Abstract
Microcin E492, a channel-forming bacteriocin with the ability to form amyloid fibers, is exported as a mixture of two forms: unmodified (inactive) and posttranslationally modified at the C terminus with a salmochelin-like molecule, which is an essential modification for conferring antibacterial activity. During the stationary phase, the unmodified form accumulates because expression of the maturation genes mceIJ is turned off, and microcin E492 is rapidly inactivated. The aim of this work was to demonstrate that the increase in the proportion of unmodified microcin E492 augments the ability of this bacteriocin to form amyloid fibers, which in turn decreases antibacterial activity. To this end, strains with altered proportions of the two forms were constructed. The increase in the expression of the maturation genes augmented the antibacterial activity during all growth phases and delayed the loss of activity in the stationary phase, while the ability to form amyloid fibers was markedly reduced. Conversely, a higher expression of microcin E492 protein produced concomitant decreases in the levels of the modified form and in antibacterial activity and a substantial increase in the ability to form amyloid fibers. The same morphology for these fibers, including those formed by only the unmodified version, was observed. Moreover, seeds formed using exclusively the nonmodified form were remarkably more efficient in amyloid formation with a shorter lag phase, indicating that the nucleation process is probably improved. Unmodified microcin E492 incorporation into amyloid fibers was kinetically more efficient than the modified form, probably due to the existence of a conformation that favors this process.
INTRODUCTION
Microcin E492 (MccE492) belongs to a family of low-molecular-weight channel-forming bacteriocins produced by Klebsiella pneumoniae RYC492 (1, 2). The group of genes necessary for the production of active MccE492 has been cloned and expressed in Escherichia coli (3, 4). This bacteriocin is found in two forms, unmodified (7,887 Da) and posttranslationally modified by the covalent linkage of salmochelin-like molecules of different molecular masses (5). Glycosylation of enterochelin [the cyclic trimer of N-(2,3-dihydroxybenzoyl)-l-serine (DHBS)] and its derivatives give rise to molecules collectively known as salmochelin-like molecules (6, 7). Thus, MccE492 can be modified with a linear trimer of DHBS linked to the serine-84 carboxylate through a β-d-glucose moiety (8,718 Da) and also with derivatives involving a DHBS dimer (8,495 Da) and monomer (8,272 Da) (5). MccE492 modified with the diglycosylated DHBS trimer is also found (8,880 Da). Posttranslational modification is carried out by the maturation gene products MceC and MceIJ encoded in the MccE492 gene cluster (4). MceC glycosylates enterochelin and its derivatives generating salmochelin-like molecules, while the covalent linkage of the salmochelin-like molecules to the C-terminus of MccE492 is accomplished by MceIJ (8). MceC, MceI, and MceJ mutants and mutant enzymes in the enterochelin synthesis pathway produce exclusively unmodified MccE492 with no detectable antibacterial activity (9). The C-terminal modification is the moiety recognized by the MccE492 outer membrane receptors FepA, Fiu, and Cir (10).
Another salient feature of MccE492 is its ability to form amyloid fibrils (11, 12). Besides MccE492, the few well-characterized cases of in vitro amyloid formation by bacterial cells include those formed by curli and chaplins (13, 14). In amyloid fibrils formed by curli and chaplins, the amyloid structure plays a physiological role, such as biofilm formation and lowering the water surface tension to allow aerial growth, respectively. However, amyloid formation in bacteria seems to be a widespread feature throughout many bacterial phyla that has only recently started to be unveiled. Thus, Larsen et al. (15) identified isolates producing extracellular amyloid adhesins that belonged to several phyla: Proteobacteria (Alphaproteobacteria, Betaproteobacteria, Gammaproteobacteria, and Deltaproteobacteria), Bacteriodetes, Chloroflexi, and Actinobacteria.
Currently, there is no assigned function for amyloid fibers formed by MccE492, although they play a role in modulating antibacterial activity. Amyloid formation in vitro as well as in vivo is associated with the loss of MccE492 antibacterial activity (11), and some pieces of evidence indicate that MccE492 oligomers with antibacterial activity can be recovered from mature fibers, suggesting that such macromolecular structures can act as a toxin reservoir (16). Although MccE492 aggregation kinetics and fiber structure were previously characterized (11, 12), nothing is known about how the posttranslational modification influences amyloid formation. In this regard, previous work showed that amyloid fibrils are produced in vivo mainly during the stationary phase (11) and that the mceIJ maturation genes are not transcribed at that stage (17). Considering that these two facts may be related, it is plausible that posttranslational modification of MccE492 could somehow delay or interfere with the amyloid formation process. Accordingly, unmodified MccE492 accumulation during stationary phase could promote aggregation of the modified and unmodified forms, accounting for the rapid inactivation of active MccE492 synthesized in the exponential phase of growth.
To test the hypothesis mentioned above, experiments in vivo as well as in vitro to alter the proportions of both forms were performed. To this end, we used the following plasmids. pJAM434, a wild-type plasmid derivative that produces a moderately low activity, has an internal 7-kb XhoI fragment (in which the mceJIHG gene cluster is located) that is inverted respect to its orientation in K. pneumoniae RYC492 chromosome (18). This arrangement truncated orfX (19), an open reading frame that appears to be a positive regulator of mceJI, situated upstream of mceJ (S. Gutiérrez, P. Lobos, Y. Argandoña, R. Lagos, unpublished results). The lack of the original promoter of the gene cluster in which the maturation and export genes are located, plus the interruption of orfX lowers the production of active MccE492. In contrast, pJEM15, which is also a wild-type plasmid derivative, has duplicated an internal 2.5-kb SalI fragment encoding mceF and orfK, and for unknown reasons produces highly active MccE492. These two plasmids were used to generate poorly producing and overproducing strains. To increase the gene dosage of either the maturation genes or the MccE492 structural gene, the poorly producing (pJAM434) and the overproducing (pJEM15) strains were transformed with compatible plasmids carrying the mceA, mceIJ, or mceC gene. In addition, two strains that produce only unmodified MccE492 carrying the following plasmids were used: npB4, mutated in mceC (9), and np45, mutated in mceI.
Thus, the incremental expression of the maturation genes led to the production of higher proportions of modified MccE492 with a higher specific activity, and the inactivation observed in stationary phase was substantially delayed. Under this condition, the MccE492 produced had a reduced ability to form amyloid fibers in vitro. The opposite effect was observed when augmenting the expression of the structural gene of MccE492: the cultures exhibited lower antimicrobial activity that dramatically decreased in the stationary phase, and MccE492 isolated in this stage had a strong tendency to form amyloid fibers. Despite such differences, electron microscopy observations indicated that mature fibers formed by MccE492 isolated from all the different strains were indistinguishable. The in vitro kinetics experimental results showed that unmodified MccE492 seeds were more effective in accelerating fiber formation than seeds prepared from modified MccE492, suggesting that posttranslational modification affects fiber nucleation. Additionally, mass spectrometry analysis revealed that unmodified MccE492 is kinetically more efficient for the incorporation into fibers than the modified form, although both are finally incorporated. Both forms have a distinct secondary structure pattern, suggesting that such different conformation can account for the distinct propensity to amyloid aggregation.
MATERIALS AND METHODS
Bacterial strains, plasmids, and plasmid construction.
