, that is essential to the function of the pilin. Since this domain is conserved in many pilins that have been identified in other archaeal species, our results have important implications for these species. Finally, two of these six H. volcanii pilins have negative charges at the −2 position. Only positive charges had been observed at the −2 position in the initial flagellin training set for the FlaFind prepilin prediction program (30). Therefore, our results have also allowed us to refine FlaFind, which may lead to more-accurate predictions of archaeal PilA pilins.
MATERIALS AND METHODS
Reagents.
All enzymes used for standard molecular biology procedures, except iProof high-fidelity DNA polymerase, which was purchased from Bio-Rad, were purchased from New England BioLabs. The ECL Plus Western blotting system detection and horseradish peroxidase-linked sheep anti-mouse antibodies were purchased from GE Healthcare. The polyvinylidene difluoride membrane, MF membrane filters (0.025 μm), and the Ultracel-3K membrane were purchased from Millipore. DNA and plasmid purification kits and anti-His antibodies were purchased from Qiagen. NuPAGE gels, buffers, and reagents were purchased from Invitrogen. Difco agar and Bacto yeast extract were purchased from Becton, Dickinson, and Company. Peptone was purchased from Oxoid. 5-Fluoroorotic acid (5-FOA) was purchased from Toronto Research Biochemicals. All other chemicals and reagents were purchased from either Fisher or Sigma.
Strains and growth conditions.
The plasmids and strains used in this study are listed in Table 1. H. volcanii strains were grown at 45°C in liquid or on solid semidefined Casamino acid (CA) medium (38) or complex modified growth medium (MGM) (38). Plates contained 1.5% (wt/vol) agar unless otherwise noted. To ensure equal concentrations of agar in all plates, the agar was completely dissolved prior to autoclaving, and the autoclaved medium was stirred before plates were poured. Strain H53 (39) was grown in CA medium supplemented with tryptophan and uracil (50-μg/ml final concentration) or MGM without any supplements. For the selection of the ΔpilA deletion mutants (see below), 5-FOA was added at a final concentration of 150 μg/ml in CA medium and uracil was added at one-fifth of its normal concentration, i.e., a 10-μg/ml final concentration, during 5-FOA selection. Strain H53 and the pilA deletion mutants transformed with pTA963 were grown in CA medium supplemented with tryptophan. Escherichia coli strains were grown at 37°C in NZCYM medium supplemented with ampicillin (200 μg/ml) (40).
Table 1.
Strains and plasmids
| Strain or plasmid | Relevant characteristic(s)a | Reference or source |
|---|---|---|
| Plasmids | ||
| pTA131 | Ampr; pyrE2 under a ferredoxin promoter | 39 |
| pTA963 | Ampr, pyrE2 and hdrB markers, inducible ptna promoter | 48 |
| pRE1 | pTA131 containing chromosomal pilA2 flanking regions 700 bp upstream and 700 bp downstream | This study |
| pRE3 | pTA963 containing pilA2His | This study |
| pRE8 | pTA131 containing chromosomal pilA1 flanking regions 700 bp upstream and 700 bp downstream | This study |
| pRE32 | pTA963 containing pilA3His | This study |
| pRE33 | pTA963 containing pilA4His | This study |
| pRE34 | pTA963 containing pilA5His | This study |
| pRE35 | pTA963 containing pilA6His | This study |
| pRE36 | pTA963 containing pilA1hybridHis | This study |
| pMT1 | pTA131 containing chromosomal flgA1-flgA2 flanking regions 700 bp upstream and 700 bp downstream | 37 |
| pMT24 | pTA963 containing pilA1His | This study |
| Strains | ||
| E. coli | ||
| DH5α | F− 80dlacZΔM15 Δ(lacZYA-argF)U169 recA1 endA enda1 hsdR17(rK− mK−) supE44 thi-1 gyrA relA1 | Invitrogen |
| DL739 | MC4100 recA dam-13::Tn9 | 47 |
| H. volcanii | ||
| H53 | ΔpyrE2 Δtrp | 39 |
| RE10 | H53 ΔpilA2 | This study |
| RE12 | H53 ΔpilA2 ΔpilA3 ΔpilA4 ΔpilA5 ΔpilA6 | This study |
| RE13 | H53 containing pMT24 | This study |
| RE17 | MT4 containing MT24 | This study |
| RE19 | H53 containing pRE3 | This study |
| RE23 | MT4 containing pRE3 | This study |
| RE27 | H53 ΔpilA1 | This study |
| RE29 | H53 ΔpilA1 ΔpilA3 ΔpilA4 ΔpilA5 ΔpilA6 | This study |
| RE30 | RE43 containing pTA963 | This study |
| RE37 | H53 containing pRE34 | This study |
| RE38 | MT4 containing pRE34 | This study |
| RE39 | MT4 containing pRE35 | This study |
| RE40 | H53 containing pRE32 | This study |
| RE41 | H53 containing pRE33 | This study |
| RE42 | MT4 containing pRE33 | This study |
| RE43 | H53 ΔpilA1 ΔpilA2 ΔpilA3 ΔpilA4 ΔpilA5 ΔpilA6 | This study |
| RE44 | RE43 containing pRE34 | This study |
| RE45 | RE43 containing pMT24 | This study |
| RE46 | H53 ΔpilA1 ΔpilA2 ΔpilA3 ΔpilA4 ΔpilA5 ΔpilA6 ΔflgA1 ΔflgA2 | This study |
| RE47 | H53 ΔpilA1 ΔpilA2 | This study |
| RE48 | H53 containing pRE35 | This study |
| RE49 | MT4 containing pRE32 | This study |
| RE50 | RE43 containing pRE35 | This study |
| RE51 | RE43 containing pRE32 | This study |
| RE52 | RE43 containing pRE33 | This study |
| RE53 | RE43 containing pRE3 | This study |
| RE55 | RE43 containing pRE36 | This study |
| RE58 | H53 ΔpilA3 ΔpilA4 | This study |
| RE59 | H53 ΔpilA3 ΔpilA4 ΔpilA5 ΔpilA6 | This study |
| RE61 | MT4 containing pRE36 | This study |
| RE71 | RE46 containing pRE36 | This study |
| RE72 | RE46 containing pMT24 | This study |
| RE73 | RE46 containing pRE3 | This study |
| RE74 | RE46 containing pRE32 | This study |
| RE75 | RE46 containing pRE33 | This study |
| RE76 | RE46 containing pRE34 | This study |
| RE77 | RE46 containing pRE35 | This study |
| RE78 | RE46 containing pTA963 | This study |
| RE79 | H53 containing pRE36 | This study |
| MT2 | H53 ΔflgA1 ΔflgA2 | 37 |
| MT4 | H53 ΔpibD | 37 |
| MT13 | H53 containing pTA963 | 43 |
| MT44 | MT2 containing pTA963 | M. Tripepi, unpublished data |
| MT60 | H53 ΔpilA5 ΔpilA6 | This study |
Ampr, ampicillin resistant.
