Abstract
The role of the multisubunit sodium/proton antiporter (Mrp) of Methanosarcina acetivorans was investigated with a mutant deleted for the gene encoding the MrpA subunit. Antiporter activity was 5-fold greater in acetate-grown versus methanol-grown wild-type cells, consistent with the previously published relative levels of mrp transcript. The rate, final optical density, and dry weight/methane ratio decreased for the mutant versus wild type when cultured with a growth-limiting concentration of acetate. All growth parameters of the mutant or wild type were identical when grown with methanol in medium containing a growth-limiting Na+ concentration of 1.04 M. The lag phase, growth rate, and final optical density for growth of the mutant were suboptimal compared to the wild type when cultured with acetate in medium containing either 0.54 or 1.04 M Na+. The addition of 25 mM NaCl to resting cell suspensions stimulated ATP synthesis driven by a potassium diffusion potential. ATP synthesis was greater in wild-type than mutant cells grown with acetate, a trend that held for methanol-grown cells, albeit less pronounced. Both sodium and proton ionophores reduced ATP synthesis in the wild type grown with either substrate. The results indicated that the Mrp complex is essential for efficient ATP synthesis and optimal growth at the low concentrations of acetate encountered in the environment.
INTRODUCTION
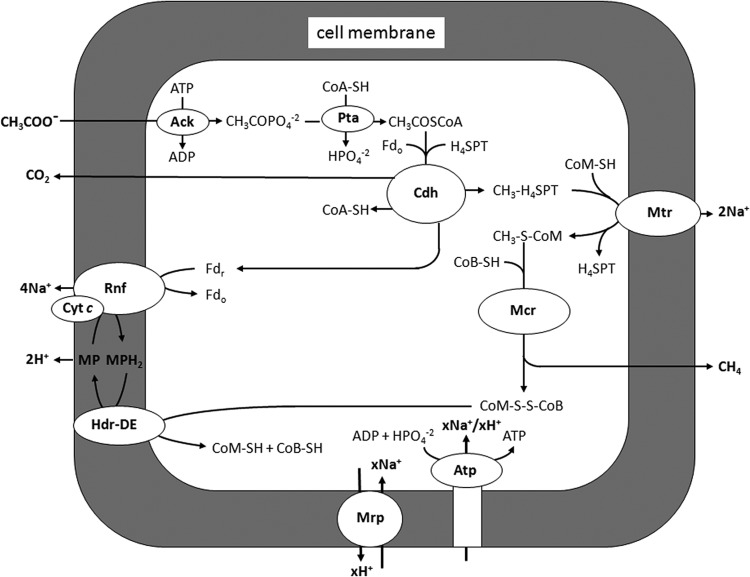
Acetate is the major source of biological methane in both freshwater and marine environments (1, 2). Only two genera (Methanosarcina and Methanosaeta) of acetate-utilizing methane-producing microbes are known, of which Methanosarcina species have been researched to a greater extent. Most investigations have focused on Methanosarcina barkeri and Methanosarcina mazei, for which electron transport in the pathway of acetate conversion to methane is dependent on the production and consumption of H2, although the majority of Methanosarcina species are unable to metabolize H2 (3). On the other hand, all Methanosarcina species investigated transfer the methyl group of acetate to methane similarly, beginning with the CO dehydrogenase/acetyl coenzyme A (CoA) complex (Cdh), which cleaves activated acetate into methyl and carbonyl groups (4). The Cdh transfers the methyl group to tetrahydrosarcinapterin (THSPT), followed by transfer from CH3-THSPT to coenzyme M (HS-CoM), a reaction catalyzed by a membrane complex (Mtr). The exothermic reaction producing CH3-S-CoM is coupled to generation of a Na+ gradient with the potential to drive ATP synthesis. A methylreductase (Mcr) catalyzes reduction of the methyl group of CH3-S-CoM to methane, with electrons donated by coenzyme B (HS-CoB). The heterodisulfide CoM-S-S-CoB, a product of the CH3-S-CoM demethylation reaction, is reduced to the active sulfhydryl forms of the cofactors by heterodisulfide reductase (HdrDE). The two electrons required for this reduction are derived from oxidation of the carbonyl group of acetate, which is catalyzed by the Cdh for which ferredoxin (Fd) is the electron acceptor.