The plasmids and bacterial strains used in this work are shown in Table 1. pACYC184 was the vector used to make the constructions compatible with pHC79, the plasmid in which the MccE492 system is cloned (pJAM434 and pJEM15) (3). The mceAB, mceC, and mceIJ genes cloned in pACYC184 under the control of the T7 promoter were cloned following the same strategy used previously for the cloning of iroB in this vector (9). Basically, these genes were amplified with Pfu DNA polymerase using as the templates the pT7-7 derivatives that included these genes. The amplification included the T7 promoter, and the T7pro primer (9) was used as the forward primer in all cases. The reverse primer for mceAB was ABcarboxi (5′ATTAGGATCCTTTAACTACCCACTACCGGAACTGGA3′). For mceC, the reverse primer was C2 (4), and for mceIJ, it was I2 (4). The resulting DNAs were cloned into the EcoRV site of pACYC184, and the recombinants were selected by resistance to chloramphenicol and sensitivity to tetracycline. All the constructions were fully sequenced. Standard techniques for cloning, electroporation, restriction analysis, and plasmid DNA preparation were performed as described previously (20, 21, 22). The pT7-7 promoter (23) corresponds to the first generation of T7 RNA polymerase promoters that does not exhibit high stringency as the pET series does (24). The basal activity is around 10 to 20% of the full induced activity. We considered that this level of expression was enough to keep the cells healthy with an increased level of the maturation proteins, probably in the soluble form, based on the fact that MceC and MceIJ cloned under a pET promoter have been purified from the soluble fraction with a yield of 4 to 5 mg per liter of culture (8).
Table 1.
Bacterial strains and plasmids used in this work
| Bacterial strain or plasmid | Relevant genotype or phenotype or use | Source or reference |
|---|---|---|
| E. coli strains | ||
| VCS257 | DP50 supF supE44 supF58 hsd53(rB mB) dapD8 lacY1 glnV44 (gal-uvrB)47 tyrT58 gyrA29 tonA53(thyA57) | Stratagene |
| VCS257(DE3) | Lysogenization with λDE3 Novagen kit | This work |
| BL21(DE3) | F− ompT (rB mB) | Novagen |
| Plasmids | ||
| pJAM434 | Moderately low production of MccE492; AMPr | 3 |
| pJEM15 | Overproduces MccE492; AMPr | 3 |
| npB4 | pJEM15mceC::Tn5; insertion in codon 60; AMPr KANr | 9 |
| np45 | pJEM15mceI::Tn5; insertion in codon 133; AMPr KANr | This work |
| pT7-mceAB | mceAB cloned in pACYC184 under the control of the T7 promoter; CHLr | This work |
| pT7-mceC | mceC cloned in pACYC184 under the control of the T7 promoter; CHLr | This work |
| pT7-mceIJ | mceIJ cloned in pACYC184 under the control of the T7 promoter; CHLr | This work |
Growth conditions and MccE492 activity assay on plates.
Bacterial growth was performed as previously described (3, 4, 25) with shaking (220 rpm) at 37°C. The overnight culture was diluted 10,000 to 100,000 in M9 minimal medium supplemented with citrate and glucose (26) with the following antibiotics and concentrations: ampicillin (AMP), 100 μg/ml; kanamycin (KAN), 50 μg/ml; tetracycline (TET), 50 μg/ml; and chloramphenicol (CHL), 50 μg/ml.
MccE492 activity in plates was determined by mixing an aliquot of 0.2 ml of 2 × 107 cells/ml grown in LB (with the corresponding antibiotic) of the sensitive E. coli strain with 3 ml soft agar and overlaying the resulting mixture onto LB plates. MccE492 activity (either purified or from culture supernatants) was determined by the critical dilution method (27) by serially diluting the MccE492 preparations in 50% sterile nanopure water and seeding 3 μl onto these plates. MccE492 antibacterial activity was detected by the formation of growth inhibition halos and expressed in arbitrary units. A lawn of the sensitive E. coli BL21(DE3) was routinely used as the indicator strain. The culture supernatant for activity determination was obtained as follows: a 1-ml aliquot culture was centrifuged at 15,000 × g for 10 min, and 0.8 ml of the supernatant was treated with 30 μl chloroform, vortexed for 10 s, and centrifuged at 15,000 × g for 10 min. The top layer was serially diluted as mentioned above.
MccE492 purification.
For matrix-assisted laser desorption ionization–time of flight mass spectrometry (MALDI-TOF MS) determinations and immunoblotting, MccE492 was extracted from 500 ml of the supernatant from cultures of E. coli VCS257(DE3) carrying the corresponding plasmid using a procedure similar to that described previously (5). The cells were diluted 10,000 to 100,000 and grown at 37°C in M9 minimal medium supplemented with citrate and glucose (26) with 220 rpm shaking for 20 h, and the supernatant was collected by centrifugation at 17,000 × g for 30 min. The supernatant containing MccE492 was loaded onto a Sep-Pak C8 cartridge (Waters), which had been previously equilibrated with 5 ml of 0.1% trifluoroacetic acid (TFA) in nanopure water. The cartridge was washed with 5 ml of 30% acetonitrile (ACN)–0.1% TFA, and MccE492 was eluted with 5 ml of 40% ACN–0.1% TFA. MccE492 used for amyloid formation was purified as described by Bieler et al. (11) using E. coli BL21(DE3) or VCS257 as the host system. For circular dichroism and soluble modified MccE492 fraction analysis, an optimized large-scale purification procedure was used. Four liters of cell-free medium from a late-exponential (optical density at 600 nm [OD600] of 0.6 to 0.8) MccE492-producing bacterial culture was incubated (4°C) with 50 g of previously ACN-activated Bondapak C18 resin (Waters). After 2 h, the resin was filtered through a Buchner funnel, washed with 100 ml of 40% methanol, 100 ml of 25% ACN, and then eluted with a 30% to 100% ACN stepwise gradient. MccE492-enriched fractions (usually 50% and 60%) were dialyzed for 4 h against 40 volumes of 5 mM Tris (pH 8.5) and then were lyophilized and stored at −20°C. For circular dichroism analysis, lyophilized MccE492 was suspended in 50 mM phosphate buffer (pH 6.5)–100 mM NaCl and then was further purified by size exclusion chromatography in a Sephacryl S-100 (GE Healthcare) column (4°C), eluting with the same phosphate buffer. Fractions containing MccE492 were pooled, lyophilized, and suspended in nanopure H2O. Further desalting was carried out by gel filtration in a Biogel P4 column, previously equilibrated with 5 mM phosphate buffer, pH 6.5.
PAGE and immunoblotting.
PAGE was carried out under the conditions described by Schägger and von Jagow (28), and nitrocellulose membranes (Millipore) were used in immunoblot transfer (2 h, 100 V at −20°C, using chilled 25 mM Tris-HCl, 190 mM glycine, and 20% methanol as the transfer buffer). MccE492 was detected with a polyclonal antibody prepared in rabbits as described previously (9) (antiserum dilution 1:500) and horseradish peroxidase-linked anti-rabbit goat antibody (Pierce) (dilution 1:20,000). The chemiluminescence reaction was performed in 100 mM Tris-HCl (pH 8.5), 1.25 mM luminol, and 0.2 mM p-coumaric acid. The reaction was started by the addition of an aliquot of 30% H2O2 (final concentration of 0.01%). The membrane was exposed to X-OMAT AR film (Kodak) for 2 to 5 min depending on the signal obtained. Alternatively, MccE492 was detected using a goat anti-rabbit alkaline phosphatase-conjugated secondary antibody, washing the membrane in FAL buffer (100 mM Tris-HCl [pH 9.5], 100 mM NaCl, 5 mM MgCl2), and then soaking it in a mixture of BCIP (5-bromo-4-chloro-3′-indolylphosphate p-toluidine salt) and NBT (nitroblue tetrazolium chloride) (0.3 and 0.15 mg/ml in FAL buffer, respectively) until enough color appeared.