Generation of chromosomal pilA deletions in H53.
Chromosomal deletions were generated by using a homologous recombination (pop-in/pop-out) method previously described by Allers and Ngo (41). Plasmid constructs for use in the pop-in/pop-out knockout process were generated using overlap PCR as described previously by Tripepi et al. (37) (primers used are listed in Table S1 in the supplemental material). These methods result in an in-frame markerless deletion. Thus, to confirm that the chromosomal replacement event occurred at the proper location on the chromosome, the genomic DNA isolated from colonies derived using these techniques was screened by PCR (primers used are listed in Table S1 in the supplemental material). The identities of the PCR products were verified by sequencing using the primers lying outside the gene of interest (see Table S1 in the supplemental material). The pilA deletion mutants generated in strain H53 are listed in Table 1.
Surface adhesion assay.
Surface adhesion was assayed using a modified air-liquid interface (ALI) assay protocol (42) as follows: 3 ml of culture in CA medium, supplemented with tryptophan and/or uracil as necessary, at an optical density at 600 nm (OD600) of ∼0.3 was incubated in each well of a 12-well plate. Plastic coverslips (22 by 22 mm; 0.19 to 0.25 mm thick) were inserted into each well at an angle of 90°. Lids were placed over the wells; the plates were then placed on wet paper towels to limit evaporation. The wrapped plates were incubated at 45°C without shaking. Coverslips were incubated overnight and then removed from the wells using forceps, submerged for 3 min in 2% (vol/vol) acetic acid, and allowed to air dry. Dry coverslips were stained in 0.1% (wt/vol) crystal violet for 10 min. The coverslips were then washed three times with distilled water. Stained coverslips were air dried and then examined using light microscopy.
Protein extraction, LDS-PAGE, and Western blotting.
Liquid cultures were grown until the mid-log phase (OD600 = ∼0.5). Cells were collected by centrifugation at 4,300 × g for 10 min at 4°C. Cell pellets were resuspended and lysed in 1% (vol/vol) NuPAGE lithium dodecyl sulfate (LDS) supplemented with 50 mM dithiothreitol (DTT) and stored at −20°C. The electrophoresis of protein samples was performed with 12% (vol/vol) Bis-Tris NuPAGE gels under denaturing conditions using morpholinepropanesulfonic acid (MOPS) buffer at pH 7.7. Proteins were transferred from the gel onto a polyvinylidene difluoride membrane using a Bio-Rad Transblot-SD semidry transfer cell at 15 V for 30 min. Western blots of whole-cell lysates grown to early log phase (OD600 = ∼0.3) of strains expressing C-terminally His-tagged constructs with a 3-amino-acid linker sequence were probed with an anti-His antibody at a dilution of 1:1,000, followed by a secondary anti-mouse antibody at a dilution of 1:10,000. Antibody-labeled protein bands were identified using the Amersham ECL Plus Western blotting detection system.
Electron microscopy.
A total of 500 μl of cell culture was fixed in CA medium with 2% (vol/vol) glutaraldehyde and 1% (vol/vol) paraformaldehyde for 1 h. A total of 10 μl was put onto glow-discharged carbon grids for 10 min. The grids were rinsed two times in double-distilled water (ddH2O) and negatively stained using 1% (wt/vol) uranyl formate. Grids were then analyzed using a Philips Tecnai 12 operating at 120 kV and a Gatan US1000 2K-by-2K (2,024- by 2,024-pixel) camera. Ten-microliter samples of CsCl gradient density fractions were applied onto copper grids coated with carbon-Formvar. The grids were left for 5 min at room temperature, washed with water, blotted with a filter paper, and stained with 2% (wt/vol) uranyl acetate for 10 s. Grids were then analyzed using a Philips Tecnai 12 instrument operating at 120 kV and a Gatan US1000 2K-by-2K (2,024- by 2,024-pixel) camera.
Phase-contrast microscopy.
Liquid cultures were grown to early log phase (OD600 = ∼0.3). A 1-ml sample of cell culture was put onto a 35-mm petri dish with a glass coverslip over a hole in the bottom of the plate for optimal visualization of the cells. The cells were maintained at 45°C and viewed in the same position at ×100 magnification using phase-contrast microscopy on an Eclipse TE2000-U inverted microscope (Nikon) with a Plan Apo ×100 1.0 Na objective and Cascade 512B charge-coupled device (CCD) camera (Photometrics) driven by Metamorph imaging software (Molecular Devices) over the course of 10 h.
Duf1628 sequence characterization.
Sequences of predicted Duf1628 proteins from the 28 archaeal species listed in Table 2 were downloaded from the Pfam v26.0 database entry for Duf1628 (http://pfam.sanger.ac.uk/family/DUF1628). The set of Duf1628 proteins (from here on referred to as PilA pilins) was scanned with the PERL program FlaFind 1.2 (http://signalfind.org/flafind.html), searching for the signal sequence motif [KRDE][GA][ALIFQMVED][ILMVTAS]. This program was extended from the original program FlaFind 1.0 (30) to allow for glutamate and aspartate at the −2 position.
Colocalization of pilA genes with putative pilin and pilus biosynthesis genes.
A gene was considered a putative pilin and pilus biosynthesis gene if it (i) was annotated as a PibD, PilB, or PilC homolog, (ii) was FlaFind 1.2 positive, and (iii) contained Duf1628. Using the NCBI Sequence Viewer graphical display on the gene report page, the annotation of the two genes upstream and downstream of pilin genes was determined. Moreover, these four local genes were analyzed with FlaFind 1.2 and further screened with Pfam to determine whether the genes encoded a Duf1628-containing protein not recognized by FlaFind 1.2.
Prepilin prediction and sequence analysis.
Complete genome protein sequences from the 28 archaeal species listed in Table S2 in the supplemental material were downloaded from the NCBI website (http://www.ncbi.nlm.nih.gov) and analyzed using FlaFind 1.2.