Transfer of electrons from Fd to HdrDE involves a membrane-bound electron transfer chain remarkably different in H2-utilizing M. barkeri and M. mazei compared to the majority of Methanosarcina species, which are unable to metabolize H2. In M. barkeri and M. mazei, Fd donates electrons to a hydrogenase complex (Ech) that produces H2 and generates a proton gradient for ATP synthesis (5–8). A hypothesis has been advanced wherein H2 is reoxidized by another membrane-bound hydrogenase (Vho), depositing protons outside the cell membrane and transferring electrons to methanophenazine (MP), a quinone-like electron carrier (9). Finally, the reduced MP donates electrons to HdrDE, which reduces CoM-S-S-CoB to HS-CoM and HS-CoB and is accompanied by translocation of protons, which further contributes to ATP synthesis.
An electron transport chain has been proposed for Methanosarcina acetivorans, the only non-H2-metabolizing Methanosarcina species for which the genome has been sequenced. The genome does not encode Ech hydrogenase (10), further excluding H2 in electron transport. In contrast to M. barkeri and M. mazei, M. acetivorans synthesizes a six-subunit complex (Rnf) with high identity to membrane-bound Rnf (Rhodobacter nitrogen fixation) complexes from the domain Bacteria (11). The genes (MA0657-MA0664), encoding the Rnf complex of M. acetivorans, are cotranscribed with a gene (MA0658) that encodes a heme cytochrome c and another gene (MA0665) that encodes a hypothetical membrane integral protein with unknown function (11). The cytochrome c is synthesized at high levels in acetate-grown cells, where it dominates the UV-visible spectrum of purified membranes (11). Reduction of purified membranes from acetate-grown cells with Fd leads to reduction of the cytochrome c, which is reoxidized by the addition of either CoM-S-S-CoB or an analog of MP, 2-hydoxyphenazine (12). Reduced 2-hydoxyphenazine is reoxidized by membranes, and this is dependent on the addition of CoM-S-S-CoB, which implies the translocation of protons. Furthermore, it has been reported that a Δ(MA0658-MA0665) mutant of M. acetivorans fails to grow with acetate as the sole substrate (13). The combined evidence supports a H2-independent membrane-bound electron transport chain originating with Fd and culminating with reduction of CoM-S-S-CoB that involves the Rnf complex, cytochrome c, and MP. More recently, it was reported that inverted membrane vesicles of M. acetivorans catalyze Na+ transport coupled to the oxidation of Fd and reduction of CoM-S-S-CoB (14). It was further reported that a Δrnf mutant is unable to grow with acetate and that Na+ transport coupled to Fd:CoM-S-S-CoB oxidoreductase activity of membranes is abolished (14), which supports a role for the Rnf complex in generation of a Na+ gradient supplementing the Na+ gradient generated by Mtr. Thus, it is anticipated that both Na+ and H+ gradients are generated during acetate-dependent growth of M. acetivorans. This conjecture is consistent with the concurrently coupled translocation of both Na+ and H+ by the ATP synthase of M. acetivorans (15, 16); however, the Na+/H+ ratio for optimal ATP synthesis is yet to be investigated.
The genome of M. acetivorans encodes a homolog of the multisubunit Na+/H+ antiporter Mrp, which was first characterized from species in the domain Bacteria. The homolog is upregulated in M. acetivorans in response to growth with acetate (11, 17), a result consistent with an Mrp requirement for optimal acetotrophic growth. Mrp homologs from species in the domain Bacteria function in Na+ resistance, pH homeostasis of alkaliphiles, bile salt resistance, and energy-yielding metabolism (18–22). Although Mrp homologs are encoded in the genomes of species from the domain Archaea (18), none has been investigated. Here, we report the phenotypic characteristics of a ΔmrpA mutant of the archaeon M. acetivorans that indicate an essential role for MrpA in efficient ATP synthesis and optimal growth at the low concentrations of acetate encountered in the environment.
MATERIALS AND METHODS
Cell growth.