MALDI-TOF MS.
Samples were analyzed in a MALDI-TOF mass spectrometer (Microflex MALDI-TOF mass spectrometer; Bruker Daltonics, Inc.) with α-cyano-4-hydroxycinnamic acid as the matrix (10 mg/ml of α-cyano-4-hydroxycinnamic acid in a mixture of ACN and 0.1% formic acid [1:1, vol/vol]). Data acquisition was performed in positive polarity and reflection mode, and the final spectra corresponded to 10 scans of 40 laser shots in different points selected randomly. Calibration was carried out with an external standard using a mix of peptides (5,000 to 20,000 Da; Bruker Daltonics, Inc.). Data analysis and area quantification of the spectra were performed using the mMass package software tools described by Strohalm et al. (29) and ImageJ 1.41 (http://rsbweb.nih.gov/ij/download.html), respectively.
MccE492 aggregation and Congo red binding assay.
Amyloid formation was initiated with samples devoid of aggregates. To this end, the acetonitrile fraction containing MccE492 was concentrated in a CentriVap concentrator (Labconco) to 0.8 mg/ml in a volume of 0.1 ml, and 1 μl of 10 N NaOH was added, incubating the mixture for 15 min at 37°C. Alternatively, an appropriate amount of lyophilized microcin powder was suspended in 5 mM Tris (pH 8.5). Concentrated MccE492 was centrifuged for 30 min at 16,000 × g to eliminate aggregates. The supernatant was diluted to 0.4 mg/ml in the aggregation buffer (100 mM PIPES-NaOH, 0.5 M NaCl [pH 6.5]) and incubated with agitation (800 rpm) for the indicated time. Amyloid formation was quantified by the diminution of free Congo red using the following protocol. Aggregated MccE492 samples were incubated at 37°C for 15 min with 22 μM Congo red and centrifuged at 16,000 × g for 15 min at 4°C. The supernatant absorbance was registered at 490 nm, and the free Congo red fraction was determined as the ratio between the absorbance registered at each time and the absorbance at time zero. MccE492 seeds were prepared by incubating 0.8 mg/ml of MccE492 in aggregation buffer for 48 h at 37°C. Then, the fibrils produced were sonicated three times for 10 s each time at 3 W in a sonicator (Misonix 3000).
Determination of soluble modified MccE492 during the aggregation curve.
To determine the proportion of the remaining modified MccE492 at each time during the amyloid formation kinetics, MccE492 from the pJEM15/pT7-IJ system was purified and then incubated in aggregation buffer at 37°C in the presence of 10% npB4 seeds (isolated from a strain that produces only unmodified MccE492). At the times indicated in the figures, aliquots were extracted and mixed with Congo red (final concentration of 22 μM) and then centrifuged for 1 h at 20,000 × g (4°C). The supernatant was analyzed by SDS-PAGE and immunoblotting to detect MccE492 and assayed for Congo red binding, and an aliquot was diluted 10-fold with methanol and stored at −20°C. The methanol-diluted supernatants from all the samples were dialyzed against 1,000 volumes of cold methanol using a 3,000-Da molecular-weight-cutoff (MWCO) membrane (Spectrapore) during 2 h. After dialysis, samples were analyzed by MALDI-TOF MS as described previously in this section.
Electron microscopy.
Aggregate-free MccE492 samples were incubated at 37°C with agitation (300 rpm) in phosphate-buffered saline (PBS) (Roche) with the pH adjusted to 7.4 for at least 15 h or until aggregation was observed (as determined by Congo red binding). The aggregated samples were placed onto 300-square-mesh copper-rhodium grids coated with carbon and negatively stained with 2% uranyl acetate. Micrographs were taken under conditions of minimal dose on Kodak SO-163 film, in a JEOL JEM1200EXII microscope with a wolframium filament operated at 100 kV and with 20,000 to 60,000× magnification.
Circular dichroism analysis.
Spectra were collected in a Jasco-600 spectropolarimeter controlled by J-700 standard analysis software for Windows (Jasco, Tokyo, Japan) at room temperature, using cells with 1-mm path length for a protein concentration of 10 to 20 μM. Each sample and buffer were measured with five scans, using a 10-nm/min scan speed, 2-nm bandwidth, and a step resolution of 0.2 nm.
RESULTS
The augmented expression of the mceCIJ maturation genes increases antimicrobial activity and delays inactivation in the stationary phase.
In order to study the effect of augmented expression of the maturation genes on antibacterial activity at different growth phases, two MccE492 producer strains with different activity levels (overproducing [pJEM15] or poorly producing [pJAM434]) were used as hosts to receive the plasmids that express mceC and mceIJ genes. To achieve moderate expression from these plasmids, no isopropyl-β-d-thiogalactopyranoside (IPTG) was added to the culture (see Materials and Methods). Figure 1 shows that an increment in the expression of these genes resulted in an increase in the antibacterial activity present in the culture supernatant during the whole growth curve. The same qualitative behavior was observed in both host strains, although the activity levels showed important differences (Fig. 1A and B). A delay in the loss of activity in the stationary phase in both cases was also observed. To demonstrate that the increment in antibacterial activity was not due to an increase in the amount of MccE492 produced, immunoblotting of MccE492 samples was performed. Figure 1C shows that the amounts of MccE492 present in purified preparations obtained from the culture supernatants with augmented expression of the maturation genes in early stationary phase (17 h) and in late stationary phase (40 h) are roughly the same as that of the control. This behavior was observed in both the poorly producing (pJAM434) and overproducing (pJEM15) host strains. Antibacterial activity (expressed in arbitrary units) was also determined, and as expected, the preparations obtained from the strains with increased expression of the maturation genes presented a higher specific activity (Fig. 1C). The same behavior was observed in the strain with pJEM15 expressing more MceC (not shown).
Fig 1.
Antibacterial activity during the growth of different MccE492 producer strains. E. coli VCS257(DE3) carrying the MccE492 producer system (●) pJAM434 (A) or pJEM15 (B) transformed with a compatible plasmid expressing the mceC maturation gene (▼), the mceIJ maturation genes (○), or the MccE492 structural gene mceA (△) were grown in M9 minimal medium, and the antibacterial activity was determined from the culture supernatants at the indicated times, as described in Materials and Methods. Activity is expressed in arbitrary units (AU)/ml, and the scale used in panel B is logarithmic. The growth phase of the culture is indicated by the bar below the activity curve. The behavior showed in this figure is representative of several experiments repeated independently. (C) SDS-PAGE followed by immunoblotting of MccE492 purified from culture supernatants after 17 h (late exponential [LE]) or 40 h (late stationary [LS]) of growth under the conditions described above. All lanes were loaded with equivalent numbers of cells, as determined by optical density. The antibacterial activity for each preparation is indicated.