RESULTS
H. volcanii expresses type IV pili.
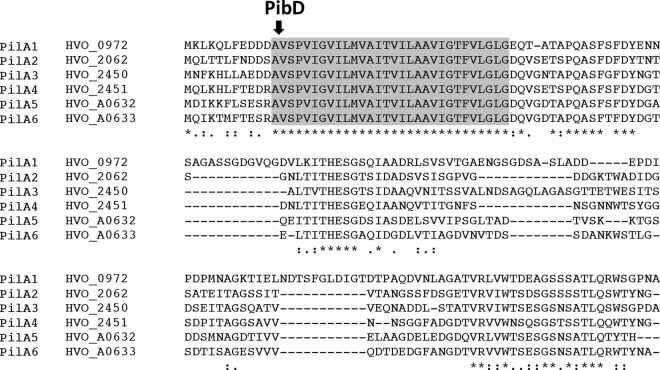
Based on mass spectrometry, flagellum biosynthesis is inhibited in a deletion mutant lacking the oligosyltransferase aglB. However, CsCl purification studies revealed that an aglB deletion strain unable to make flagella still has surface filaments (43). To determine whether these structures are type IV pili, we analyzed cells of H. volcanii H53, the parent strain used in this study, from here on referred to as the wild type (wt), as well as H. volcanii strains lacking either the flagellin genes flgA1 and flgA2 or pibD by electron microscopy (Fig. 1). Consistent with these filaments being type IV pili, while ΔflgA[1-2] cells have surface filaments, the ΔpibD strain lacks surface structures (Fig. 1). Similarly, CsCl gradient density purification of filaments in supernatants of these strains confirms the absence of any detectable filaments in the ΔpibD strain (data not shown). Fitting with the observation of type IV pili on the ΔflgA[1-2] cells, microarray data identified a set of predicted type IV pilins that, unlike the majority of H. volcanii FlaFind positives, appeared to be expressed, albeit at varied levels, under the growth conditions used in the adhesion assays (C. Daniels, unpublished data). These pilins (from here on referred to as PilA[1-6]) contained a highly conserved domain of unknown function (Duf1628) that starts at the predicted −2 position of the PibD motif and has an identical amino acid sequence across the whole 30-amino-acid H domain (Fig. 2). H. volcanii has three additional genes encoding proteins containing the Duf1628 domain, two of which have a predicted PibD processing site (Hvo_2704 as well as Hvo_0308 upon reannotation of the start codon) while the third protein lacks an amino acid sequence that resembles a PibD motif (see Fig. S1 in the supplemental material). The amino acid sequences of the H domains of these proteins, while sharing significant homology, are not identical to the signal peptide/pilin core domains of PilA[1-6].
Fig 1.
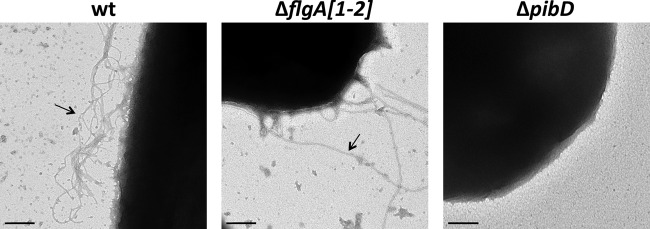
H. volcanii expresses nonflagellum type IV pili. Electron micrograph of wild-type (wt), ΔflgA[1-2], and ΔpibD cells fixed in 2% glutaraldehyde and 1% paraformaldehyde and stained with 1% uranyl formate. Surface filaments are observed on the wt and ΔflgA[1-2] cells (arrows). Bars, 200 nm.
Fig 2.
Six predicted pilins contain a highly conserved Duf1628. Amino acid sequence alignment of PilA[1-6] was formulated by Clustal Omega (46). The amino acid sequence of the H domain (gray shading) starting at the −1 position of the PibD processing site (arrow) is completely conserved.
PilA pilins are processed by PibD.
Unlike previously characterized subunits of archaeal flagella and other type IV pilus-like structures, the −2 positions in the pilins predicted to contain Duf1628 do not appear to be limited to arginine and lysine; rather, they include the aspartate and serine amino acid residues that are allowed in bacterial PilD processing motifs (44). Consistent with the inclusion of aspartate in PibD processing motifs, we previously showed that PilA5His, which contains an arginine at the −2 position (−2R), and PilA3His (−2D) migrate more slowly under electrophoresis when expressed in a ΔpibD mutant, indicating the lack of signal peptide processing (37). The expression of PilA1His (−2D), PilA4His (−2R), and PilA6His (−2R) shows a similar change, while a size shift is not clearly observed for PilA2His (−2S) (Fig. 3).
Fig 3.
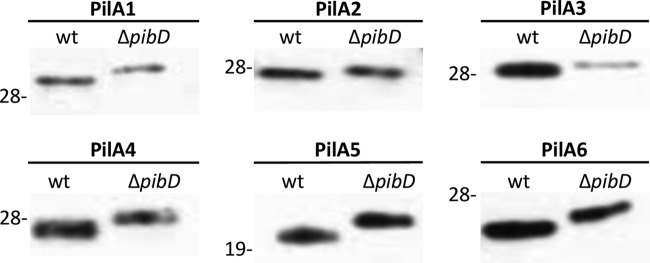
PibD processes PilA pilins. Western blot analysis was performed on PilA1His protein extracts from cell lysates of the wt or ΔpibD strain expressing a pilAHis gene under the regulation of a trp-inducible promoter. His-tagged proteins were detected using anti-His antibodies. Comparable amounts of protein were loaded into each lane. The migration of molecular mass standards is indicated on the left (in kDa).
PilA pilins are required for surface adhesion.
Previous data have shown that ΔpibD mutants cannot adhere to glass or plastic surfaces (37). To determine which of the PibD-dependent PilA pilins were required for adhesion, deletion mutants were constructed for each pilA gene, pilA1 and pilA2, or the colocalized genes pilA[3-4] and pilA[5-6]. Knockouts were confirmed by PCR (data not shown). Since none of these deletion mutant strains exhibits significant adhesion defects on glass (data not shown) or plastic (Fig. 4), we also constructed mutants that contained multiple deletions of pilA genes (Fig. 5A and B). Interestingly, while ΔpilA[2-6], a strain that expresses only PilA1, can adhere, adhesion is completely abolished only in ΔpilA[1-6], a strain that lacks all six pilins, or ΔpilA[1,3-6], a strain expressing only PilA2. This suggests that at least PilA1 and PilA[3-6] pilins have functions that partially overlap (Fig. 4). Expression of any of the six pilins in trans under the control of an inducible trp promoter in the ΔpilA[1-6] strain restores adhesion, albeit to various degrees (Fig. 6). These data support the hypothesis that the proteins containing Duf1628 are required for adhesion and also demonstrate that each pilin-like protein can, by itself, complement the adhesion defect of the ΔpilA[1-6] strain.
Fig 4.
