Abstract
Thermus thermophilus exhibits hypersensitivity to a lysine analog, (S)-2-aminoethyl-cysteine (AEC). Cosmid libraries were constructed using genomes from two AEC-resistant mutants, AT10 and AT14, and the cosmids that conferred AEC resistance on the wild-type strain were isolated. When the cosmid library for mutant AT14 was screened, two independent cosmids, conferring partial AEC resistance to the wild type, were obtained. Two cosmids carried a common genomic region from TTC0795 to TTC0810. This region contains genes encoding an ATP-binding cassette (ABC) transporter consisting of TTC0806/TTC0795, using TTC0807 as the periplasmic substrate-binding protein. Sequencing revealed that AT14 carries mutations in TTC0795 and TTC0969, causing decreases in the thermostability of the products. TTC0969 encodes the nucleotide-binding protein of a different ABC transporter consisting of TTC0967/TTC0968/TTC0969/TTC0970 using TTC0966 as the periplasmic substrate-binding protein. By similar screening for cosmids constructed for the mutant AT10, mutations were found at TTC0807 and TTC0969. Mutation in either of the transporter components gave partial resistance to AEC in the wild-type strain, while mutations of both transporters conferred complete AEC resistance. This result indicates that both transporters are involved in AEC uptake in T. thermophilus. To elucidate the mechanism of AEC uptake, crystal structures of TTC0807 were determined in several substrate-binding forms. The structures revealed that TTC0807 recognizes various basic amino acids by changing the side-chain conformation of Glu19, which interacts with the side-chain amino groups of the substrates.
INTRODUCTION
Resistance to the structural analogues of amino acids can be due to different mechanisms in cells, such as increased degradation or reduced uptake of the analogue and decreased sensitivity of the target enzyme. (S)-2-aminoethyl-cysteine (AEC) (Fig. 1) is known to be a lysine analogue. In Escherichia coli and Bacillus subtilis, lysine-binding L-box, a regulatory leader sequence, is present upstream of the corresponding lysine biosynthetic genes, and mutations in L-box provide resistance to AEC by increasing the production of aspartate kinase (AK) (1, 2), which catalyzes the first step in the lysine biosynthetic pathway. In addition to the regulatory leader regions, mutations are found in the regulatory domain of AK, which causes decreased sensitivity to AEC-mediated feedback inhibition (3). In both cases, mutations widen the flow toward lysine biosynthesis to cause the accumulation of a large amount of lysine, which in turn titrates out AEC to avoid its misincorporation into protein. As a different target of AEC, lysyl-tRNA synthetase is proposed based on the isolation of mutants that contain lysyl-tRNA synthetases with lower affinity to AEC (4).
Fig 1.
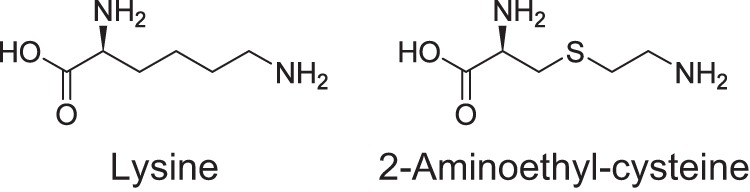
Chemical structures of lysine and AEC.
An extremely thermophilic bacterium, Thermus thermophilus, exhibits hypersensitivity to AEC. The growth of T. thermophilus was inhibited by AEC at 10 μM, which contrasts with other bacteria, such as Escherichia coli and Corynebacterium glutamicum, which grow even in the presence of 5,000 μM AEC. In most bacteria, lysine is synthesized through the diaminopimelate (DAP) pathway; however, it is synthesized through the α-aminoadipate (AAA) pathway in T. thermophilus (5, 6). In T. thermophilus, in addition to the repression by lysine of the expression of genes involved in lysine biosynthesis (7), lysine biosynthesis is regulated through feedback inhibition by lysine of homocitrate synthase (HCS), which catalyzes the first step in lysine biosynthesis (8). Lysine at μM levels inhibits HCS of T. thermophilus (8). As in the instances described above, it is possible that the growth inhibition of T. thermophilus by AEC is due to the inhibition of HCS; however, we also found that the growth of a T. thermophilus mutant possessing HCS with H72L replacement, which is feedback resistant to high concentrations of lysine (9), was inhibited by AEC with the same sensitivity as that of the wild-type strain. This observation suggests that AEC acts on a target other than HCS to inhibit the growth of T. thermophilus. In this study, we isolated AEC-resistant mutants of T. thermophilus and tried to elucidate the mechanism of the hypersensitivity to AEC of T. thermophilus by identification of the mutated genes, and we found that there are two different systems that transport AEC into cells. To elucidate the mechanism of AEC uptake in detail, crystal structures of a periplasmic substrate-binding protein were determined.
MATERIALS AND METHODS
Strains, media, and chemicals.
E. coli DH5α (10) was used for DNA manipulation, and E. coli BL21-Codon-Plus (DE3)-RIL {F− ompT hsdS (rB− mB−) dcm+ Tetr gal (DE3) endA Hte [argU ileY leuW Camr]} (Stratagene, La Jolla, CA) was used as the host to express TTC0795, TTC0806, TTC0807, and TTC0969 genes. The 2× YT medium (10) generally was used for cultivation of E. coli cells, whereas TM (nutrient medium) (11) and MM (minimal medium) (12) were used for cultivation of T. thermophilus HB27 and mutant strains. Antibiotics and isopropyl β-d-thiogalactopyranoside (IPTG) were added to the medium when required. All chemicals were purchased from Sigma-Aldrich (St. Louis, MO), Wako Pure Chemical (Osaka, Japan), and Kanto Chemicals (Tokyo, Japan). Enzymes for DNA manipulation were purchased from TaKaRa Shuzo (Kyoto, Japan) and Toyobo (Osaka, Japan).
Chemical mutagenesis of T. thermophilus and screening of AEC-resistant strains.
To identify the genes responsible for AEC hypersensitivity, we isolated AEC-resistant strains of T. thermophilus by the following procedures. T. thermophilus cells cultured in TM (72 ml) were washed and suspended in 48 ml buffer I (500 mM Tris-HCl, pH 8.0, 1 mg ml−1 N-methyl-N′-nitro-N-nitrosoguanidine [NTG]). Suspended cells were cultured at 70°C for 60 min without shaking. The cells were harvested by centrifugation at 10,000 × g for 10 min at 4°C, washed with sterile water, and suspended in 20 ml MM. The cells were spread on an MM gellan gum plate containing 500 μM AEC. After 4 days of cultivation at 70°C, eight first-growing colonies were isolated as AEC-resistant mutants.
Isolation of DNA fragments responsible for AEC resistance.
Genomic DNAs from AEC-resistant mutants were purified and partially digested with Sau3AI. DNA fragments larger than 20 kb were ligated to BamHI- and phosphatase-treated pOJ446 cosmid vector (13), packaged with a Lambda Inn packaging kit (Nippon-Gene, Tokyo, Japan), and introduced into E. coli XL1-Blue MRF′ cells according to the manufacturer's instructions. For every AEC mutant, about 200 colonies were obtained, sufficient to cover the whole genome of T. thermophilus. Cosmid DNA was purified from each E. coli colony and used as the cosmid library, which was pooled to transform T. thermophilus HB27 (11). T. thermophilus HB27 cells transformed with the cosmid library were grown in liquid MM supplemented with 500 μM AEC at 70°C. Cosmids that gave AEC resistance to the wild-type strain were selected as candidates that carry mutations responsible for showing AEC resistance.
Thermostability of mutated ABC transporter components.