M. acetivorans C2A (DSM 2834) was previously isolated from marine sediment in the Summer Branch of Scripps Canyon near La Jolla, CA (23). Both the wild type and ΔmrpA mutant, described below, were cultured at 37°C in marine medium (0.54 M total Na+) containing, per liter: 25.4 g NaCl, 3.8 g NaHCO3, 1.8 g KCl, 11 g MgCl2, 0.2 g CaCl2, 1 g NH4Cl, 0.5 g cysteine. The medium was supplemented with vitamin and mineral solutions (24) at 0.0001% (wt/vol), resazurin as a redox indicator, and 100 mM acetate or 100 mM methanol as growth substrate where indicated. The medium was prepared anaerobically in an atmosphere that contained N2-CO2 (4:1), generated by a modification of the Hungate technique (25), and to which was added 10 ml of 2.5% (wt/vol) Na2S per liter. All gases were passed through a column of reduced copper turnings at 350°C to remove traces of oxygen. The pH was adjusted to 6.8 as previously described (11, 23). Media with different concentrations of total Na+ were prepared by adjusting the NaCl concentration. Media with different concentrations of sodium acetate were maintained at 0.54 M Na+ by adjusting the NaCl concentration. Media with different pHs (7.5, 8.0, and 8.5) were buffered with NaHCO3 (45 mM), HEPES (45 mM), or bicine (45 mM), respectively, and maintained at 0.54 M total Na+ by adjusting the NaCl concentration. All growth experiments used inocula grown under standard conditions of 0.54 M NaCl and pH 6.8.
Construction of a ΔmrpA deletion in M. acetivorans.
The gene encoding M. acetivorans mrpA (MA4572) was PCR amplified approximately 300 bp upstream and downstream of the structural gene with primers 236 (5′-GCTTGATACTGAAGATGCGTCCAG-3′) and 237 (5′-CAAACAGACCTTCGCGATTGT-3′). A reaction was set up with an Invitrogen AmpliTaq kit using 1× PCR buffer II, 1.5 mM MgCl2, 1.6 mM deoxynucleoside triphosphate mix, 1 μg M. acetivorans C2A genomic DNA, 2 pmol of each primer, 2.5 U AmpliTaq DNA polymerase (Invitrogen) in a 50-μl total volume. Conditions for PCR were as follows: initial denaturation at 94°C for 5 min, followed by 30 amplification cycles of 94°C for 30 s, 55°C for 1 min, and 72°C for 3 min. The PCR product was ligated into the PCR 4.0 cloning vector (Invitrogen TOPO cloning kit) to generate pEA 157. A serCP::proC cassette from pJK88 (26) PCR amplified with primers 253 (5′-CCCTCTAGAAGCTTGCGATCTCAACC-3′; HindIII site underlined) and 254 (5′-CGTAACCGCACCGAAGACGCGCCATGAC-3′; BbsI site underlined) as described above was ligated into the unique HindIII and BbsI restriction sites in mrpA, which deleted 1,331 bp of the mrpA gene, generating pEA163. The deletion insert was PCR amplified with primers 236 and 237, and the product (4 μg) was used as the DNA transformed into M. acetivorans WWM24 strain as described previously (27). Transformants were selected for growth on solidified medium without proline and screened for PCR amplification of product with primers 265 (CCAAACTAAACCTCTTGATCCACAGTTTTAATGG) and 269 (CACCGGGAGGCACCACCATTGAAGC) as described above, which included the mrpAproC junction (28). The correct constructs were confirmed in four clones by sequencing, and one clone (KSC63) was used for further experiments.
Antiporter assays.
Proton uptake in cells preloaded with NaCl, LiCl, or KCl was determined as described previously (29) with modifications. Briefly, cells grown to stationary phase in a 100-ml culture were harvested and washed twice with 50 ml of buffer A containing 15 mM MgCl2, 3.0 mM CaCl2, 1.7 mM cysteine, 15 mM NH4Cl, 0.5 mM Na2HPO4, 0.01% (wt/vol) Na2S, and 0.0001% resazurin in 5 mM HEPES (pH 7.5). The cells were resuspended in the same buffer except with 2.5 mM Tris (pH 7.8) substituted for HEPES (buffer B). The cell suspension (12 to 15 mg of protein/ml) was incubated with 0.2 M NaCl, LiCl, or KCl for 2 h under a N2 atmosphere and then washed and resuspended in 10 ml of buffer B. The cell suspension was gently stirred at 125 rpm on a rotary shaker under aerobic conditions at 37°C. An HCl pulse was added (2,000 nmol of H+), and realkalinization of the medium was recorded.