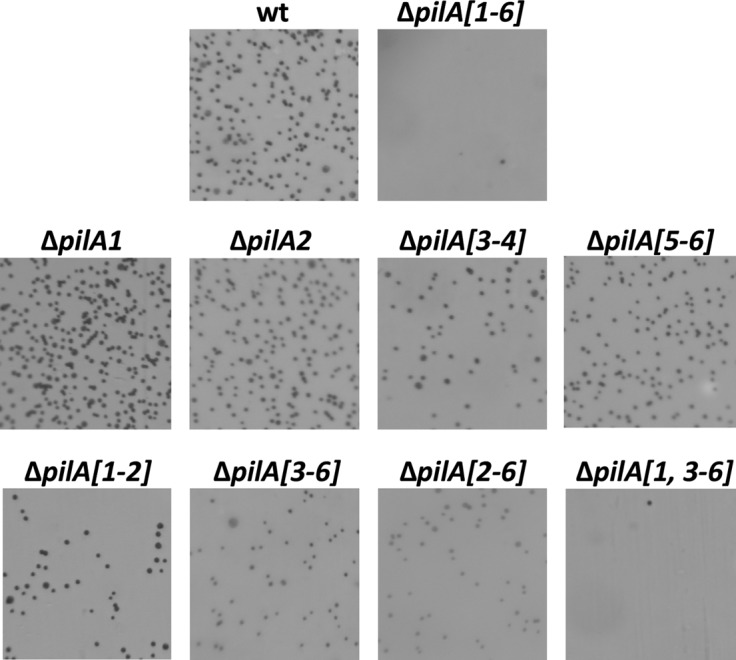
H. volcanii requires the presence of at least one pilin gene for adhesion. Adhesion to plastic coverslips was tested using a modified ALI assay (42). Light micrographs of the coverslips taken at ×35 magnifications are shown. Coverslips were placed in individual wells of 12-well plates, each containing 3 ml of a mid-log-phase liquid CA culture (wt, ΔpilA[1-6], ΔpilA[1], ΔpilA[2], ΔpilA[3-4], ΔpilA[5-6], ΔpilA[1-2], ΔpilA[3-6], ΔpilA[2-6], and ΔpilA[1,3-6]). After overnight incubation, cells were fixed with 2% acetic acid, stained with 0.1% crystal violet, and observed by light microscopy. Adhesion is abolished in the ΔpilA[1,3-6] and ΔpilA[1-6] strains.
Fig 5.
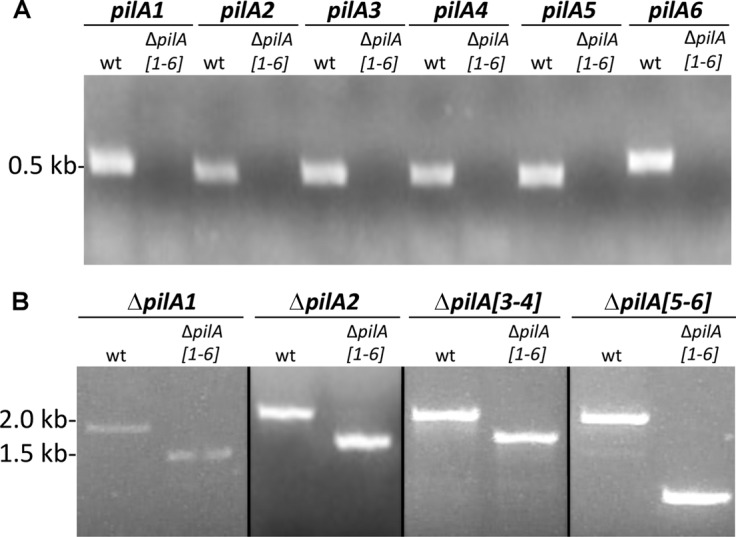
Deletion of the six pilA genes in the wt. (A) Intragenic primers were made for each of the pilA genes. PCR was performed using genomic DNA isolated from the wt or the ΔpilA[1-6] strain as template DNA. (B) PCR was performed as stated above. For amplification of ΔpilA[1-6] genes, the primers were against the region 700 bp upstream and 700 bp downstream of ΔpilA[1-6]. As expected, no amplicons were obtained from ΔpilA[1-6] when the DNA was amplified using ΔpilA[1-6]-specific primers. The amplicon sizes obtained when the ΔpilA[1-6] flanking regions were amplified were approximately 500 nucleotides smaller when DNA from the ΔpilA[1-6] strain was used as the template than the amplicons obtained when using the wt as a template.
Fig 6.
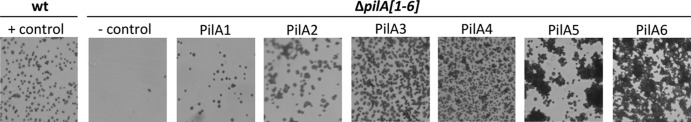
One PilA paralog is sufficient for adhesion. The wt with pTA963 (+ control) and ΔpilA[1-6] with pTA963 (− control) or pTA963 expressing one of the six pilin genes (pilA[1-6]) were incubated overnight using the modified ALI assay (42) as described in Fig. 4 (×35 magnification).
The expression of individual pilins results in differing adhesion phenotypes. Although expression of pilA1 and pilA2 under the control of the inducible promoter in the ΔpilA[1-6] strain improves adhesion compared to expressing these genes individually from the chromosome, PilA1 and PilA2 rescue the adhesion defect least effectively. Conversely, strains expressing the PilA3 or PilA4 pilin in trans adhere more strongly than the wild type does. Most surprisingly, expression of either PilA5 or PilA6 not only allows cells to adhere to a plastic surface, it also leads to the formation of microcolonies (Fig. 6). These data suggest that although the pilins appear to have some overlapping function, cell-cell interactions are promoted by specific pilins.
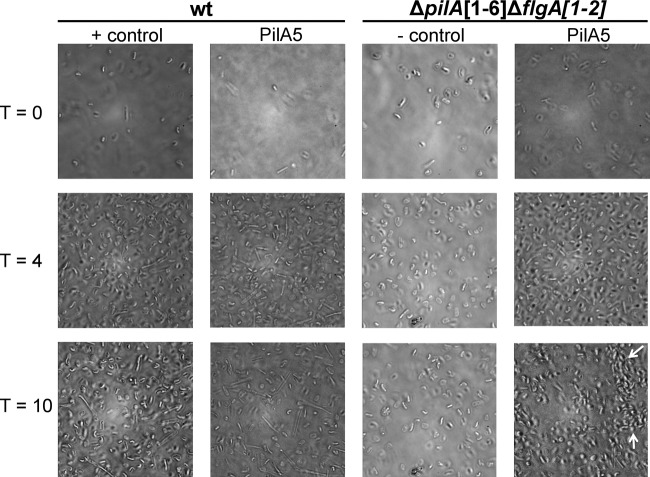
PilA5-dependent microcolony formation does not occur in a wild-type background and does not require the flagella.
As shown in Fig. 6, wild-type cells do not form detectable microcolonies. While we do see minor cell clusters in the wt cells, the movement into microcolonies over time is not observed. To determine whether this is due to low expression of PilA5 and/or PilA6 from the chromosome, we expressed PilA5His in trans in a wild-type background. Surprisingly, expression of this construct in the presence of the chromosomally expressed pilins does not promote the formation of microcolonies, as is observed in the ΔpilA[1-6] background. It is possible that other pilins may inhibit microcolony formation (Fig. 7).
Fig 7.
PilA5 allows microcolony formation in a flagellum-independent manner. Phase-contrast microscopy images at ×100 magnification of the wt transformed with pTA963 (+ control) or pTA963 expressing PilA5 and ΔpilA[1-6]ΔflgA[1-2] transformed with pTA963 (− control) or pTA963 expressing PilA5 and incubated at 45°C for 10 h. One milliliter of mid-log-phase cultures was transferred into a 35-mm petri dish with a glass coverslip at the bottom. PilA5 expressed in ΔpilA[1-6]ΔflgA[1-2] forms microcolonies (arrows) that become strongest after 10 h.