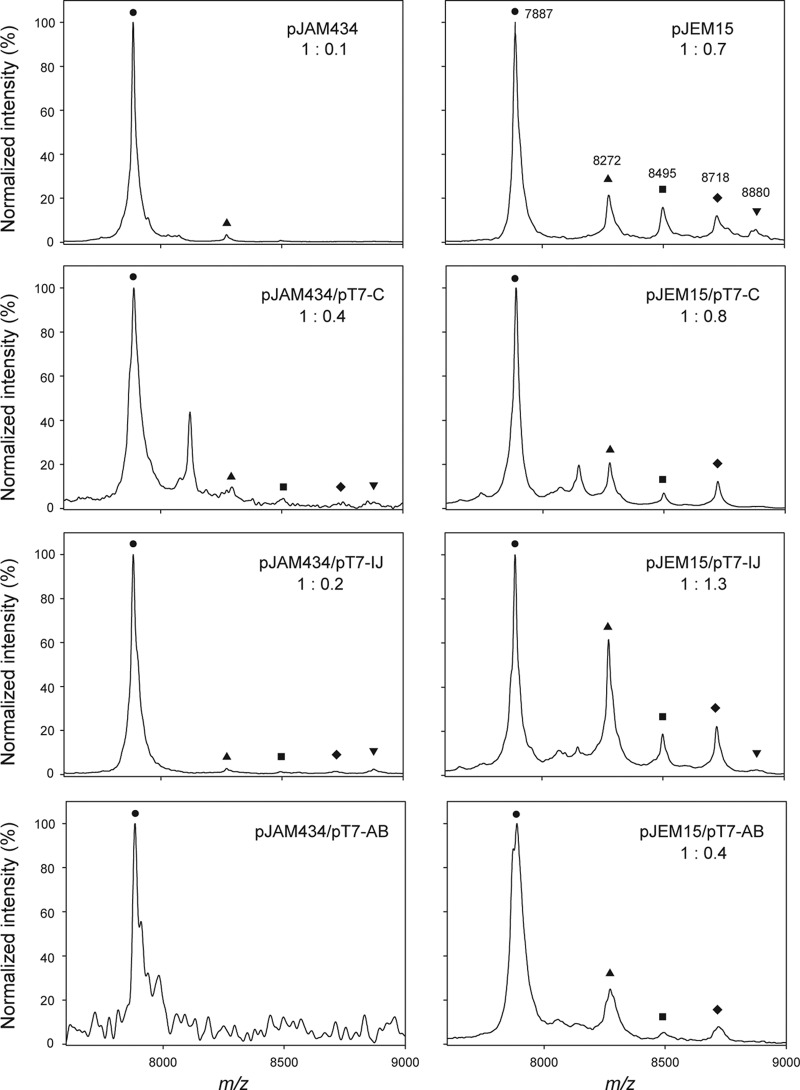
To demonstrate that the increase in antibacterial activity was related to a higher percentage of the modified form, the proportion of unmodified versus modified MccE492 isolated from the different strains was measured. MccE492 was purified after 20 h of culture growth, and the samples were analyzed by MALDI-TOF MS (Fig. 2). The MccE492 forms that were identified from a preparation by mass analysis are as follows: unmodified (7,887 Da), modified with glucose plus a DHBS monomer (8,272 Da), dimer (8,495 Da), and trimer (8,718 Da). A modification with diglucosyl enterobactin (the DHBS trimer plus two glucose moieties) was also observed (8,880 Da). The “modified form” was defined as the sum of all the forms presenting the posttranslational modifications mentioned above and was calculated as described in Materials and Methods. As shown in Fig. 2, the MccE492 produced by the strains that produce more maturation proteins express more modified MccE492. A strain with pJAM434, the poor producer strain in terms of activity, produced 10-fold-more unmodified MccE492 with respect to the modified form, while there was an increase of 2 and 4 times of the modified form when mceIJ and mceC expression was augmented, respectively. On the other hand, the untransformed strain carrying pJEM15 produced marginally more unmodified MccE492 than the modified version (1:0.7). Higher expression of mceC did not significantly change this proportion (1:0.8), but a substantial increase of the modified form was obtained when mceIJ expression was higher (1:1.3). These results correlated well with the activity curves presented in Fig. 1, and the effects were particularly pronounced when the expression of mceC in the pJAM434 strain and the expression of mceIJ in cells with pJEM15 was increased. Some strains presented an additional peak (8,110 Da) that was not identified.
Fig 2.
A higher level of expression of maturation genes increases the proportion of modified MccE492. MccE492 from culture supernatants of strains carrying the indicated plasmid(s) was purified as described in Materials and Methods and analyzed by MALDI-TOF MS. The proportion of unmodified versus modified forms was calculated using the program ImageJ version 1.42. The area under the curve of the unmodified form (7,887 Da [●]) was normalized to 1, and the proportion of the modified forms was calculated as the sum of MccE492 modified with glycosyl-DHBS (8,272 Da [▲]), glycosyl-(DHBS)2 (8,495 Da [■]), glycosyl-(DHBS)3 (8,718 Da [⧫]) and di-glycosyl-(DHBS)3 (8,880 Da [▼]).
Conversely, when the MccE492 structural gene was produced in larger amounts, there was a significant reduction in the antibacterial activity (Fig. 1), which correlated with a decrease of the modified form (Fig. 2). A strain with pJAM434 and expressing more mceAB, the structural gene for MccE492 and its immunity protein, did not produce detectable antibacterial activity, and no modified MccE492 was discernible. Consistently, a marked decrease in antibacterial activity occurred when the host carrying pJEM15 was used, which correlated with a decrease of the modified form (1:0.4 [unmodified to modified]).
The amyloid-like fibers produced by preparations of MccE492 with different proportions of the modified form have the same morphology.
In order to determine whether the modification of MccE492 has any effect on the morphology of the amyloid-like fibers, samples from different preparations aggregated in vitro until fibril formation were analyzed by negative-stain electron microscopy. For comparison purposes, MccE492 preparations isolated from strains with mutations in the mceC (npB4) and mceI (np45) maturation genes that produce only unmodified MccE492 (9) were included. Figure 3 shows that the fibers formed by preparations ranging from highly modified (pJEM15–pT7-IJ and pJEM15–pT7-C) to unmodified MccE492 (npB4 and np45) have the same morphology; mainly helical fibrils with a width between 123 and 144 Å were produced. No fibrils were detected in soluble samples that were not incubated under the aggregation conditions.
Fig 3.
Electron micrographs of negatively stained amyloids formed by MccE492 isolated from different strains. MccE492 (0.4 mg/ml) isolated from the indicated strains was incubated at 37°C in PBS (adjusted to pH 7.4) for at least 15 h or until fibril formation. The magnification used was ×60,000. Bars, 100 nm.
MccE492 preparations with a high proportion of the unmodified form have an increased capacity to form amyloid-like fibers.
In order to quantify the ability of preparations with different quantities of modified MccE492 to form amyloid-like fibers in vitro, aggregation was assessed by binding of Congo red. Congo red is a compound widely used to detect amyloid aggregation and has been previously employed to monitor amyloid formation by MccE492 (11, 12). Figure 4 indicates that most of the MccE492 isolated from the pJEM15 strain with augmented expression of the mceIJ maturation genes remained soluble after 15 h of incubation, while MccE492 from a preparation that expressed a larger amount of mceAB is mostly bound to Congo red, indicative of fiber formation. The antibacterial activity of these preparations after the incubation was proportional to the amount of soluble MccE492, i.e., not bound to Congo red. For a control for aggregation of unmodified MccE492, a preparation obtained from the npB4 strain was performed. This preparation has no antibacterial activity and presented a higher tendency to form amyloid fibrils.