All PCR primers used are listed in Table S1 in the supplemental material. TTC0795 and TTC0969 of mutant AT14 (AEC-resistant T. thermophilus no. 14) were prepared as follows. TTC0795 was prepared with a Strep tag at the N terminus in E. coli BL21 RIL-Codon Plus (DE3) cells using pET26b(+) as the expression vector. Harvested cells were suspended in 8 ml buffer II (20 mM Tris-HCl, pH 8.0, 150 mM NaCl), washed, and disrupted by sonication. The supernatant, prepared by centrifugation at 40,000 × g for 20 min, was applied to a Strep-Tactin column preequilibrated with buffer III (100 mM Tris-HCl, pH 8.0, 150 mM NaCl). After washing with the same buffer, adsorbed proteins were eluted with buffer III supplemented with 2.5 mM desthiobiotin. TTC0969 with a His8 tag was purified using an Ni2+ affinity column. Buffer II supplemented with 20 or 500 mM imidazole was used for column preequilibration and protein elution, respectively. Protein concentrations were determined by the Bradford method using a Bio-Rad protein assay kit (Bio-Rad, Tokyo, Japan). After the protein concentration had been adjusted to 1 mg ml−1, protein samples were heated at appropriate temperatures for 30 min. These samples were then cooled on ice for 10 min and centrifuged at 20,000 × g for 10 min to remove aggregates. The supernatants were applied to SDS-PAGE.
Preparation of knockout mutants of T. thermophilus.
To verify the involvement of the mutation found in the isolated cosmid in AEC resistance, T. thermophilus mutants were constructed that lacked TTC0807, TTC0969, or both TTC0807 and TTC0969. The plasmid to delete the TTC0807 gene was constructed as follows. The first PCR was performed with primers ttc0807-up-fw and ttc0807-up-rv to amplify the upstream region of the TTC0807 gene of 700 bp. The second PCR was performed with primers htk-fw and htk-rv to amplify the HTK (hyperthermostable kanamycin nucleotidyltransferase) gene (14). The third PCR was performed with primers ttc0807-down-fw and ttc0807-down-rv to amplify the downstream region (700 bp) of the TTC0807 gene. Each of the three amplified fragments was digested with XbaI/SpeI, SpeI/BamHI, and BamHI/XhoI, respectively, and cloned separately into pBlueScript II KS(+) for sequence verification. Three fragments with the correct sequences were ligated together with pBlueScript II KS(+) digested with XbaI/XhoI. The resulting plasmid, named pKS-d0807, was used to knock TTC0807 out of T. thermophilus HB27. Colonies that were resistant to 200 μg/ml kanamycin on TM were picked up, and the knockout was confirmed by PCR using ttc0807 check-fw and ttc0807 check-rv. Other genes (TTC0795, TTC0806, and TTC0969) were knocked out in a similar way. In some cases, hyg10, the thermostable hygromycin resistance gene (15), was used in place of the HTK gene. All PCR primers used for gene disruption are listed in Table S2 in the supplemental material. Complete cloning methods (thermocycling conditions, polymerase enzymes used, etc.) are available from the corresponding author upon request.
AEC sensitivity analysis.
T. thermophilus cells were precultured in TM at 70°C overnight. The cells were washed 2 times with MM and suspended in an equal volume of MM. A 0.1% volume of the culture was transferred into fresh MM left unsupplemented or supplemented with 500 μM AEC. The strains were grown aerobically at 70°C, and the optical density at 600 nm (OD600) of the cultures was monitored every 3 h.
Pulldown assay.
TTC0795 with a Strep tag at the N terminus, TTC0806 with an His8 tag at the N terminus, and TTC0807 with a FLAG tag at the C terminus were separately produced in E. coli BL21 RIL-Codon Plus (DE3) cells. All PCR primers used for the pulldown assay are listed in Table S3 in the supplemental material. Harvested cells were suspended in 32 ml buffer II, washed, and disrupted by sonication. The supernatant was prepared by centrifugation at 40,000 × g for 20 min. The precipitate containing TTC0806 was suspended in 20 ml buffer IV (20 mM Tris-HCl, pH 8.0, 300 mM NaCl, 20% glycerol, 0.8% dodecyl maltoside), agitated for solubilization at 4°C for 2 h, and centrifuged at 40,000 × g for 30 min. TTC0795 was purified with a Strep-Tactin column (Novagen) according to the manufacturer's instructions. The supernatant containing TTC0806 was applied to an Ni2+-nitrilotriacetic acid (NTA) column preequilibrated with buffer II supplemented with 20 mM imidazole and 0.03% dodecyl maltoside. After washing with buffer II containing 100 mM imidazole and 0.03% dodecyl maltoside, adsorbed proteins were eluted with buffer II supplemented with 500 mM imidazole and 0.03% dodecyl maltoside. TTC0807 with FLAG tag was purified as follows. The supernatant containing TTC0807 was applied to an anti-FLAG M2 affinity column (Sigma-Aldrich) preequilibrated with buffer V (50 mM Tris-HCl, pH 7.4, 150 mM NaCl). After washing with the same buffer, adsorbed TTC0807 was eluted with buffer V supplemented with FLAG peptide (Sigma-Aldrich) at 0.1 mg ml−1. Purified TTC0795, TTC0806, and TTC0807 were dialyzed with buffer II, buffer II supplemented with 0.03% dodecyl maltoside, and buffer V, respectively. TTC0806 (His8 tag) was mixed with either TTC0795 (Strep tag) or TTC0807 (FLAG tag) and incubated for 1 h at room temperature. TTC0806 (His8 tag) mixed with TTC0795 (Strep tag) was applied to an Ni2+-NTA column, and adsorbed proteins were eluted with buffer II supplemented with 500 mM imidazole and 0.03% dodecyl maltoside. A similar experiment was performed for a mixture from which TTC0806 (His8 tag) was omitted. TTC0806 (His8 tag) mixed with TTC0807 (FLAG tag) was applied to an anti-FLAG M2 affinity column, and adsorbed protein was eluted in the way described above using FLAG peptide at 0.1 mg ml−1. A similar experiment was performed for a mixture from which TTC0807 (FLAG tag) was omitted. Both eluates were separated on 12% SDS-PAGE.
Preparation of TTC0807 for crystallization.
Because TTC0807 was predicted to encode a periplasmic substrate-binding protein with a signal peptide (Met1 to Ala18) for periplasmic localization by the SignalP 4.0 server (16), a DNA fragment corresponding to the mature protein (Gln19 to Lys254) was amplified by PCR with a pair of synthetic oligonucleotides listed in Table S4 in the supplemental material, digested with EcoRI/HindIII, and cloned into pBluescript II KS(+). A DNA fragment with the desired sequence was cloned into the EcoRI/HindIII site of pHIS8 (17) to yield pHIS8-0807, and it was introduced into E. coli BL21-CodonPlus (DE3)-RIL cells. Using this construct, TTC0807 was designed to be produced in a form with an His8 tag at the N terminus. E. coli cells harboring pHIS8-0807 were cultured in 2× YT medium supplemented with 50 μg/ml kanamycin and 30 μg/ml chloramphenicol at 37°C. When the E. coli cells were grown to an OD600 of 0.6, IPTG was added at a final concentration of 0.1 mM. Culture was continued at 25°C for an additional 12 h. TTC0807 was purified as follows. E. coli cells harboring pHIS8-0807 from a 1.6-liter culture were harvested, washed, and suspended in 32 ml buffer II. Suspended cells were disrupted by sonication. The supernatant, prepared by centrifugation at 40,000 × g for 20 min, was heated at 70°C for 30 min, and denatured proteins from E. coli cells were removed by centrifugation at 40,000 × g for 20 min. Supernatant was applied to an Ni2+-NTA resin column, which was preequilibrated with buffer II supplemented with 20 mM imidazole. After washing the column with a sufficient volume of the same buffer, TTC0807 was eluted with buffer II supplemented with 500 mM imidazole. The eluted fraction was concentrated with a Vivaspin concentrator (molecular weight cutoff [MWCO], 10,000 Da; Sartorius Stedim Biotech, Bohemia, NY) and then applied to a Superdex 200-pg HiLoad 26/60 gel filtration chromatography column (GE Healthcare) equilibrated with buffer II. The TTC0807-containing fractions were concentrated with a Vivaspin concentrator (MWCO, 10,000 Da). The purity of the recombinant protein was verified by 12% SDS-PAGE. The protein concentration was adjusted to 10 mg ml−1 and used for crystallization.
Crystallization of TTC0807.