We previously showed that adhesion of H. volcanii does not require functional flagella (37). To determine whether microcolony formation requires flagella, PilA5 was expressed in a ΔpilA[1-6]ΔflgA[1-2] background. As determined by phase-contrast microscopy, the flagella do not appear to inhibit or enhance the microcolony formation, as microcolonies can be observed within 8 h in both the ΔpilA[1-6] strain and the ΔpilA[1-6]ΔflgA[1-2] strain when PilA5 is expressed in trans (Fig. 7).
Each of the six adhesion pilins can be assembled into a functional pilus.
The adhesion data presented above suggest that the ΔpilA[1-6] strain can form functional pili when any one of the six highly conserved adhesion pilins is expressed in trans. To confirm pilus biosynthesis, we performed electron microscopy (EM) using the ΔpilA[1-6]ΔflgA[1-2] strain, as H. volcanii flagella are indistinguishable from type IV pili under our experimental conditions (Fig. 1). Consistent with PilA pilins being the only subunits of observed type IV pili in the ΔflgA[1-2] strain, EM analysis of the strain lacking flgA[1-2] and the pilA[1-6] genes indeed shows a lack of observable filaments on cell surfaces (Fig. 8). Moreover, complementation of the ΔpilA[1-6]ΔflgA[1-2] strains with just one of the PilA[1-6] pilins results in observable cell surface filaments (Fig. 8). However, the numbers of pili on the surface are lower than seen in the ΔflgA[1-2] strain.
Fig 8.

One PilA paralog is sufficient to promote pilus synthesis. Electron micrograph of ΔflgA[1-2] transformed with pTA963 (+ control) and ΔpilA[1-6]ΔflgA[1-2] transformed with pTA963 (− control) or pTA963 expressing one of the six pilin genes (pilA[1-6]). Pili are observable when at least one pilin gene is expressed (arrows). Similar numbers of pili were seen on the surfaces of PilA[1-6] mutants. Cells were fixed in 2% glutaraldehyde and 1% paraformaldehyde and stained with 1% uranyl formate. Bars, 200 nm.
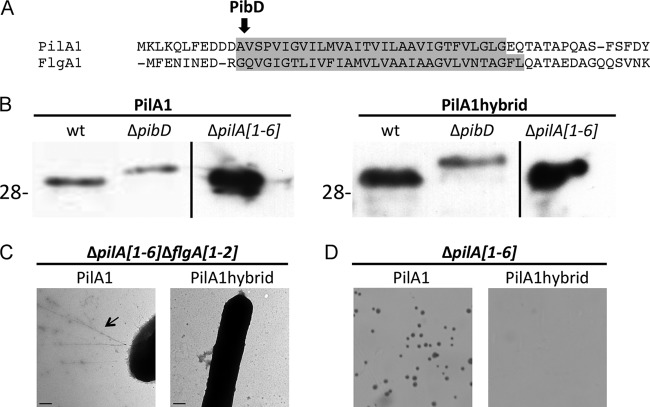
The conserved hydrophobic domain is essential for pilus biosynthesis and function.
To investigate the role of the conserved H domain in pilus biosynthesis and function, we replaced the H domain of PilA1, starting at the PibD processing site, with the H domain of the PibD signal peptide of FlgA1 (Fig. 9A). Although the His-tagged PilA1hybrid still requires PibD for processing and can be expressed in the ΔpilA[1-6] strain, as determined by Western blotting (Fig. 9B), pili are not observed on the surface of ΔpilA[1-6]ΔflgA[1-2] cells (Fig. 9C). Consistent with the requirement of a type IV pilus structure for surface adhesion, adhesion is not observed when this hybrid protein construct is expressed in trans (Fig. 9D). It should be noted that a PilA3hybrid was also constructed, but attempts to express the construct in H. volcanii were unsuccessful, further underscoring the importance of the H domain sequence (data not shown).
Fig 9.
The conserved H domain sequence is required for PilA pilus assembly and function. (A) N-terminal amino acid sequence of H. volcanii PilA1 and FlgA1. Replaced amino acids are highlighted in gray. (B) Western blot analysis of wt, ΔpibD, and ΔpilA[1-6] cell lysates expressing PilA1His or PilA1hybridHis under the regulation of a trp-inducible promoter. His-tagged proteins were detected using anti-His antibodies. Comparable amounts of protein were loaded into each lane. The migration of molecular mass standards is indicated on the left (in kDa). (C) EM, as described in Fig. 8, of ΔpilA[1-6]ΔflgA[1-2] expressing PilA1His or PilA1hybridHis. Pili are seen on the surface of ΔpilA[1-6]ΔflgA[1-2] expressing PilA1His (arrow). (D) Adhesion assays (42), as described in Fig. 4, of ΔpilA[1-6] expressing PilA1His or PilA1hybridHis (×35 magnification).
Homologs of PilA pilins are identified predominantly in euryarchaea.
Pfam lists proteins with a Duf1628 from a diverse set of organisms. In this study, we examined the presence of putative PilA homologs in 28 genomes representing a diverse range of archaeal phyla, including the thaumarchaeal species. All but two of the predicted pilins containing the conserved PilA N terminus (one crenarchaeal and one thaumarchaeal sequence) were identified in euryarchaeal species (Table 2).
Table 2.