Fig 4.

MccE492 preparations with a lower content of the modified form have an increased capacity to form amyloid-like fibrils. MccE492 aggregation before incubation (0 h) and after 15 h of incubation was assessed by Congo red binding. The experiment was initiated with pure soluble MccE492 (time zero); therefore, this value was normalized to 100%, representing the free Congo red present in the solution. The numbers above the bars are the antibacterial activity expressed in arbitrary units (AU)/ml. The results presented in this figure are representative of at least five independent experiments performed with different MccE492 preparations.
MccE492 amyloid formation is more efficient when seeds of unmodified MccE492 are used.
In vitro MccE492 amyloid formation follows the typical kinetics for disease-associated amyloids, with a nucleation-dependent process that presents a characteristic lag phase in which no amyloid structures can be detected, followed by fast growth and then a stationary phase (11). The lag phase could be bypassed by the addition of preformed seeds (11), in a manner similar to that observed for disease-associated amyloids. In order to test whether the seeds formed by pure unmodified MccE492 are more efficient in evading the lag phase, aggregation kinetics using seeds obtained from fibers formed by the wild type containing modified MccE492 and from a mutant in the maturation gene that contains only the unmodified form were performed using the same assay conditions described in the legend to Fig. 4. The top graph in Fig. 5 shows the aggregation kinetics of highly modified MccE492 isolated from the overproducing strain carrying the pJEM15 plasmid and the unmodified form isolated from the strain carrying the npB4 plasmid. It can be observed that the unmodified form started to aggregate after 6 h of incubation, and after 24 h, it is fully aggregated, while modified MccE492 exhibits modest aggregation after 24 h of incubation. The modified MccE492 preparation used for this set of experiments substantially aggregated after 48 h of incubation, and for this reason, this was the condition selected to produce the fibrils that were utilized to generate the seeds. The bottom graph in Fig. 5 clearly shows that there is significant acceleration in the aggregation of the modified form of MccE492 in the presence of seeds formed either by modified or unmodified MccE492. However, the acceleration induced by unmodified MccE492 seeds is significantly higher with noticeable aggregation after 2 h of incubation and full aggregation at 8 h, while its modified seed counterpart showed full aggregation after 24 h of incubation. The shortening of the lag phase observed with unmodified MccE492 strongly suggests that this form favors the nucleation process.
Fig 5.
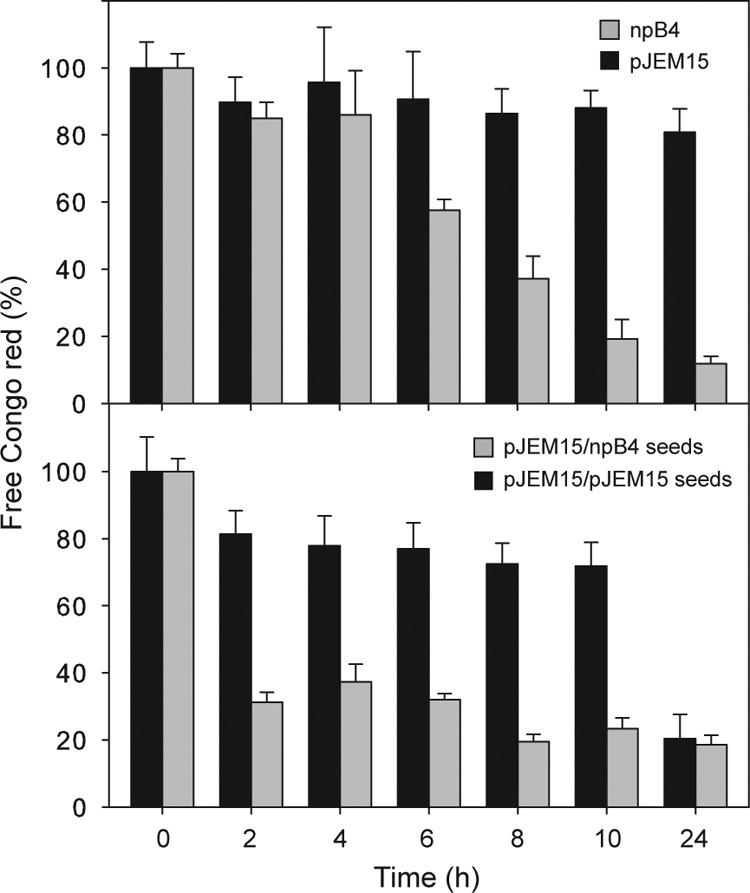
Seeds formed exclusively with unmodified MccE492 are more efficient in accelerating in vitro amyloid polymerization. MccE492 aggregation was performed as described in the legend to Fig. 4. Modified and unmodified MccE492 were purified from E. coli BL21(DE3) carrying plasmid pJEM15 and npB4, respectively. The seeding experiments shown in the bottom panel were performed using 10% seeds. For each time point, three independent samples were used. The error bars show the standard deviations.
Unmodified MccE492 is kinetically more efficient in the incorporation into amyloid fibers than the modified form.
The results presented above showed important differences in the aggregation kinetics observed with unmodified MccE492 preparations compared with those containing modified MccE492. There are different alternatives to explain this result: one possibility is that posttranslationally modified MccE492 has a reduced capacity to be incorporated into amyloid fibers. Alternatively, it is also possible that amyloid fibers are formed only by unmodified MccE492, so in a mixture of both modified and unmodified forms, only the last type would be incorporated into the fiber, while modified MccE492 would remain soluble. To test these possibilities, we designed an experiment to measure the proportion of modified MccE492 that remained soluble at distinct times during the aggregation kinetics. To this end, highly modified MccE492 was purified from the pJEM15–pT7-IJ production system and subjected to amyloid aggregation, as described in Materials and Methods. MccE492 prepared from this system contains over 55% modified form (Fig. 2) and has the longest lag phase during amyloid aggregation (Fig. 4), which can be over 96 h (not shown). To overcome the long lag phase of MccE492 purified from the pJEM15–pT7-IJ strain, MccE492 was incubated at 37°C in aggregation buffer in the presence of 10% npB4 seeds. Amyloid formation was followed measuring Congo red binding, and after 4 h of incubation, there was aggregation with 30% free Congo red (Fig. 6A). Simultaneously, at each time point, an aliquot was extracted from the reaction mixture and centrifuged at 20,000 × g for 1 h to sediment MccE492 incorporated into amyloids. The supernatant was analyzed by SDS-PAGE and immunoblotting to detect the presence of soluble MccE492 (Fig. 6B). A gradual decrease in soluble MccE492 was observed over time, concomitant with the incorporation into amyloid fibers. Importantly, after 20 h of incubation, no soluble MccE492 could be detected, indicating that nearly all the protein present at the beginning of the reaction was aggregated. From the same supernatants, an aliquot was extracted, microdialyzed, and analyzed by MALDI-TOF MS to determine the proportion of unmodified versus modified MccE492 that remained soluble at each time point. If the unmodified form were preferably incorporated into amyloid fibers, an enrichment of modified MccE492 should be observed over time, due to the unmodified MccE492 depletion as a consequence of incorporation into the fibers. Data quantification of the MALDI-TOF mass spectrometry shown in Fig. 6C indicates that the incorporation of the unmodified form is more efficient than incorporation of modified MccE492, which remains soluble over 8 h after incubation. Nevertheless, both soluble modified and unmodified MccE492 cannot be detected by immunoblotting after 20 h of incubation (Fig. 6B), and both forms were nearly undetected by MALDI-TOF MS (not shown). This indicates that, although with different kinetics efficiency, both unmodified and modified forms can be incorporated into amyloid fibers. It was not possible to perform experiments directly analyzing the composition of the fibers through MALDI-TOF MS, because different treatments used to solubilize amyloids destroyed modification of the control samples.