Crystallization conditions of TTC0807 with AEC, lysine, ornithine, and arginine were screened by the hanging drop vapor diffusion method using screening kits that are commercially available. Drops of 2 μl consisting of reservoir solution (1 μl) and 10 mg/ml TTC0807 solution (1 μl) supplemented with 10 mM lysine, AEC, ornithine, or arginine were equilibrated against 500 μl reservoir solution at 293 K. Crystals of TTC0807 with AEC, lysine, ornithine, and arginine were obtained from droplets using a solution (0.2 M ammonium sulfate and 20% polyethylene glycol [PEG] 4000). Crystals were grown under each condition in a few days.
Structure determination of TTC0807 complexed with AEC, lysine, ornithine, and arginine.
Prior to data collection, crystals were briefly soaked in reservoir solution supplemented with 20% (vol/vol) PEG 400 as a cryoprotectant, flash-cooled in a nitrogen gas stream at 95 K, and stored in liquid nitrogen. Diffraction data of TTC0807 with lysine, AEC, and ornithine were collected at the NW12 station of the Photon Factory, High Energy Accelerator Research Organization (KEK) (Tsukuba, Japan). Diffraction data of TTC0807 with arginine were collected at the NE-3A station of the Photon Factory. Diffraction images were indexed, integrated, and scaled using the HKL-2000 program suite (18). Details of data collection statistics are summarized in Table 1. The data of TTC0807 crystals collected at a wavelength of 1.0 Å were used for subsequent molecular replacement and crystallographic refinement. Molecular replacement was performed with Phaser (19) in the CCP4 program suite (20) using the model of LAO-binding protein (StLAO-BP) from Salmonella enterica serovar Typhimurium (Protein Data Bank [PDB] code 1LST) (21). Refinement was performed with Refmac 5.5 (22), and model correction in the electron density map was performed with Coot (23). Data collection, refinement statistics, and the results of Ramachandran plots, produced by the program PROCHECK (24), are summarized in Table 1. The figures were prepared using PyMOL (available online).
Table 1.
X-ray data collection and refinement statistics
| Parameter | Value(s) forc: |
|||
|---|---|---|---|---|
| TTC0807/AEC | TTC0807/Lys | TTC0807/Orn | TTC0807/Arg | |
| Data collection | ||||
| Beamline | PF-NW12 | PF-NW12 | PF-NW12 | PF-NE3A |
| Wavelength (Å) | 1.0 | 1.0 | 1.0 | 1.0 |
| Space group | P212121 | P212121 | P212121 | P212121 |
| Cell dimensions | ||||
| a (Å) | 68.6 | 68.8 | 68.3 | 68.9 |
| b (Å) | 68.6 | 82.0 | 71.1 | 68.8 |
| c (Å) | 98.5 | 105.8 | 105.7 | 104.6 |
| Resolutiona (Å) | 1.90 (1.90–1.97) | 2.00 (2.00–2.07) | 2.07 (2.07–2.14) | 1.90 (1.90–1.93) |
| Total no. of reflections | 256,649 | 264,459 | 229,528 | 283,817 |
| No. of unique reflections | 37,509 | 36,243 | 32,657 | 40,039 |
| Completenessa (%) | 99.9 (100.0) | 99.9 (100.0) | 99.5 (100.0) | 99.2 (100.0) |
| Avg I/σ(I)a | 14.6 (2.2) | 27.6 (4.3) | 24.5 (3.5) | 18.5 (2.4) |
| Rmergea,b (%) | 14.0 (69.9) | 7.5 (46.0) | 8.7 (44.4) | 12.0 (64.6) |
| Refinement | ||||
| Resolution (Å) | 30.67–1.90 | 27.31–2.00 | 30.79–2.07 | 32.69–1.90 |
| R factor/Rfree (%) | 19.1/24.3 | 20.4/26.0 | 20.0/24.9 | 19.3/24.0 |
| No. of protein atoms | 3,777 | 3,806 | 3,806 | 3,806 |
| No. of amino acids | 2 | 2 | 2 | 2 |
| No. of SO42− molecules | 1 | 2 | 0 | 2 |
| No. of waters molecules | 275 | 340 | 300 | 179 |
| Avg B factor (Å2) | ||||
| Protein | 18.5 | 27.2 | 29.4 | 22.9 |
| Amino acid | 19.2 | 18.2 | 22.0 | 16.3 |
| SO42− | 54.2 | 35.6 | 46.9 | |
| Water | 31.7 | 37.1 | 39.4 | 30.4 |
| RMSD from ideal values | ||||
| Bond length (Å) | 0.013 | 0.012 | 0.014 | 0.011 |
| Bond angles (°) | 1.4 | 1.5 | 1.5 | 1.3 |
| Ramachandran plot (%) | ||||
| Favored | 91.6 | 91.2 | 92.9 | 92.6 |
| Additional allowed | 8.4 | 8.8 | 7.1 | 7.4 |
| Generously allowed | 0 | 0 | 0 | 0 |
| Disallowed | 0 | 0 | 0 | 0 |
Values in parentheses are for the highest-resolution shell.
Rmerge = Σ|Ii − <I>|/Σ<I>.
Lysine, ornithine, and arginine are abbreviated as Lys, Orn, and Arg, respectively.
Crystallographic data accession numbers.
The atomic coordinates and structure factors for TTC0807/AEC, TTC0807/ornithine, TTC0807/lysine, and TTC0807/arginine complexes have been deposited in the Protein Data Bank (PDB codes 3VV5, 3VVD, 3VVE, and 3VVF, respectively).
RESULTS
Isolation of AEC-resistant mutants.
To elucidate the mechanism of hypersensitivity to AEC of T. thermophilus HB27, we isolated mutants that exhibited AEC resistance by NTG mutagenesis. The mutagenized cells were spread on an MM plate supplemented with 500 μM AEC. Although the wild-type strain showed no growth under this condition (Fig. 2), eight colonies could grow on the plate containing AEC. From these eight mutants, two strains, named AT10 and AT14, were chosen for further analyses.
Fig 2.
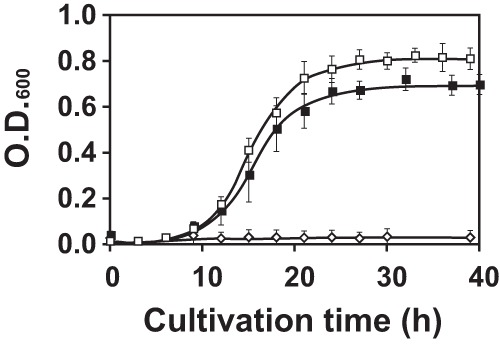
Growth profiles of AT10 and AT14 in MM containing 500 μM AEC. Open square, AT14; closed square, AT10; open diamond, wild type.
Identification of mutations responsible for AEC resistance. (i) AT14.
Since T. thermophilus exhibits natural competence, DNA can be easily taken up into the cells and integrated into the genome by double-crossover homologous recombination. We obtained approximately 200 cosmids carrying DNA from an AT14 mutant and screened for the DNA fragment that conferred AEC resistance to the wild-type strain. We isolated two cosmids from AT14, cos14-204 with a 23-kb insert and cos14-277 with a 33 kb insert, which gave AEC resistance to T. thermophilus. Both of the cosmids carried the same DNA region from TTC0795 to TTC0810, suggesting that this region should contain a mutation(s) responsible for the AEC resistance of AT14. Sequencing of the region revealed that there were two in-frame deletions of 3 bp, causing a loss of Gln143 of TTC0795 in the AT14 genome. AT14 also carries another in-frame deletion of 3 bp, causing a loss of Leu175 of TTC0969 in the genome (Fig. 3A).
Fig 3.