ATP synthesis driven by a K+ diffusion potential.
ATP synthesis was determined under anaerobic conditions (75% N2, 20% CO2, and 5% H2 atmosphere) as described previously (30) with modifications. Briefly, cell suspensions (0.9 ml) containing 0.5 to 1.0 mg of protein were incubated at 25°C for 3 min in 25 mM NaCl in the presence or absence of the protonophore carbonyl cyanide-m-chlorophenyl-hydrazone (CCCP) or the sodium ionophore ETH157. Valinomycin was added to a final concentration of 50 μM, and after 2 min the reaction was stopped by addition of precooled perchloric acid at a 3% (vol/vol) final concentration. The ATP concentration was determined as previously described (31).
Analytical methods.
Culture densities were determined spectrophotometrically at 600 nm (1-cm path length). For dry weight determinations, 10 to 30 ml of cells was collected on a preweighed 0.22-μm-pore-size filter (Millipore). The filters containing cells were washed twice with fresh medium without growth substrate and dried at 80 to 85°C overnight before weighing. Protein was determined by the Biuret method (32) using bovine serum albumin as the standard. The methane concentration was determined by gas chromatography as described previously (33).
RESULTS
Growth parameters of the wild type versus ΔmrpA mutant of M. acetivorans.
Roles for Mrp of M. acetivorans of the domain Archaea were addressed by comparing growth parameters of the wild type versus the ΔmrpA mutant cultured with either methanol or acetate. The MrpA subunit of the Mrp complex was targeted, since it had been shown that the homologous MrpA of Bacillus subtilis is essential for Na+/H+ exchange activity and the proposed site of Na+ translocation (21, 34, 35).
Figure 1 shows similar growth rates and final optical densities between the wild type and mutant grown with methanol, although the lag phase for both strains was lengthened when cultured at pH 8.0 compared to pH 6.8, indicating a required adaptation period for optimal growth at the higher pH for both the mutant and wild type. These results indicated MrpA is not essential for maintaining pH homeostasis for optimal growth when M. acetivorans is cultured with methanol. The lag phase of wild-type and mutant strains grown with 100 mM acetate was prolonged (Fig. 2) relative to that for methanol-grown cells (Fig. 1). However, in contrast to methanol-grown strains (Fig. 1), the mutant reached a final optical density that was lower than the wild type, with similar differences at both pHs. These results suggested that, although not important for pH homeostasis, MrpA is important for optimal growth with 100 mM acetate.
Fig 1.
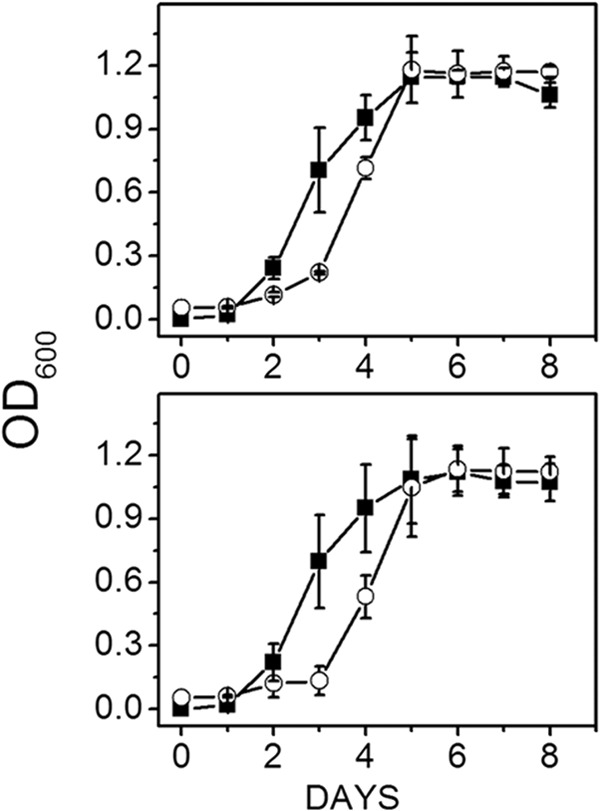
Effect of pH on growth of wild-type versus ΔmrpA mutant strains of M. acetivorans cultured with methanol and 0.54 M Na+. Data shown are the means ± standard deviations of four or five replicate experiments. Top panel, wild type; bottom panel, ΔmrpA mutant. Symbols: ■, pH 6.8; ○, pH 8.0. The final optical densities for the wild type and mutant at pH 6.8 were 1.11 ± 0.06 and 1.12 ± 0.09, respectively. The final optical densities for the wild type and mutant at pH 8.0 were 1.17 ± 0.09 and 1.12 ± 0.07, respetively.