Fig 6.
Unmodified MccE492 is incorporated into amyloid fibers more efficiently than the modified form. Modified MccE492 purified from E. coli BL21(DE3) carrying pJEM15–pT7-IJ was aggregated in the presence of 10% npB4 seeds as described in the legend to Fig. 5. (A) The aggregation kinetics was monitored by Congo red binding. Samples were taken at 0, 4, 8, and 20 h. (B and C) The remaining soluble MccE492 was analyzed at each time point by SDS-PAGE and immunoblotting (B), and by mass spectrometry (C), as described in Materials and Methods. The behavior showed in this figure is representative of two experiments.
Modified and unmodified MccE492 have different secondary structure patterns assessed by circular dichroism.
The difference in the propensity of modified and unmodified MccE492 to incorporate into amyloid fibers could be explained by different structural conformations. In a previous work, we characterized the structure of MccE492 amyloid fibers using electron microscopy and image processing techniques and X-ray diffraction (12). Based on the observed structures, it was proposed that MccE492 has two different conformations: a native conformation that would assemble mainly into a pentameric structure, which likely functions as a pore, and an amyloid conformation, which results in the formation of different types of amyloid filaments. In this model, a transition between native and amyloidogenic conformation must occur before the fiber formation, and probably the propensity of such a conformational change depends on the structure of both native and amyloid conformers. In this context, it is possible that the salmochelin-like moiety linked at the C terminus of MccE492 pushes toward a different molecular structure that has a lower propensity to undergo the transition to the amyloid form. To gain information about this possibility, circular dichroism (CD) spectra were measured for both npB4-derived (unmodified) and pJEM15–pT7-IJ-derived (highly modified) MccE492, following the procedure described in Materials and Methods. Figure 7 shows that the CD spectra of unmodified and modified MccE492 under the conditions tested are distinct, indicative of a different secondary structure content. However, the addition of trifluoroethanol (TFE), a structure-forming solvent, induced in both forms CD spectra with characteristics of helical secondary structure, indicating that under this condition it is possible to convert these two different forms into a new conformation with similar features.
Fig 7.
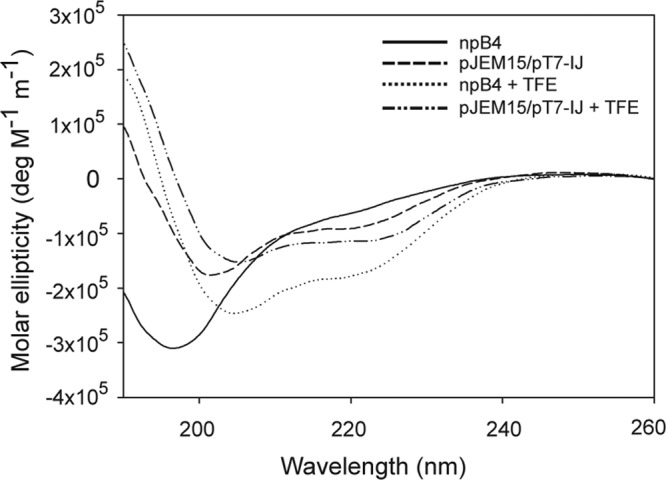
Unmodified and modified MccE492 exhibit different CD spectra. Circular dichroism spectra of MccE492 prepared from npB4 (unmodified) and pJEM15–pT7-IJ (modified) systems. Data were recorded in 5 mM phosphate buffer (pH 6.5) with or without the secondary structure inductor trifluoroethanol (TFE) (33%). The spectra were constructed from the means of at least 5 measurements.
DISCUSSION
The present work demonstrates that the degree of MccE492 modification influences specific antibacterial activity and the ability to polymerize into amyloid fibers in vitro. As predicted, a higher level of expression of the maturation genes produced an increase in MccE492 activity and in the proportion of the modified form. The observation that in the late stationary phase there is a decrease in the antibacterial activity even when the expression of the maturation genes is augmented may indicate that a factor necessary for posttranslational modification may become limiting. A possible candidate is EntF, a protein that is essential for the production of active MccE492 (9) and that is regulated by iron availability through the Fur repressor (30). The explanation that the decrease in activity may be related to iron availability is supported by the fact that inactive MccE492 is produced under iron-limiting conditions (31). The results presented in Fig. 1C also exclude the possibility of MccE492 degradation.
The experimental results presented in this work also indicate that unmodified MccE492 is more efficient in forming amyloid fibers in both their nucleation (the rate-limiting step) and elongation. Thus, when seeds prepared from unmodified MccE492 are used, the process of elongation with either a preparation containing modified or only unmodified MccE492 is substantially accelerated. The evidence presented in this work indicates that both modified and unmodified MccE492 are finally incorporated into the fibrils, albeit with a different kinetics efficiency. A direct effect of salmochelin-like molecules on aggregation was discarded, because the addition of different salmochelin concentrations did not have a significant effect on the kinetics of amyloid formation of the unmodified form (A. Marcoleta, C. Muñoz, and R. Lagos, unpublished results). The observation that modified and unmodified MccE492 seem to have different conformations would account for the difference in the kinetics efficiency of amyloid formation. Here, the transition to an amyloidogenic conformation of unmodified MccE492 would occur faster, shortening both the nucleation and elongation processes. Additionally, fibers produced in vitro using a preparation that contains modified MccE492, although formed less efficiently, are morphologically indistinguishable from those formed using exclusively the unmodified version. This strongly suggests that the amyloidogenic conformation reached by these two MccE492 forms is the same.
The amyloid fold has been seen as a useful feature in microbial amyloids, because it would lower the concentration of toxic precursors (32). Among the mechanisms to prevent toxicity would be the rapid transition to aggregation, for instance through a nucleator protein such as CsgB, the crucial role of some particular residues, or the control of amyloid formation through chaperones, all exemplified in the curli system (32). The MccE492 system provides another mechanism, in which the abundance of the unmodified form would speed up the amyloidogenic process.