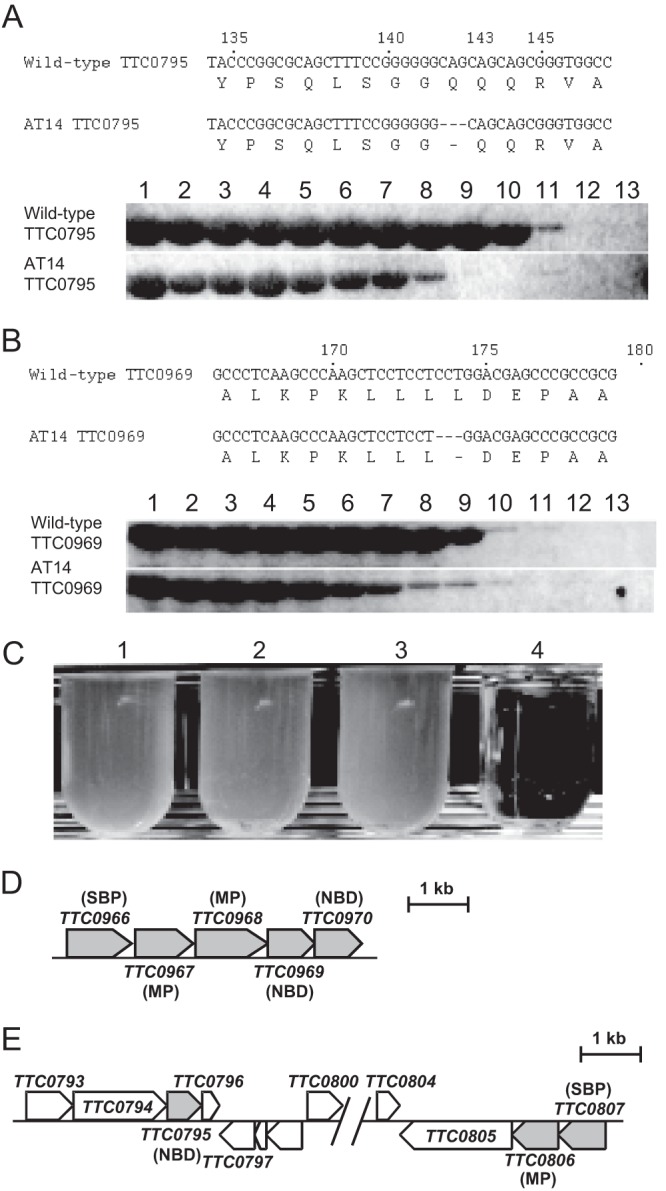
Mutation sites in AT14. (A) Deletion of Gln143 in TTC0795 (upper panel) and comparison of heat stability between TTC0795 proteins from the wild-type strain and AT14 (lower panel). Lane 1, not heated; 2, 31.0°C; 3, 34.1°C; 4, 36.1°C; 5, 39,0°C; 6, 41.0°C; 7, 42.4°C; 8, 45.2°C; 9, 50.2°C; 10, 56.0°C; 11, 60.8°C; 12, 63.5°C; 13, 65°C. (B) Deletion of Leu175 in TTC0969 (upper panel) and comparison of heat stability between TTC0969 proteins from wild-type strain and AT14 (lower panel). Lanes are the same as those for panel A. (C) Growth of AT14 in MM at permissive and nonpermissive temperatures. Lanes 1 and 2, cultured at 70°C; lanes 3 and 4, cultured at 50°C. Lanes 1 and 3, culture in the absence of 500 μM AEC; lanes 2 and 4, culture in the presence of 500 μM AEC. (D) TTC0966-TTC0970 region in wild-type T. thermophilus HB27. (E) TTC0793-TTC0807 region in wild-type T. thermophilus HB27. Periplasmic substrate-binding protein is abbreviated as SBP.
These results suggest that loss of function of either of these genes or both is a cause of the AEC resistance of AT14. Interestingly, TTC0795 and TTC0969 both are components of ABC transporters. ABC transporters are composed of two transmembrane domains and two cytoplasmic nucleotide-binding domains that hydrolyze ATP to drive transport. In Gram-negative bacteria, ABC transporters require a binding protein that binds the substrates to be imported and transfers them to the transporters. TTC0969 is annotated as a gene encoding the nucleotide-binding subunit (NBD) of ABC transporters. TTC0969 forms a cluster with four flanking genes, TTC0966, TTC0967, TTC0968, and TTC0970 (Fig. 3D). TTC0966 is annotated to encode a periplasmic substrate-binding protein for ABC transporters. Both TTC0967 and TTC0968 are annotated to code for membrane subunits (MPs) of ABC transporters, while TTC0970 is annotated to encode the NBD of ABC transporters, suggesting that these five proteins are involved in solute uptake. On the other hand, TTC0795 is also annotated as a gene encoding an NBD component of ABC transporters. As described below, TTC0795 is suggested to be an NBD component that interacts with TTC0806, a transmembrane subunit, of ABC transporters. TTC0806 forms a gene cluster with TTC0807 (Fig. 3E), which is annotated as a gene encoding periplasmic amino acid-binding protein for ABC transporters.
These two missing residues are located away from the substrate-binding residues on the respective proteins based on the crystal structures of PDB code 2OLJ (NBD of arginine transporters from Geobacillus stearothermophilus) for TTC0795 and PDB code 1G6H (NBD of ABC transporter from Methanocaldococcus jannaschii) for TTC0969 (25); therefore, we hypothesized that loss of the amino acid residues affects protein stability. In order to examine this possibility, TTC0795-ΔGln143 was heated at elevated temperatures, and undenatured fractions were subjected to SDS-PAGE (Fig. 3A and B). Analysis revealed that the mutant was completely inactivated at 50.2°C, which was considerably lower than the 63.5°C needed for wild-type TTC0795. Similar analysis was performed for TTC0969-ΔLeu175. The mutant was also inactivated at a lower temperature than wild-type TTC0969. These observations indicate that two proteins involved in ABC transporter function are heat labile in AT14. In accordance with this, AT14 exhibited AEC sensitivity in culture at 50°C, which contrasts with the culture at 70°C (Fig. 3C).
(ii) AT10.
Similar experiments were conducted for the AT10 cosmid library. Unexpectedly, the simultaneous introduction of two cosmids, cos10-3 and cos10-5, was required to confer AEC resistance on T. thermophilus HB27. Sequence analyses of the inserts on cos10-3 (38 kb) and cos10-5 (34 kb) revealed that there was an inversion of a locus between TTC0807 and TTC0969 on the AT10 genome, which explains the necessity of simultaneous introduction of the two cosmids to cause inversion of the wild-type genome. Similar nucleotide sequences were found around the junction points of TTC0807 and TTC0969, and this might assist genome inversion between these loci (Fig. 4A to C). At the junction points of the AT10 genome, two TTGAT sequences were inverted.
Fig 4.
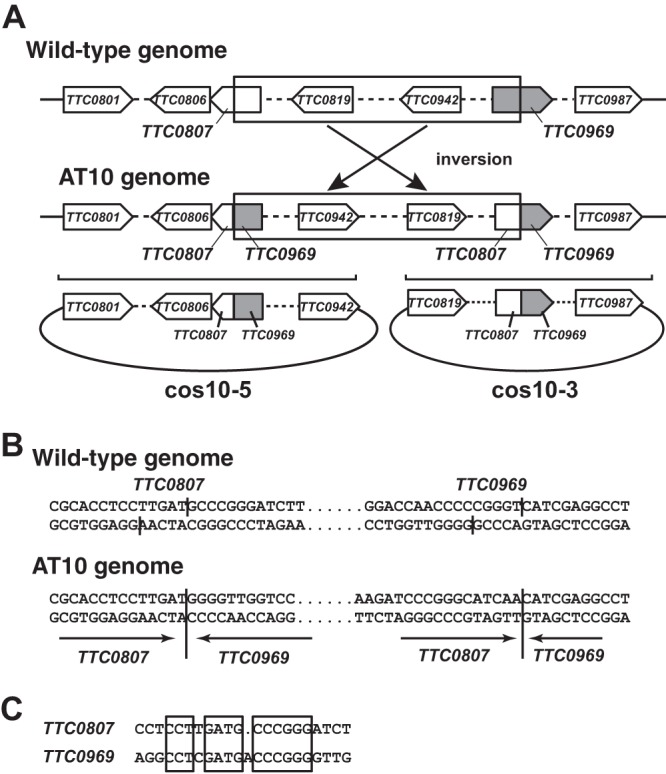
Two cosmids from AT10 necessary for conferring AEC resistance to wild-type strain. (A) Inversion of a locus between TTC0807 and TTC0969 on the AT10 genome and schematic structures of inserts in cos10-3 and cos10-5. Only a few open reading frames are shown; portions containing other open reading frames are shown as broken lines. (B) Nucleotide sequences around the recombination sites in TTC0807 and TTC0969. (C) Sequence comparison of the recombination sites.