In silico analysis of archaeal PilA and non-PilA pilins
| Species | No. of sequences containing PilA (Duf1628) |
No. of FlaFind 1.2 positives (no. with D/no. with E) at −2 | ||
|---|---|---|---|---|
| Pfam | No. of FlaFind 1.2 positives (no. with D/no. with E) at −2 | Colocalized with putative pilins | ||
| Crenarchaeota | ||||
| Ignicoccus hospitalis KIN4/I | 0 | 18/3 | ||
| Acidianus hospitalis W1 | 0 | 13/2 | ||
| Thermoproteus neutrophilus (Pyrobaculum neutrophilum V24Sta) | 1a | 0/0 | 1 | 17/3 |
| Sulfolobus acidocaldarius DSM 639 | 0 | 20/1b | ||
| Sulfolobus solfataricus P2 | 0 | 28/0 | ||
| Sulfolobus tokodaii strain 7 | 0 | 25/1 | ||
| Aeropyrum pernix K1 | 1 | 1/0 | 0 | 19/1 |
| Pyrobaculum aerophilum strain IM2 | 0 | 20/2 | ||
| Euryarchaeota | ||||
| Archaeoglobus fulgidus DSM 4304 | 1 | 1/0 | 0 | 19/4c |
| Ferroglobus placidus DSM 10642 | 3 | 3/0 | 0 | 31/6 |
| Haloarcula marismortui ATCC 43049 | 8 | 3/2 | 1 | 48/11 |
| Haloferax volcanii DS2d | 9 | 7/2 | 4 | 47/9 |
| Natronomonas pharaonis DSM 2160 | 7 | 6/0 | 3 | 34/5 |
| Methanocaldococcus jannaschii DSM 2661 | 0 | 19/5 | ||
| Methanococcus maripaludis C5 | 0 | 16/1 | ||
| Methanosarcina mazei Go1 | 7 | 5/2 | 5e | 18/5 |
| Methanospirillum hungatei JF-1 | 19 | 8/6 | 16 | 20/12 |
| Picrophilus torridus DSM 9790 | 0 | 9/1 | ||
| Pyrococcus furiosus DSM 3638 | 0 | 18/1 | ||
| Pyrococcus horikoshii OT3 | 0 | 23/3 | ||
| Thermococcus kodakaraensis KOD1 | 0 | 38/0 | ||
| Thermoplasma acidophilum DSM 1728 | 2 | 2/0 | 0 | 6/2 |
| Nanoarchaeota | ||||
| Nanoarchaeum equitans Kin-4-M | 0 | 6/0 | ||
| Korarchaeota | ||||
| “Candidatus Korarchaeum cryptofilum” OPF8 | 0 | 12/3 | ||
| Thaumarchaeota | ||||
| Cenarchaeum symbiosum A | 0 | 15/3 | ||
| Nitrosopumilus maritimus SCM1 | 0 | 10/0 | ||
| “Candidatus Caldiarchaeum subterraneum” | 1 | 1/0 | 1 | 18/3 |
| “Candidatus Nitrosoarchaeum limnia” SFB1 | 0 | 19/3 | ||
Only FlaFind 1.2 positive with the second predicted transmembrane (TM).
Additional putative motif with −2K/R predicted for one of these proteins.
Additional putative motif with −2K/R predicted for two of these proteins.
Hvo_0308 is FlaFind 1.2 positive after reannotation.
One gene is colocalized with a pilB homologue.
To provide corroboration that these proteins are indeed type IV pilins, we used FlaFind 1.2 with an extended signal sequence motif to additionally allow for aspartate or glutamate at position −2 of the PibD motif to analyze the PilA homologs and also determined whether the pilA genes were colocalized with putative pilin and/or pilus biosynthesis genes. Of the 59 Duf1628 proteins that Pfam identified, 37 are also predicted by FlaFind 1.2. Moreover, 31 of the 59 Duf1628 proteins and 21 of the 37 Duf1628 and FlaFind 1.2 positives colocalize with other predicted pilin or pilus biosynthesis genes. These data strongly suggest that these proteins are indeed pilins and that our studies of the PilA pilins will lead to a better understanding of type IV pili of other archaea (Table 2; see also Table S2 in the supplemental material).
Finally, while PilA1 and PilA3, the two confirmed pilins with a negative charge observed at the −2 position of the PibD motif, are PilA paralogs, FlaFind 1.2 analysis revealed that PibD signal sequence motifs with D or E at the −2 position are not limited to the subset of pilins containing Duf1628. In the 28 archaeal genomes, we identified 90 pilin-like proteins containing negatively charged amino acid residues at the −2 position, only 12 of which were determined to contain Duf1628 by Pfam. It also appears that this motif can be found in phyla of all archaeal kingdoms, but interestingly, there are drastic differences between species (Table 2; see also Table S3 in the supplemental material).
DISCUSSION
In silico analyses have predicted that, similar to bacteria, archaeal genomes contain a diverse set of genes that encode pilin-like proteins. However, the functions of most of these putative archaeal pilus subunits remain unknown (30). While type IV pilins generally share little sequence homology, based on highly conserved domains or motifs, clusters of pilins have been identified across a diverse range of archaeal phyla. The clusters include two subsets of proteins, each containing a unique, highly conserved domain identified by Pfam: PF04021 and Duf1628. Both domains begin at the predicted prepilin peptidase processing motif. Previously, we showed that PF04021, which is found mainly in euryarchaeal proteins, is a class III signal peptide containing a unique prepilin peptidase processing motif that is specifically recognized by EppA, a PibD paralog found only in species that encode proteins having this domain (30). We have now demonstrated that the second domain, Duf1628, which unlike PF04021 extends significantly past its signal peptide, is a PilA adhesion pilin. These pilins, which require PibD for processing, contain a highly conserved H domain. While signal peptide recognition requires merely a stretch of hydrophobic amino acids, interactions with this pilin domain also provide the means through which the alpha-helical core of the pilus forms. Therefore, the highly conserved nature of this domain in PilA pilins indicates that these pilins are likely involved in the biosynthesis of a unique type IV pilus structure. Indeed, a mutant in which this H domain is replaced with that of the major flagellin, which is also an archaeal type IV pilus-like structure (37), is insufficient for the synthesis of a pilus, even one that is nonfunctional. We note that the conservation of this region continues beyond the H domain in many of these proteins, albeit to a much lesser degree. Hence, interactions between these pilins may also occur in regions other than the H domains, and these interactions may depend on the specific alpha-helical core structure.
While the crenarchaea seem limited to just one PilA paralog, many euryarchaeal genomes encode numerous PilA paralogs. For example, Pfam analysis identified 19 Duf1628-containing proteins in Methanospirillum hungatei, eight of which were FlaFind 1.2 positive and an additional four of which contain a serine at −2. A serine residue, which is allowed at the −2 position of bacterial pilins, is also likely to be allowed in haloarchaea. However, a shift in the apparent size of the PilA2 (−2S) pilin was not observed in a pibD deletion mutant (37), possibly due to differential posttranslational modifications of the mature pilins and the pilin precursors. Interestingly, there appears to be variability in the prepilin peptidase recognition site sequence between phyla. For example, a negative charge at the −2 position of the Sulfolobales PibD motif appears to be excluded, while aspartate and glutamate are readily identified at this position in other phyla.
Undoubtedly, the identification of adhesion pilins, as well as those identified in Sulfolobales and methanogens, underscores the value of prediction programs. This is particularly evident considering that neither the gene encoding the major Methanococcus maripaludis adhesion pilin (31) nor the pilA genes are coregulated with pilB and pilC, which encode highly conserved components of the pilus biosynthesis machinery (18, 19). Pilin genes had previously been identified through proximity to these two genes, since pilins typically lack a high degree of similarity. However, these studies clearly show the value of combining in silico predictions with in vivo confirmations and also demonstrate the need to apply these tools to more than one or two model systems.
The N terminus of other proteins, such as Hvo_0875, that have an H domain that displays significant homology to the H domain of PilA also seems to lack a PibD processing motif (see Fig. S1 in the supplemental material). While these proteins likely are not processed by PibD, the conserved H domain may play an important role in pilus biosynthesis or regulation.