Fig 2.
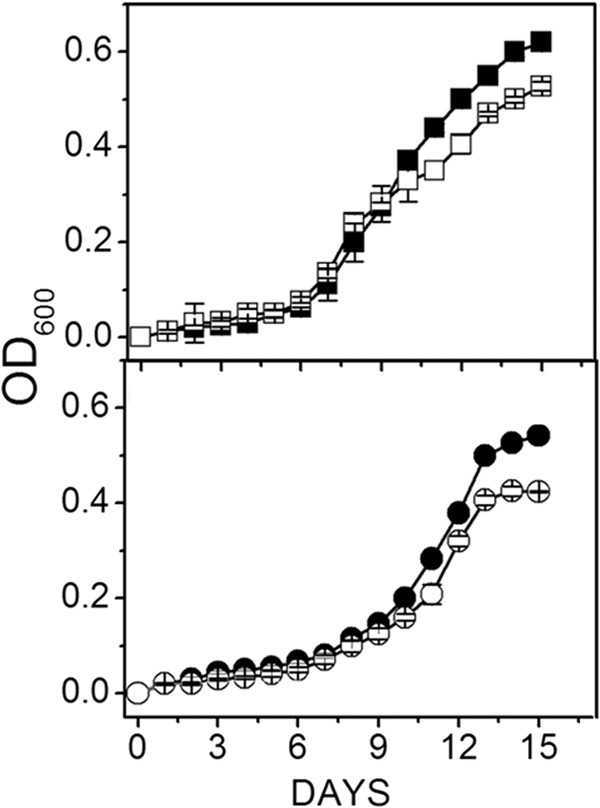
Effect of pH on growth of wild-type versus ΔmrpA mutant strains of M. acetivorans cultured with acetate and 0.54 M Na+. Data shown are the means ± standard deviations of four or five replicate experiments. (Top panel) Results at pH 6.8. Symbols: ■, wild type; □, ΔmrpA mutant. (Bottom panel) Results at pH 8.0. Symbols: ●, wild type; ○, ΔmrpA mutant. The final optical densities for the wild type and mutant at pH 6.8 were 0.62 ± 0.01 and 0.54 ± 0.01, respectively. The final optical densities for the wild type and mutant at pH 8.0 were 0.53 ± 0.01 and 0.42 ± 0.00, respectively.
Figure 3 shows similar growth rates and final optical densities for both the wild-type and mutant strains cultured at pH 6.8 for optimal growth of the wild type and either 0.54 M Na+, within the salinity range for optimal growth of the wild type, or the less-optimal 1.04 M Na+ (23). However, the lag phase for both strains was lengthened when cultured with 1.04 M Na+ compared to 0.54 M Na+, indicating a required adaptation period for optimal growth at the higher salinity for both mutant and wild type. The results indicated that MrpA is not essential to protect against salt stress when cells are cultured with methanol. Figure 4 shows that the final optical density of the mutant was only slightly less than for the wild type when both were cultured with acetate and 0.54 M Na+. When cultured with 1.04 M Na+, the lag phase of the wild type was lengthened, similar to methanol-grown cultures, and the final optical density was significantly less than when cultured with 0.54 M Na+, indicating stress at the higher concentration of Na+. In contrast to methanol-grown cultures, all growth parameters (lag phase, growth rate, and final optical density) of the mutant were suboptimal relative to wild type, suggesting a role for MrpA in combating Na+ stress when cultured with 100 mM acetate.