Deletion of genes for proposed AEC transporters.
To confirm further the involvement of the mutations in AEC resistance, we constructed single and double mutants of TTC0807 and TTC0969 and examined the growth of the T. thermophilus mutants lacking these genes. When ΔTTC0807 and ΔTTC0969 single-site mutants were cultured in MM containing 500 μM AEC, they grew very slowly, which contrasts with the ΔTTC0807 ΔTTC0969 double mutant, which was capable of growing rapidly, with a growth rate similar to that of AT10 (Fig. 5). These results indicate that two different transporters, in each of which either TTC0807 or TTC0969 is involved, contribute to AEC uptake in T. thermophilus HB27. Here, we call the two ABC transporters AecT-I and AecT-II. TTC0806 and TTC0807 are involved in AecT-I function, and TTC0967 to TTC0970 are involved in AecT-II function. In addition to TTC0807 and TTC0969, we also constructed mutants lacking either TTC0795 or TTC0806 (see Fig. S1 in the supplemental material). The ΔTTC0806 strain grew with a profile similar to that of the ΔTTC0807 strain, suggesting that these products are used as mechanisms in the same AEC uptake system. The growth profile of the ΔTTC0795 mutant is similar to those of the ΔTTC0806 and ΔTTC0807 strains, suggesting that TTC0795 is the subunit of ABC transporters working with TTC0806.
Fig 5.
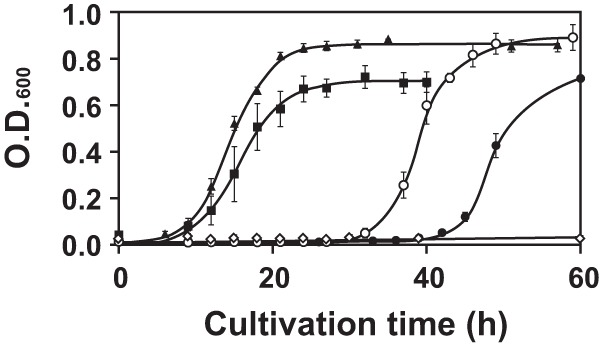
Effect of deletion of TTC0807 and/or TTC0969 on growth in MM containing 500 μM AEC. Closed square, AT10; open diamond, wild type; open circle, ΔTTC0807 mutant; closed circle, ΔTTC0969 mutant; closed triangle, ΔTTC0807 ΔTTC0969 double mutant.
Interaction of TTC0806 with TTC0795 and TTC0807.
As described above, mutations in both AT10 and AT14 resulted in malfunctioning of two AEC uptake systems. AecT-II is composed of TTC0967 to TTC0970, using TTC0966 as a periplasmic substrate-binding protein. The other transporter, AecT-I, contains TTC0806 as an MP, using TTC0807 as a periplasmic substrate-binding protein. The ΔTTC0795 mutant responded to AEC similarly to ΔTTC0806 and ΔTTC0807 mutants. Therefore, we hypothesized that TTC0795 functions as a component of AecT-I, with TTC0806 using TTC0807 as a periplasmic substrate-binding subunit. To examine this hypothesis, a pulldown assay was performed using the three components with different tags. TTC0806 with an N-terminal His tag, TTC0807 with a C-terminal FLAG tag, and TTC0795 with an N-terminal Strep tag were separately produced in E. coli cells and purified as described in Materials and Methods. When TTC0806 (His8 tag) was mixed with TTC0795 (Strep tag) and applied to an Ni2+-NTA column, TTC0795 (Strep tag) was coeluted with TTC0806 (His8 tag) by addition of 500 mM imidazole (Fig. 6A). TTC0795 (Strep tag) alone was not adsorbed to the Ni2+-NTA column (Fig. 6A), which indicates that TTC0795 can interact with TTC0806. This supports the above-mentioned assumption that TTC0795 is an NBD component of AecT-I that takes up AEC. When TTC0806 (His8 tag) was mixed with TTC0807 (FLAG tag) and applied to an anti-FLAG M2 column, addition of FLAG peptide unexpectedly caused elution of only TTC0807 (FLAG tag) (Fig. 6B).
Fig 6.
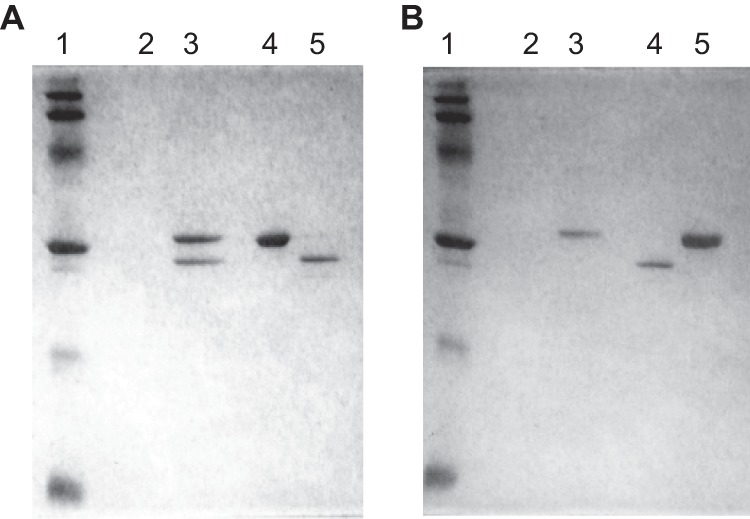
Pulldown assay for TTC0806, TTC0807, and TTC0795. (A) Pulldown assay for TTC0795 and TTC0806. Lane 1, molecular size marker; lane 2, eluate from Ni2+-NTA column for TTC0795 only (Strep tag); lane 3, eluate from Ni2+-NTA column for mixture of TTC0795 (Strep tag) and TTC0806 (His8 tag); lane 4, purified TTC0795 (Strep tag); lane 5, purified TTC0806 (His8 tag). (B) Pulldown assay for TTC0806 and TTC0807. Lane 1, molecular size marker; lane 2, eluate from anti-FLAG M2 affinity column for TTC0806 only (His8 tag); lane 3, eluate from anti-FLAG M2 affinity for mixture of TTC0806 (His8 tag) and TTC0807 (FLAG tag); lane 4, purified TTC0806 (His8 tag); lane 5, purified TTC0807 (FLAG tag).
Crystal structure of TTC0807 complexed with amino acids.