Our most striking observation was that six of the nine PilA paralogs contained identical 30-amino-acid hydrophobic domains starting at the −1 position of the PibD site recognition motif. While it is conceivable that these proteins include major and minor pilins of pili having cores that require conserved H domain sequences to facilitate proper pilus assembly, it is intriguing that expression of any one of these PilA paralogs in trans in a ΔpilA[1-6] strain leads to the assembly of a functional pilus. Yet these pilins do not appear to have completely redundant functions, as the ability of the ΔpilA[1-6] strain to adhere varies significantly depending on which pilin is expressed. Interesting insights may be gleaned using these PilA pilins in adhesion studies with surfaces encountered in more-natural environments. Moreover, while each pilin can rescue the adhesion defect of the ΔpilA[1-6] strain, the ΔpilA[1-6] strain can establish microcolonies only when expressing the highly conserved pilins PilA5 and PilA6. The expression of these pilins, including PilA5 and PilA6, in the wild type does not result in microcolony formation, suggesting that some pilins may inhibit the aggregation necessary to form microcolonies. The requirement of a specific pilin to promote microcolony formation has not been previously reported in archaea or bacteria. In Gram-negative bacteria, this cell aggregation requires PilT-dependent pilus retraction, allowing cells to move along a surface to sites where new microcolonies can be formed and to move within those microcolonies (20, 21). Thus far, this twitching motility has not been observed in archaea, which lack an ATPase having significant homology to a PilT. However, it has been hypothesized that a PilB paralog may facilitate the extension and retraction functions (45). Microcolony formation of the ΔpilA[1-6] strain complemented with PilA5 expressed in trans provides a simple system that can be used to determine which factors promote and which inhibit this step in biofilm formation.
Finally, although the results presented here strongly suggest that each pilin can be assembled into a pilus, we cannot exclude the possibility that other pilins are contained in these pili. Future mass spectrometric analyses, combined with studies of deletion mutants for additional PilA1 homologs, should aid in deciphering between these possibilities.
Supplementary Material
ACKNOWLEDGMENTS
M.P., R.X., and R.E. were supported by National Aeronautics and Space Administration grant NNX10AR84G. R.E. was also supported by the National Institutes of Health Cell and Molecular Biology Training grant TM32 GM-07229.
We thank Tanya Svitkina and Dewight Williams for advice on microscopy and Dieter Schifferli, Jay Zhu, and the Pohlschroder lab for helpful discussions. We also thank Zhongqiang Chen for updating the SignalFind webpage and Chuck Daniels for providing the microarray data.
Footnotes
Published ahead of print 21 June 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JB.00572-13.
REFERENCES
- 1.Tyson GW, Chapman J, Hugenholtz P, Allen EE, Ram RJ, Richardson PM, Solovyev VV, Rubin EM, Rokhsar DS, Banfield JF. 2004. Community structure and metabolism through reconstruction of microbial genomes from the environment. Nature 428:37–43 [DOI] [PubMed] [Google Scholar]
- 2.Matz C, McDougald D, Moreno AM, Yung PY, Yildiz FH, Kjelleberg S. 2005. Biofilm formation and phenotypic variation enhance predation-driven persistence of Vibrio cholerae. Proc. Natl. Acad. Sci. U. S. A. 102:16819–16824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hansen SK, Rainey PB, Haagensen JA, Molin S. 2007. Evolution of species interactions in a biofilm community. Nature 445:533–536 [DOI] [PubMed] [Google Scholar]
- 4.Monds RD, O'Toole GA. 2009. The developmental model of microbial biofilms: ten years of a paradigm up for review. Trends Microbiol. 17:73–87 [DOI] [PubMed] [Google Scholar]
- 5.O'Toole GA, Kolter R. 1998. Flagellar and twitching motility are necessary for Pseudomonas aeruginosa biofilm development. Mol. Microbiol. 30:295–304 [DOI] [PubMed] [Google Scholar]
- 6.Higashi DL, Lee SW, Snyder A, Weyand NJ, Bakke A, So M. 2007. Dynamics of Neisseria gonorrhoeae attachment: microcolony development, cortical plaque formation, and cytoprotection. Infect. Immun. 75:4743–4753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jurcisek JA, Bookwalter JE, Baker BD, Fernandez S, Novotny LA, Munson RS, Bakaletz LO. 2007. The PilA protein of non-typeable Haemophilus influenzae plays a role in biofilm formation, adherence to epithelial cells and colonization of the mammalian upper respiratory tract. Mol. Microbiol. 65:1288–1299 [DOI] [PubMed] [Google Scholar]
- 8.Varga JJ, Therit B, Melville SB. 2008. Type IV pili and the CcpA protein are needed for maximal biofilm formation by the gram-positive anaerobic pathogen Clostridium perfringens. Infect. Immun. 76:4944–4951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Burrows LL. 2012. Pseudomonas aeruginosa twitching motility: type IV pili in action. Annu. Rev. Microbiol. 66:493–520 [DOI] [PubMed] [Google Scholar]
- 10.Arts J, van Boxtel R, Filloux A, Tommassen J, Koster M. 2007. Export of the pseudopilin XcpT of the Pseudomonas aeruginosa type II secretion system via the signal recognition particle-Sec pathway. J. Bacteriol. 189:2069–2076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Francetic O, Buddelmeijer N, Lewenza S, Kumamoto CA, Pugsley AP. 2007. Signal recognition particle-dependent inner membrane targeting of the PulG pseudopilin component of a type II secretion system. J. Bacteriol. 189:1783–1793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Strom MS, Nunn DN, Lory S. 1993. A single bifunctional enzyme, PilD, catalyzes cleavage and N-methylation of proteins belonging to the type IV pilin family. Proc. Natl. Acad. Sci. U. S. A. 90:2404–2408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Albers SV, Szabo Z, Driessen AJ. 2003. Archaeal homolog of bacterial type IV prepilin signal peptidases with broad substrate specificity. J. Bacteriol. 185:3918–3925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ng SY, VanDyke DJ, Chaban B, Wu J, Nosaka Y, Aizawa S, Jarrell KF. 2009. Different minimal signal peptide lengths recognized by the archaeal prepilin-like peptidases FlaK and PibD. J. Bacteriol. 191:6732–6740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Craig L, Volkmann N, Arvai AS, Pique ME, Yeager M, Egelman EH, Tainer JA. 2006. Type IV pilus structure by cryo-electron microscopy and crystallography: implications for pilus assembly and functions. Mol. Cell 23:651–662 [DOI] [PubMed] [Google Scholar]
- 16.Hansen JK, Forest KT. 2006. Type IV pilin structures: insights on shared architecture, fiber assembly, receptor binding and type II secretion. J. Mol. Microbiol. Biotechnol. 11:192–207 [DOI] [PubMed] [Google Scholar]