Amyloid fibrils are typically rigid, straight, unbranched, with a diameter of 5 to 13 nm, are resistant to proteases, produce an increase in the fluorescence of thioflavin T, and cause a red shift in the absorbance spectrum of Congo red (33, 34). These structural features are shared by MccE492 amyloids, whose fibers are indistinguishable from those formed by proteins associated with misfolding diseases (11). Although the fibers formed by different proteins have similar structures, the functionality of these fibers can be diverse. The recent discovery that bacterial amyloid fibrils have specific functions and as such, are a productive part of the microorganism biology has led to the coining of the term “functional amyloids” (13, 34, 35, 36, 37). The best-characterized bacterial amyloid is curli, whose formation in vivo requires at least six specific proteins dedicated to curli biogenesis (38). For nucleation, the major curli protein CsgA requires the minor fiber subunit CsgB, which contains an amyloidogenic domain that directs CsgA polymerization (39). CsgA also has an amyloidogenic domain and can polymerize in vitro in the absence of CsgB (40) or can polymerize more efficiently on the surfaces of CsgB+ cells (41). The efficient nucleation in vivo however is more complex and requires other proteins (42). This system is a very useful model for studying the initial stages of amyloid fiber formation, because it is possible to molecularly dissect the nucleation and elongation phases (discussed in reference 35). As proposed, MccE492 may also be a helpful model for studying the toxic structure of an amyloid, because a conserved folding pathway with oligomeric intermediates characterizes most amyloid fibers (35).
There are other reported cases in which posttranslational modification negatively influences amyloid formation. For example, glycosylation of the prion protein PrPc has a significant effect on decelerating the rate of fibril formation (43). A similar effect is observed in the bacterial autotransporter Ag43, which has the ability to aggregate into amyloid-like structures. Glycosylation reduced the ability of Ag43 to form bacterial amyloids, indicating that this modification may play a significant role in the structural and functional properties of this autotransporter (44). Another example in which glycosylation prevents amyloid formation is chicken cystatin expressed in yeast (45). In this case, a consensus sequence for N-linked glycosylation was introduced in the amyloidogenic form of cystatin, that when expressed in Pichia pastoris was glycosylated and unable to form amyloid fibrils.
A general mechanism of aggregation to form amyloid fibers has been proposed by Lashuel and Lansbury, Jr. (46). The process is initiated by the formation of soluble oligomers in which ring-shaped structures can be found. These elements would undergo further assembly into protofilaments that in turn would form the mature fibrils, mediated by lateral interactions. In this respect, it is possible to observe ring-shaped preamyloid oligomers formed by MccE492 (12). These structures, reminiscent of pores, are probably the toxic structure of MccE492, which is associated with membrane depolarization by ion-conducting channels (2). This feature resembles the toxic structures that have been postulated for protein misfolding diseases, in which this type of oligomer would also act through a mechanism of pore formation (46). This attribute lends more value to the in vitro study of the intermediate structures of MccE492 in fiber formation as a model for the polymerization of amyloids associated with misfolding diseases. In addition, MccE492 has a cytotoxic effect on malignant cells (47), and the MccE492 fibrils are the most toxic form (discussed in reference 19). Thus, characterization of MccE492 amyloid formation and the oligomeric toxic forms and study of the amyloidogenic region will help in understanding the general processes of amyloidosis and toxicity.
ACKNOWLEDGMENTS
This work was supported by grants 1061128, 7080177, and 1100141 from the Fondo Nacional de Desarrollo Científico y Tecnológico and CSIC/Universidad de Chile 04/09-10. Mass spectrometry was done thanks to a MECESUP UCH-0115 project, in the Mass Spectrometry Unit of the University of Chile. G.M. received a predoctoral CONICYT fellowship, and her stay in J.M.V.'s lab was funded by MECESUP UCH-0604.
We are very grateful to Michael Handford for critically reading the manuscript and for the advice given by the members of J.M.V.'s lab for the preparation of the electron microscopy samples. We also thank Verónica García, Claudia Estévez, and Gonzalo Rojas for constructing plasmid pT7-AB, pT7MceIJ, and pT7-MceC, respectively, and Jonathan Arias for advice in the use of ImageJ. The technical assistance of Roselyn Orellana is also acknowledged.
Footnotes
Published ahead of print 8 July 2013
REFERENCES
- 1.de Lorenzo V. 1984. Isolation and characterization of microcin E492 from Klebsiella pneumoniae. Arch. Microbiol. 139:72–75 [DOI] [PubMed] [Google Scholar]
- 2.Lagos R, Wilkens M, Vergara C, Cecchi X, Monasterio O. 1993. Microcin E492 forms ion channels in phospholipid bilayer membranes. FEBS Lett. 321:145–148 [DOI] [PubMed] [Google Scholar]
- 3.Wilkens M, Villanueva JE, Cofré J, Chnaiderman J, Lagos R. 1997. Cloning and expression in Escherichia coli of genetic determinants for production of and immunity to microcin E492 from Klebsiella pneumoniae. J. Bacteriol. 179:4789–4794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lagos R, Baeza M, Corsini G, Hetz C, Strahsburger E, Castillo JA, Vergara C, Monasterio O. 2001. Structure, organization, and characterization of the gene cluster involved in the production of microcin E492, a channel-forming bacteriocin. Mol. Microbiol. 42:229–243 [DOI] [PubMed] [Google Scholar]
- 5.Thomas X, Destoumieux-Garzón D, Peduzzi J, Alfonso C, Blond A, Birlirakis N, Goulard C, Dubost L, Thai R, Tabet JC, Rebuffat S. 2004. Siderophore peptide, a new type of post-translationally modified antibacterial peptide with potent activity. J. Biol. Chem. 279:28233–28242 [DOI] [PubMed] [Google Scholar]
- 6.Hantke K, Nicholson G, Rabsch W, Winkelmann G. 2003. Salmochelins, siderophores of Salmonella enterica and uropathogenic Escherichia coli strains, are recognized by the outer membrane receptor IroN. Proc. Natl. Acad. Sci. U. S. A. 100:3677–3682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bister B, Bischoff D, Nicholson GJ, Valdebenito M, Schneider K, Winkelmann G, Hantke K, Süssmuth RD. 2004. The structure of salmochelins: C-glucosylated enterobactins of Salmonella enterica. BioMetals 17:471–481 [DOI] [PubMed] [Google Scholar]
- 8.Nolan EM, Fischbach MA, Koglin A, Walsh CT. 2007. Biosynthetic tailoring of microcin E492m: post-translational modification affords an antibacterial siderophore-peptide conjugate. J. Am. Chem. Soc. 129:14336–14347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mercado G, Tello M, Marín M, Monasterio O, Lagos R. 2008. The production in vivo of microcin E492 with antibacterial activity depends on salmochelin and EntF. J. Bacteriol. 190:5464–5471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Strahsburger E, Baeza M, Monasterio O, Lagos R. 2005. Cooperative uptake of microcin E492 by receptors FepA, Fiu, and Cir and inhibition by the siderophore enterochelin and its dimeric and trimeric hydrolysis products. Antimicrob. Agents Chemother. 49:3083–3086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bieler S, Estrada L, Lagos R, Baeza M, Castilla J, Soto C. 2005. Amyloid formation modulates the biological activity of a bacterial protein. J. Biol. Chem. 280:26880–26885 [DOI] [PubMed] [Google Scholar]