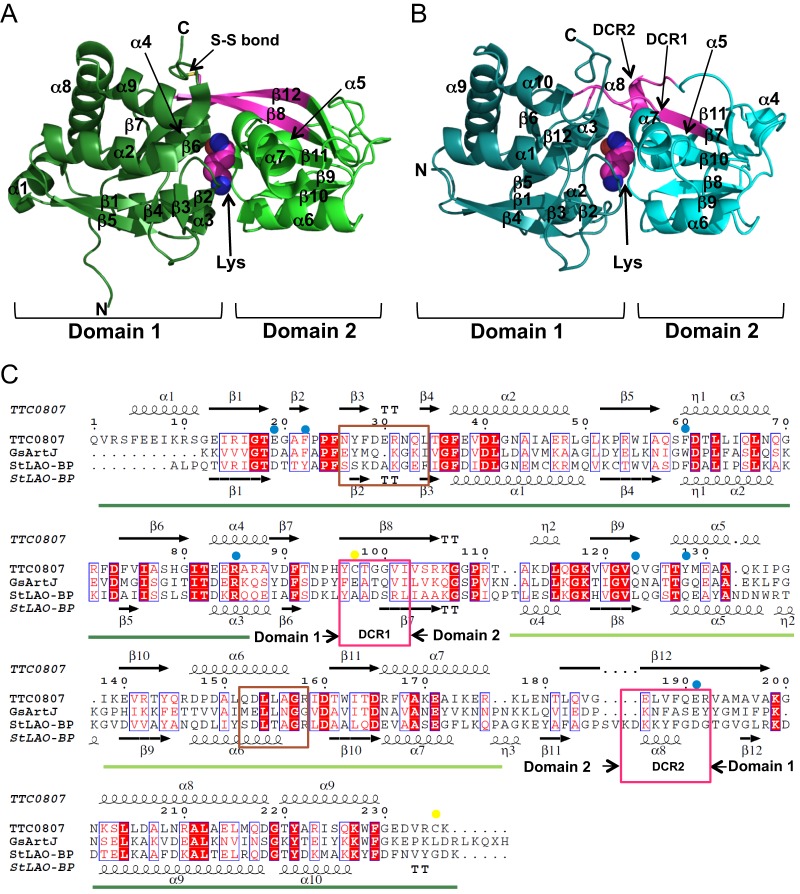
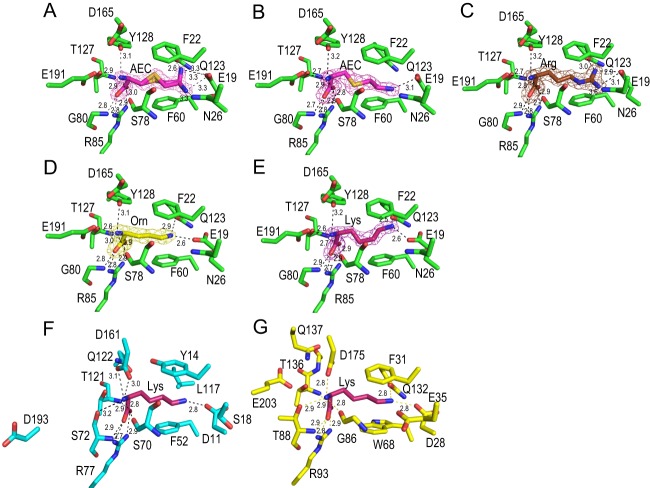
TTC0807, which is a periplasmic substrate-binding protein for AecT-I, was overexpressed, purified, and crystallized with AEC, lysine, ornithine, and arginine. Crystal structures of TTC0807 complexed with AEC, lysine, ornithine, and arginine were determined at 1.90, 2.00, 2.07, and 1.90 Å, respectively. A DALI search (26) showed that the structure of TTC0807 was most similar to that of the LAO-binding protein StLAO-BP from Salmonella Typhimurium, with a Z score of 30.6 and root mean square deviations (RMSD) of 1.3 Å for Cα atoms (Fig. 7AB). StLAO-BP is composed of domain 1 (A1-Y88 and T195-K238) and domain 2 (R93-V185) connected by the domain-connecting regions (DCR1 [A89-S92] and DCR2 [K186-G194]) (Fig. 7C). Although the structure of TTC0807 is principally similar to that of StLAO-BP, DCRs exhibit different structures. In StLAO-BP, DCR1 is flexible with a random coil structure, and DCR2 is mostly in a random coil containing a short α-helix in the middle. In contrast, in the TTC0807 structure, β8 (DCR1) and β12 (DCR2), forming a β sheet, connect the two domains. Cys97 in DCR1 forms a disulfide bond with Cys235 in TTC0807. The disulfide bond is likely a structural factor that enhances the thermostability of domain 1. Lysine and other substrates are bound in the pocket between these two domains (Fig. 7A and 8A to E). The asymmetric unit contains two TTC0807 molecules. Although AEC is bound to the B chain in a single conformation, it is bound to the A chain in alternative conformations (Fig. 8A and B). In the B chain, the side-chain amino group of AEC is recognized only by Glu19 side chains, whereas in the A chain the side-chain amino group of AEC is found at two different positions. In both conformations the amino group forms electrostatic interactions with Glu19; however, in one form it makes a hydrogen bond with Gln123, and in the other form it is hydrogen bonded with Asn26. Other portions of the AEC molecule are recognized in the same way. In both chains, the α-amino group of AEC interacts with Glu191 by electrostatic interaction and further forms hydrogen bonds with the side-chain hydroxyl group of Tyr128 and the main-chain carboxyl group of Ser78, and the α-carboxyl group forms electrostatic interactions with Arg85. The α-carboxyl group is also recognized by hydrogen bonds with the main-chain amide group of Gly80 and the side-chain hydroxyl group of Thr127 (Fig. 8A). The side-chain hydrophobic portion of AEC was stacked with two aromatic residues, Phe22 and Phe60. In arginine-, ornithine-, and lysine-bound complexes, two chains in the asymmetric unit of those crystal structures bind the substrates in a single conformation. In these complexes, the substrates are bound in a similar manner using the above-mentioned residues (Fig. 8A to E). Interestingly, in order to bind lysine and ornithine, the side chain of Glu19 was rotated 33° to form suitable electrostatic interactions with the substrates. In the TTC0807/arginine complex, arginine is bound to the enzyme with the side chain of Glu19 in a conformation similar to that for binding AEC. The position of Glu19 is necessary to accommodate the larger guanidium group of arginine in the binding pocket (Fig. 8C). It is of interest that the conformation of Glu19 in the AEC complex is very similar to that in the arginine complex but not to that in the lysine complex, although AEC is an analog of lysine. This is because AEC has a sulfur atom in the γ position. Since the C—S bond length (1.8 Å) is longer than that of the C—C bond (1.5 Å), AEC is larger than lysine by as much as 0.6 Å, although it is smaller than arginine. Regardless, TTC0807 recognizes these basic amino acids by changing the conformation of the Glu19 side chain.
Fig 7.
Structure of TTC0807. (A) Crystal structure of TTC0807/lysine complex. (B) Crystal structure of StLAO-BP/lysine complex. Domain-connecting regions (DCRs) in StLAO-BP and the corresponding portions in TTC0807 are colored magenta. N and C represent N- and C-terminal domains, respectively. The disulfide bond in TTC0807 is shown as an S—S bond. (C) Amino acid sequence alignment of TTC0807 and homologs. Secondary structures are also shown. Amino acid residues involved in substrate binding and in disulfide bond formation are marked by blue and yellow circles, respectively. DCRs are shown by magenta boxes, and the putative loop necessary for interaction with a transmembrane component of ABC transporters is indicated by a brown box.
Fig 8.
Ligand-binding site. (A) A chain of TTC0807/AEC complex. (B) B chain of TTC0807/AEC complex. (C) TTC0807/arginine complex. (D) TTC0807/ornithine complex. (E) TTC0807/lysine complex. (F) StLAO-BP/lysine complex. (G) GsArtJ/lysine complex. Nitrogen, oxygen, and sulfur atoms are shown as blue, red, and orange sticks, respectively. Carbon atoms of amino acid residues of TTC0807, StLAO-BP, and GsArtJ are shown as green, cyan, and yellow sticks, respectively. Carbon atoms of bound molecules of AEC, arginine, ornithine, and lysine are shown as light magenta, brown, yellow, and dark magenta sticks, respectively. The electron density maps (2Fo-Fc map) of AEC in A chain, AEC in B chain, arginine, ornithine, and lysine are shown as meshes contoured at 1.3, 1.3, 1.5, 1.6, and 1.6 σ, respectively.
Comparison of lysine recognition to that of other lysine-binding proteins.