- 17.Albers SV, Pohlschroder M. 2009. Diversity of archaeal type IV pilin-like structures. Extremophiles 13:403–410 [DOI] [PubMed] [Google Scholar]
- 18.Craig L, Li J. 2008. Type IV pili: paradoxes in form and function. Curr. Opin. Struct. Biol. 18:267–277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pelicic V. 2008. Type IV pili: e pluribus unum? Mol. Microbiol. 68:827–837 [DOI] [PubMed] [Google Scholar]
- 20.Merz AJ, So M, Sheetz MP. 2000. Pilus retraction powers bacterial twitching motility. Nature 407:98–102 [DOI] [PubMed] [Google Scholar]
- 21.Burrows LL. 2005. Weapons of mass retraction. Mol. Microbiol. 57:878–888 [DOI] [PubMed] [Google Scholar]
- 22.Giltner CL, Nguyen Y, Burrows LL. 2012. Type IV pilin proteins: versatile molecular modules. Microbiol. Mol. Biol. Rev. 76:740–772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Helaine S, Dyer DH, Nassif X, Pelicic V, Forest KT. 2007. 3D structure/function analysis of PilX reveals how minor pilins can modulate the virulence properties of type IV pili. Proc. Natl. Acad. Sci. U. S. A. 104:15888–15893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cisneros DA, Pehau-Arnaudet G, Francetic O. 2012. Heterologous assembly of type IV pili by a type II secretion system reveals the role of minor pilins in assembly initiation. Mol. Microbiol. 86:805–818 [DOI] [PubMed] [Google Scholar]
- 25.Kuchma SL, Griffin EF, O'Toole GA. 2012. Minor pilins of the type IV pilus system participate in the negative regulation of swarming motility. J. Bacteriol. 194:5388–5403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Watnick PI, Fullner KJ, Kolter R. 1999. A role for the mannose-sensitive hemagglutinin in biofilm formation by Vibrio cholerae El Tor. J. Bacteriol. 181:3606–3609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Krebs SJ, Taylor RK. 2011. Protection and attachment of Vibrio cholerae mediated by the toxin-coregulated pilus in the infant mouse model. J. Bacteriol. 193:5260–5270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jarrell KF, McBride MJ. 2008. The surprisingly diverse ways that prokaryotes move. Nat. Rev. Microbiol. 6:466–476 [DOI] [PubMed] [Google Scholar]
- 29.Ghosh A, Albers SV. 2011. Assembly and function of the archaeal flagellum. Biochem. Soc. Trans. 39:64–69 [DOI] [PubMed] [Google Scholar]
- 30.Szabó Z, Stahl AO, Albers SV, Kissinger JC, Driessen AJ, Pohlschröder M. 2007. Identification of diverse archaeal proteins with class III signal peptides cleaved by distinct archaeal prepilin peptidases. J. Bacteriol. 189:772–778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jarrell KF, Stark M, Nair DB, Chong JP. 2011. Flagella and pili are both necessary for efficient attachment of Methanococcus maripaludis to surfaces. FEMS Microbiol. Lett. 319:44–50 [DOI] [PubMed] [Google Scholar]
- 32.Fröls S, Ajon M, Wagner M, Teichmann D, Zolghadr B, Folea M, Boekema EJ, Driessen AJ, Schleper C, Albers SV. 2008. UV-inducible cellular aggregation of the hyperthermophilic archaeon Sulfolobus solfataricus is mediated by pili formation. Mol. Microbiol. 70:938–952 [DOI] [PubMed] [Google Scholar]
- 33.Henche AL, Koerdt A, Ghosh A, Albers SV. 2012. Influence of cell surface structures on crenarchaeal biofilm formation using a thermostable green fluorescent protein. Environ. Microbiol. 14:779–793 [DOI] [PubMed] [Google Scholar]
- 34.Zolghadr B, Klingl A, Koerdt A, Driessen AJ, Rachel R, Albers SV. 2010. Appendage-mediated surface adherence of Sulfolobus solfataricus. J. Bacteriol. 192:104–110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Henche AL, Ghosh A, Yu X, Jeske T, Egelman E, Albers SV. 2012. Structure and function of the adhesive type IV pilus of Sulfolobus acidocaldarius. Environ. Microbiol. 14:3188–3202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fröls S, Dyall-Smith M, Pfeifer F. 2012. Biofilm formation by haloarchaea. Environ. Microbiol. 14:3159–3174 [DOI] [PubMed] [Google Scholar]
- 37.Tripepi M, Imam S, Pohlschroder M. 2010. Haloferax volcanii flagella are required for motility but are not involved in PibD-dependent surface adhesion. J. Bacteriol. 192:3093–3102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dyall-Smith M. 2004. The halohandbook—protocols for haloarchaeal genetics. University of Melbourne, Victoria, Australia: http://www.haloarchaea.com/resources/halohandbook/index.html [Google Scholar]
- 39.Allers T, Ngo HP, Mevarech M, Lloyd RG. 2004. Development of additional selectable markers for the halophilic archaeon Haloferax volcanii based on the leuB and trpA genes. Appl. Environ. Microbiol. 70:943–953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Blattner FR, Williams BG, Blechl AE, Denniston-Thompson K, Faber HE, Furlong L, Grunwald DJ, Kiefer DO, Moore DD, Schumm JW, Sheldon EL, Smithies O. 1977. Charon phages: safer derivatives of bacteriophage lambda for DNA cloning. Science 196:161–169 [DOI] [PubMed] [Google Scholar]
- 41.Allers T, Ngo HP. 2003. Genetic analysis of homologous recombination in Archaea: Haloferax volcanii as a model organism. Biochem. Soc. Trans. 31:706–710 [DOI] [PubMed] [Google Scholar]
- 42.O'Toole GA, Pratt LA, Watnick PI, Newman DK, Weaver VB, Kolter R. 1999. Genetic approaches to study of biofilms. Methods Enzymol. 310:91–109 [DOI] [PubMed] [Google Scholar]
- 43.Tripepi M, You J, Temel S, Onder O, Brisson D, Pohlschroder M. 2012. N-glycosylation of Haloferax volcanii flagellins requires known Agl proteins and is essential for biosynthesis of stable flagella. J. Bacteriol. 194:4876–4887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Imam S, Chen Z, Roos DS, Pohlschröder M. 2011. Identification of surprisingly diverse type IV pili, across a broad range of gram-positive bacteria. PLoS One 6:e28919. 10.1371/journal.pone.0028919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Vignon G, Köhler R, Larquet E, Giroux S, Prévost MC, Roux P, Pugsley AP. 2003. Type IV-like pili formed by the type II secreton: specificity, composition, bundling, polar localization, and surface presentation of peptides. J. Bacteriol. 185:3416–3428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sievers F, Wilm A, Dineen DG, Gibson TJ, Karplus K, Li W, Lopez R, McWilliam H, Remmert M, Söding J, Thompson JD, Higgins DG. 2011. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol. Syst. Biol. 7:539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Blyn LB, Braaten BA, Low DA. 1990. Regulation of pap pilin phase variation by a mechanism involving differential dam methylation states. EMBO J. 9:4045–4054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Allers T, Barak S, Liddell S, Wardell K, Mevarech M. 2010. Improved strains and plasmid vectors for conditional overexpression of His-tagged proteins in Haloferax volcanii. Appl. Environ. Microbiol. 76:1759–1769 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.