- 12.Arranz R, Mercado G, Martín-Benito J, Giraldo R, Monasterio O, Lagos R, Valpuesta JM. 2012. Structural characterization of microcin E492 amyloid formation: identification of the precursors. J. Struct. Biol. 178:54–60 [DOI] [PubMed] [Google Scholar]
- 13.Chapman MR, Robinson LS, Pinkner JS, Roth R, Heuser J, Hammar M, Normark S, Hultgren SJ. 2002. Role of Escherichia coli curli operons in directing amyloid fiber formation. Science 295:851–855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Claessen D, Rink R, de Jong W, Siebring J, de Vreugd P, Boersma FGH, Dijkhuizen L, Wösten HAB 2003. A novel class of secreted hydrophobic proteins is involved in aerial hyphae formation in Streptomyces coelicolor by forming amyloid-like fibrils. Genes Dev. 17:1714–1726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Larsen P, Nielsen JL, Dueholm MS, Wetzel R, Otzen D, Nielsen PH. 2007. Amyloid adhesins are abundant in natural biofilms. Environ. Microbiol. 9:3077–3090 [DOI] [PubMed] [Google Scholar]
- 16.Shahnawaz M, Soto C. 2012. Microcin amyloid fibrils are a reservoir of toxic oligomeric species. J. Biol. Chem. 287:11665–11676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Corsini G, Baeza M, Monasterio O, Lagos R. 2002. The expression of genes involved in microcin maturation regulates the production of active microcin E492. Biochimie 84:539–544 [DOI] [PubMed] [Google Scholar]
- 18.Marcoleta A, Gutiérrez-Cortéz S, Maturana D, Monasterio O, Lagos R. 2013. Whole-genome sequence of the microcin E492-producing strain Klebsiella pneumoniae RYC492. Genome Announc. 1(3):e00178-13. 10.1128/genomeA.00178-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lagos R, Tello M, Mercado G, García V, Monasterio O. 2009. Antibacterial and antitumorigenic properties of microcin E492, a pore forming bacteriocin. Curr. Pharm. Biotechnol. 10:74–85 [DOI] [PubMed] [Google Scholar]
- 20.Sambrook J, Fritsch EF, Maniatis T. 1989. Molecular cloning: a laboratory manual, 2nd ed Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 21.Ausubel FM, Brent R, Kingston RE, Moore DD, Seidman JG, Smith JA, Struhl K. 1992. Short protocols in molecular biology, 2nd ed Greene Publishing Associates and John Wiley & Sons, New York, NY [Google Scholar]
- 22.Miller JH. 1992. A short course in bacterial genetics. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY [Google Scholar]
- 23.Tabor S, Richardson CC. 1985. A bacteriophage T7 RNA polymerase/promoter system for controlled exclusive expression of specific genes. Proc. Natl. Acad. Sci. U. S. A. 82:1074–1078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dubendorff JW, Studier FW. 1991. Controlling basal expression in an inducible T7 expression system by blocking the target T7 promoter with lac repressor. J. Mol. Biol. 219:45–59 [DOI] [PubMed] [Google Scholar]
- 25.Lagos R, Villanueva JE, Monasterio O. 1999. Identification and properties of the genes encoding microcin E492 and its immunity protein. J. Bacteriol. 181:212–217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Miller JH. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY [Google Scholar]
- 27.Mayr-Harting A, Hedges AJ, Berkeley CW. 1972. Methods for studying bacteriocins. Methods Microbiol. 7A:315–422 [Google Scholar]
- 28.Schägger H, von Jagow G. 1987. Tricine-sodium dodecyl sulfate-polyacrylamide gel electrophoresis for the separation of proteins in the range from 1 to 100 kDa. Anal. Biochem. 166:368–379 [DOI] [PubMed] [Google Scholar]
- 29.Strohalm M, Hassman M, Kosata B, Kodicek M. 2008. mMass data miner: an open source alternative for mass spectrometric data analysis. Rapid Commun. Mass Spectrom. 22:905–908 [DOI] [PubMed] [Google Scholar]
- 30.Hunt MD, Pettis GS, McIntosh MA. 1994. Promoter and operator determinants for Fur-mediated iron regulation in the bidirectional fepA-fes control region of the Escherichia coli enterobactin gene system. J. Bacteriol. 176:3944–3955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Orellana C, Lagos R. 1996. The activity of microcin E492 from Klebsiella pneumoniae is regulated by a microcin-antagonist. FEMS Microbiol. Lett. 136:297–303 [DOI] [PubMed] [Google Scholar]
- 32.DePas WH, Chapman MR. 2012. Microbial manipulation of the amyloid fold. Res. Microbiol. 163:592–606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gebbink MFBG, Claessen D, Bouma B, Dijkhuizen L, Wösten HAB. 2005. Amyloids — a functional coat for microorganisms. Nat. Rev. Microbiol. 3:333–341 [DOI] [PubMed] [Google Scholar]
- 34.Otzen D, Nielsen PH. 2008. We find them here, we find them there: functional bacterial amyloid. Cell. Mol. Life. Sci. 65:910–927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Epstein EA, Chapman MR. 2008. Polymerizing the fibre between bacteria and host cells: the biogenesis of functional amyloid fibres. Cell. Microbiol. 10:1413–1420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fowler DM, Koulov AT, Balch WE, Kelly JW. 2007. Functional amyloid - from bacteria to humans. Trends Biochem. Sci. 32:217–224 [DOI] [PubMed] [Google Scholar]
- 37.Blanco LP, Evans ML, Smith DR, Badtke MP, Chapman MR. 2012. Diversity, biogenesis and function of microbial amyloids. Trends Microbiol. 20:66–73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Barnhart MM, Chapman MR. 2006. Curli biogenesis and function. Annu. Rev. Microbiol. 60:131–147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hammer ND, Schmidt JC, Chapman MR. 2007. The curli nucleator protein, CsgB, contains an amyloidogenic domain that directs CsgA polymerization. Proc. Natl. Acad. Sci. U. S. A. 104:12494–12499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang X, Smith DR, Jones JW, Chapman MR. 2007. In vitro polymerization of a functional Escherichia coli amyloid protein. J. Biol. Chem. 282:3713–3719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wang X, Hammer ND, Chapman MR. 2008. The molecular basis of functional bacterial amyloid polymerization and nucleation. J. Biol. Chem. 283:21530–21539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nenninger AA, Robinson LS, Hultgren SJ. 2009. Localized and efficient curli nucleation requires the chaperone-like amyloid assembly protein CsgF. Proc. Natl. Acad. Sci. U. S. A. 106:900–905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bosques CJ, Imperiali B. 2003. The interplay of glycosylation and disulfide formation influences fibrillization in a prion protein fragment. Proc. Natl. Acad. Sci. U. S. A. 100:7593–7598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Knudsen SK, Stensballe A, Franzmann M, Westergaard UB, Otzen DE. 2008. Effect of glycosylation on the extracellular domain of the Ag43 bacterial autotransporter: enhanced stability and reduced cellular aggregation. Biochem. J. 412:563–577 [DOI] [PubMed] [Google Scholar]
- 45.He J, Song Y, Ueyama N, Saito A, Azakami H, Kato A. 2006. Prevention of amyloid fibril formation of amyloidogenic chicken cystatin by site-specific glycosylation in yeast. Protein Sci. 15:213–222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lashuel HA, Lansbury PT., Jr 2006. Are amyloid diseases caused by protein aggregates that mimic bacterial pore-forming toxins? Q. Rev. Biophys. 39:167–201 [DOI] [PubMed] [Google Scholar]
- 47.Hetz C, Bono MR, Barros LF, Lagos R. 2002. Microcin E492, a channel-forming bacteriocin from Klebsiella pneumoniae, induces apoptosis in some human cell lines. Proc. Natl. Acad. Sci. U. S. A. 99:2696–2701 [DOI] [PMC free article] [PubMed] [Google Scholar]