When the TTC0807/lysine complex is compared to the lysine-bound complexes of StLAO-BP and ArtJ from Geobacillus stearothermophilus (GsArtJ) (27, 28), lysine is recognized in a slightly different manner, although Arg85 (in TTC0807 numbering), which interacts with the α-carboxyl group, is conserved in StLAO-BP and GsArtJ, and similar recognition of the α-amino group of lysine is observed with the main-chain carbonyl oxygen of Ser78 and the corresponding residues in the three proteins (Fig. 7C and 8E, F, and G). In TTC0807, the α-amino group of lysine is recognized by Tyr128 and Glu191 (Fig. 8E), whereas it is recognized by Ser70 and Asp161 in StLAO-BP and Gly86, Thr88, and Asp175 in GsArtJ (Fig. 8F and G). Interestingly, although Asp161 in StLAO-BP (Asp175 in GsArtJ) is conserved as Asp165 in TTC0807, Asp165 does not interact with the α-amino group of lysine. Instead, Tyr128 recognizes the α-amino group in TTC0807. Tyr128 is replaced with Gln in StLAO-BP and GsArtJ. The α-amino group of lysine is also recognized by Glu191 from β12 in TTC0807. In StLAO-BP and GsArtJ, the corresponding residue is conserved as Asp193 and Glu203, respectively, with a similar negative charge; however, these residues do not participate in substrate binding. Thus, recognition of the α-amino group of lysine is different between TTC0807 and the other two proteins.
The ε-amino group of lysine is also recognized in a different way among these three lysine-binding proteins. The amino group is recognized by Gln123 and Glu19 in TTC0807, while it is recognized by Asp11 and a bound water molecule in StLAO-BP, in which the residue corresponding to Gln123 is replaced with Leu117 (see Fig. S3 in the supplemental material). In GsArtJ, Gln132 occupies a position similar to that of Gln123 in TTC0807; however, it is different in that the bound water molecule directly interacts with the ε-amino group of lysine. Taken together with α-amino group recognition, lysine is recognized using similar but distinct mechanisms by these related proteins.
In the AEC-bound complex of TTC0807, the side-chain amino group of AEC is recognized by Glu19 and either Asn26 or Gln123. Asn26 and Gln123 in TTC0807 are replaced with Ser18 and Leu117 in StLAO-BP, which cannot form a hydrogen bond with the amino group, suggesting weak binding of the amino group in StLAO-BP. In addition to the difference in α-amino group recognition, recognition of the side-chain amino group is stabilized by tight binding in TTC0807, suggesting that the AEC molecule is bound by TTC0807 more tightly than by StLAO-BP and possibly the ortholog in E. coli; therefore, we speculate that tight binding of AEC in TTC0807 is one of the factors responsible for the hypersensitivity of T. thermophilus to AEC.
DISCUSSION
AEC is a lysine analog that inhibits the growth of bacterial cells by reduction of AK activity in cells, resulting in decreased flow toward lysine biosynthesis, and/or by misincorporation into protein. T. thermophilus exhibits higher sensitivity to AEC than E. coli and C. glutamicum. In this study, we isolated AEC-resistant mutants of T. thermophilus to elucidate the mechanism of hypersensitivity to AEC of T. thermophilus by identification of the mutated genes.
When two AEC-resistant mutants, AT10 and AT14, were analyzed, they were found to carry mutations that resulted in malfunctioning of two different solute uptake systems. Both mutants were isolated by NTG treatment; however, NTG treatment may not be the direct cause of mutations occurring on the genomes of the mutants, because the mutagen is known to cause G·C-to-A·T transition. In AT10, genome inversion was found, while 3-bp in-frame deletions were found in two genes in AT14. A genome rearrangement with insertion of an inverted repeat sequence in AT10 is reminiscent of an insertion sequence element-mediated genome arrangement. Regardless, this inversion was not expected by NTG mutagenesis. At present, the mechanisms of this genome inversion and the in-frame deletions remain unknown.
Genes encoding AecT-II components and their cognate solute-binding proteins are clustered on the genome as TTC0966 to TTC0970; however, those for AecT-I are located in two regions. Among them, TTC0806 and TTC0807 may be transcribed with TTC0805, encoding putative valyl-tRNA synthetase in T. thermophilus (see Fig. S2 in the supplemental material). In the genome of the related microorganism, Thermus scotoductus SA-01 (29), the DNA region corresponding to TTC0793 to TTC0805 in T. thermophilus HB27 is present in the opposite direction, and an open reading frame (ORF) corresponding to TTC0795 is located downstream of TTC0806, with two intervening ORFs encoding a putative TRAP transporter and its cognate periplasmic substrate-binding protein (see Fig. S2). In another related microorganism, Marinithermus hydrothermalis (30), three ORFs, encoding MP, NBD, and substrate-binding protein, which are absent from T. thermophilus, intervened between ORFs corresponding to TTC0795 and TTC0806 (see Fig. S2). We speculate that gene inversion occurred in T. thermophilus HB27 after separation of T. thermophilus from the ancestral species. ABC transporters are composed of two MPs and two NBDs. We assume that AecT-I comprises two TTC0795 subunits and two TTC0806 subunits using TTC0807 as the periplasmic substrate-binding protein; however, there is still a possibility that other unidentified MP and NBD species are cocomponents of AecT-I with TTC0795 and TTC0806.
TTC0806 formed a gene cluster with TTC0807, and the ΔTTC0806 strain grew with a profile similar to that of the ΔTTC0807 strain. These observations suggest that TTC0806 and TTC0807 are used as mechanisms in the same AEC uptake system. In the pulldown assay, although interactions between TTC0806 and TTC0795 were observed, interactions between TTC0806 and TTC0807 were not detected. We assume that the periplasmic region of TTC0806, a membrane protein, cannot form an appropriate structure that can be recognized by TTC0807 without a membrane.
ΔTTC0807 and ΔTTC0969 single-site mutants were able to grow in MM containing 500 μM AEC; however, these mutants grew very slowly, which contrasts with the ΔTTC0807 ΔTTC0969 double mutant, which was capable of growing rapidly with a growth rate similar to that of AT10. These observations suggest that both transporting systems are involved in AEC uptake. The growth profiles of ΔTTC0807 and ΔTTC0969 strains were compared, and ΔTTC0807 strains entered the logarithmic phase before 40 h of cultivation, whereas the ΔTTC0969 strain required a longer incubation time to enter the logarithmic phase. Considering that ΔTTC0807 and ΔTTC0969 strains still possess native AecT-II and AecT-I systems, respectively, to take up AEC, we assume that AecT-I possesses a stronger ability than AecT-II to import AEC into the cytoplasm.
It is known that substrate binding causes distinct rigid body motion to close the substrate-binding pocket by bond rotation in DCR in the periplasmic substrate-binding protein (21). Since crystals of TTC0807 have not yet been obtained in an apo form, we cannot infer whether such a marked structural change is induced upon substrate binding in TTC0807. In other periplasmic substrate-binding proteins, such as StLAO-BP and GsArtJ, DCR2 forms an α-helix (α8 in StLAO-BP) that is flanked by β strands β11 and β12 (Fig. 7B and C). In contrast, in TTC0807, DCR2 takes the form of a β strand to form a long and stable β-sheet with β8 from DCR1 in every substrate-binding complex. Since Glu191 from DCR2 forms a stable salt bridge with the α-amino group of the substrates, we assume that substrate binding will induce domain closure, triggered by the displacement of Glu191, which will be elucidated experimentally in the near future.
In this study, we identified two ABC transporter-mediated AEC uptake systems in T. thermophilus and determined the crystal structures of TTC0807, a periplasmic basic amino acid-binding protein, for AecT-I. We think that these AEC uptake systems contribute to the hypersensitivity of T. thermophilus to AEC. However, we also think that it is necessary to identify a cytoplasmic molecular target of AEC other than HCS to clarify the hypersensitivity.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported in part by a Grant-in-Aid for Scientific Research from the Ministry of Education, Culture, Sports, Science and Technology, Japan, and the Asahi Glass Foundation.
Footnotes
Published ahead of print 21 June 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JB.00202-13.
REFERENCES
- 1.Grundy FJ, Lehman SC, Henkin TM. 2003. The L box regulon: lysine sensing by leader RNAs of bacterial lysine biosynthesis genes. Proc. Natl. Acad. Sci. U. S. A. 100:12057–12062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sudarsan N, Wickiser JK, Nakamura S, Ebert MS, Breaker RR. 2003. An mRNA structure in bacteria that controls gene expression by binding lysine. Genes Dev. 17:2688–2697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Shiio I, Miyajima R, Sano K. 1970. Genetically desensitized aspartate kinase to the concerted feedback inhibition in Brevibacterium flavum. J. Biochem. 68:701–710 [DOI] [PubMed] [Google Scholar]
- 4.Ataide SF, Wilson SN, Dang S, Rogers TE, Roy B, Banerjee R, Henkin TM, Ibba M. 2007. Mechanisms of resistance to an amino acid antibiotic that targets translation. ACS Chem. Biol. 2:819–827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kobashi N, Nishiyama M, Tanokura M. 1999. Aspartate kinase-independent lysine synthesis in an extremely thermophilic bacterium, Thermus thermophilus: lysine is synthesized via α-aminoadipic acid, not via diaminopimelic acid. J. Bacteriol. 181:1713–1718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nishida H, Nishiyama M, Kobashi N, Kosuge T, Hoshino T, Yamane H. 1999. A prokaryotic gene cluster involved in synthesis of lysine through the amino adipate pathway: a key to the evolution of amino acid biosynthesis. Genome Res. 9:1175–1183 [DOI] [PubMed] [Google Scholar]
- 7.Tsubouchi T, Mineki R, Taka H, Kaga N, Murayama K, Nishiyama C, Yamane H, Kuzuyama T, Nishiyama M. 2005. Leader peptide-mediated transcriptional attenuation of lysine biosynthetic gene cluster in Thermus thermophilus. J. Biol. Chem. 280:18511–18516 [DOI] [PubMed] [Google Scholar]
- 8.Wulandari AP, Miyazaki J, Kobashi N, Nishiyama M, Hoshino T, Yamane H. 2002. Characterization of bacterial homocitrate synthase involved in lysine biosynthesis. FEBS Lett. 522:35–40 [DOI] [PubMed] [Google Scholar]
- 9.Okada T, Tomita T, Wulandari AP, Kuzuyama T, Nishiyama M. 2010. Mechanism of substrate recognition and insight into feedback inhibition of homocitrate synthase from Thermus thermophilus. J. Biol. Chem. 285:4195–4205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sambrook J, Fritsch EF, Maniatis T. 1989. Molecular cloning: a laboratory manual, 2nd ed Cold Spring Harbor Laboratory Press, Cold Spring Harbor, New York [Google Scholar]
- 11.Koyama Y, Hoshino T, Tomizuka N, Furukawa K. 1986. Genetic transformation of the extreme thermophile Thermus thermophilus and of other Thermus spp. J. Bacteriol. 166:338–340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tanaka T, Kawano N, Oshima T. 1981. Cloning of 3-isopropylmalate dehydrogenase gene of an extreme thermophile and partial purification of the gene product. J. Biochem. 89:677–682 [DOI] [PubMed] [Google Scholar]
- 13.Bierman M, Logan R, O'Brien K, Seno ET, Rao RN, Schoner BE. 1992. Plasmid cloning vectors for the conjugal transfer of DNA from Escherichia coli to Streptomyces spp. Gene 116:43–49 [DOI] [PubMed] [Google Scholar]
- 14.Hoseki J, Okamoto A, Takada N, Suenaga A, Futatsugi N, Konagaya A, Taiji M, Yano T, Kuramitsu S, Kagamiyama H. 2003. Increased rigidity of domain structures enhances the stability of a mutant enzyme created by directed evolution. Biochemistry 42:14469–14475 [DOI] [PubMed] [Google Scholar]
- 15.Nakamura A, Takakura Y, Kobayashi H, Hoshino T. 2005. In vivo directed evolution for thermostabilization of Escherichia coli hygromycin B phosphotransferase and the use of the gene as a selection marker in the host-vector system of Thermus thermophilus. J. Biosci. Bioeng. 100:158–163 [DOI] [PubMed] [Google Scholar]
- 16.Nielsen H, Engelbrecht J, Brunak S, von Heijne G. 1997. Identification of prokaryotic and eukaryotic signal peptides and prediction of their cleavage sites. Protein Eng. 10:1–6 [DOI] [PubMed] [Google Scholar]
- 17.Jez JM, Ferrer JL, Bowman ME, Dixon RA, Noel JP. 2000. Dissection of malonyl-coenzyme A decarboxylation from polyketide formation in the reaction mechanism of a plant polyketide synthase. Biochemistry 39:890–902 [DOI] [PubMed] [Google Scholar]
- 18.Otwinowski Z, Minor W. 1997. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 276:307–326 [DOI] [PubMed] [Google Scholar]
- 19.McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ. 2007. Phaser crystallographic software. J. Appl. Crystallogr. 40:658–674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Collaborative Computational Project Number 4 1994. The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D 50:760–763 [DOI] [PubMed] [Google Scholar]
- 21.Oh BH, Pandit J, Kang CH, Nikaido K, Gokcen S, Ames GF, Kim SH. 1993. Three-dimensional structures of the periplasmic lysine/arginine/ornithine-binding protein with and without a ligand. J. Biol. Chem. 268:11348–11355 [PubMed] [Google Scholar]
- 22.Murshudov GN, Vagin AA, Dodson EJ. 1997. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D 53:240–255 [DOI] [PubMed] [Google Scholar]
- 23.Emsley P, Cowtan K. 2004. Coot: model-building tools for molecular graphics. Acta Crystallogr. D 60:2126–2132 [DOI] [PubMed] [Google Scholar]
- 24.Laskowski RA, MacArthur MW, Moss DS, Thornton JM. 1993. PROCHECK: a program to check the stereochemical quality of protein structures. J. Appl. Crystallogr. 26:283–291 [Google Scholar]
- 25.Karpowich N, Martsinkevich O, Millen L, Yuan YR, Dai PL, MacVey K, Thomas PJ, Hunt JF. 2001. Crystal structures of the MJ1267 ATP binding cassette reveal an induced-fit effect at the ATPase active site of an ABC transporter. Structure 9:571–586 [DOI] [PubMed] [Google Scholar]
- 26.Holm L, Kaariainen S, Rosenstrom P, Schenkel A. 2008. Searching protein structure databases with DaliLite v. 3. Bioinformatics 24:2780–2781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Oh BH, Ames GF, Kim SH. 1994. Structural basis for multiple ligand specificity of the periplasmic lysine-, arginine-, ornithine-binding protein. J. Biol. Chem. 269:26323–26330 [PubMed] [Google Scholar]
- 28.Vahedi-Faridi A, Eckey V, Scheffel F, Alings C, Landmesser H, Schneider E, Saenger W. 2008. Crystal structures and mutational analysis of the arginine-, lysine-, histidine-binding protein ArtJ from Geobacillus stearothermophilus. Implications for interactions of ArtJ with its cognate ATP-binding cassette transporter, Art(MP)2. J. Mol. Biol. 375:448–459 [DOI] [PubMed] [Google Scholar]
- 29.Gounder K, Brzuszkiewicz E, Liesegang H, Wollherr A, Daniel R, Gottschalk G, Reva O, Kumwenda B, Srivastava M, Bricio C, Berenguer J, van Heerden E, Litthauer D. 2011. Sequence of the hyperplastic genome of the naturally competent Thermus scotoductus SA-01. BMC Genomics 12:577. 10.1186/1471-2164-12-577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Copeland A, Gu W, Yasawong M, Lapidus A, Lucas S, Deshpande S, Pagani I, Tapia R, Cheng JF, Goodwin LA, Pitluck S, Liolios K, Ivanova N, Mavromatis K, Mikhailova N, Pati A, Chen A, Palaniappan K, Land M, Pan C, Brambilla EM, Rohde M, Tindall BJ, Sikorski J, Goker M, Detter JC, Bristow J, Eisen JA, Markowitz V, Hugenholtz P, Kyrpides NC, Klenk HP, Woyke T. 2012. Complete genome sequence of the aerobic, heterotroph Marinithermus hydrothermalis type strain (T1T) from a deep-sea hydrothermal vent chimney. Stand. Genomic Sci. 6:21–30 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.