Fig 3.
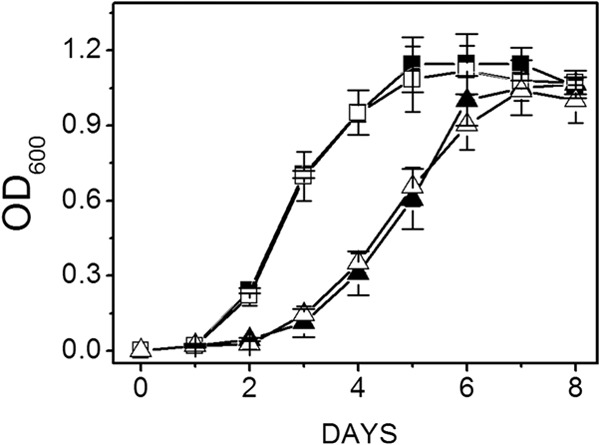
Effect of the Na+ concentration on growth of wild-type and mutant strains cultured with methanol at pH 6.8. Data shown are the means ± standard deviations of four replicate experiments. Filled symbols, wild type; open symbols, ΔmrpA mutant; squares, 0.54 M Na+; triangles, 1.04 M Na+.The final optical densities for the wild type and mutant cultured with 0.54 M Na+ were 1.12 ± 0.02 and 1.09 ± 0.08, respectively. The final optical densities for the wild type and mutant cultured with 1.04 M Na+ were 1.11 ± 0.03 and 1.11 ± 0.05, respectively.
Fig 4.
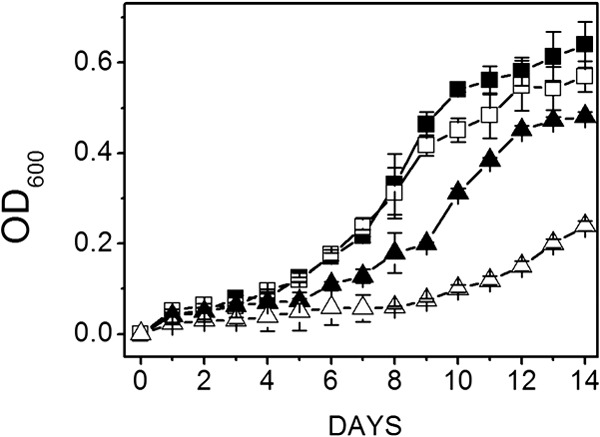
Effect of Na+ concentration on growth of wild-type and mutant strains cultured with acetate at pH 6.8. Data shown are the means ± standard deviations of four replicate experiments. Filled symbols, wild type; open symbols, ΔmrpA mutant; squares, 0.54 M Na+; triangles, 1.04 M Na+. The final optical densities for the wild type and mutant cultured with 0.54 M Na+ were 0.64 ± 0.05 and 0.48 ± 0.01, respectively. The final optical densities for the wild type and mutant cultured with 1.04 M Na+ were 0.48 ± 0.01 and 0.57 ± 0.03, respectively.
Figure 5 compares growth parameters of the wild type and ΔmrpA mutant cultured with a starting concentration of 10 mM acetate in medium buffered at pH 6.8 and containing 0.54 M Na+. The growth rate and final optical density were lower for the mutant and wild type compared with growth in medium with a starting concentration of 100 mM (Fig. 2). As the concentration of acetate steadily decreased in batch culture, the results indicated that concentrations below 10 mM were growth limiting. Furthermore, the growth rate and final optical density were substantially lower for the mutant versus wild type cultured with 10 mM acetate (Fig. 5). This trend held for wild-type and mutant strains cultured with 5 mM acetate, except the difference between growth parameters for the wild-type versus mutant were more pronounced (Fig. 6). These results establish the importance of MrpA for growth of M. acetivorans with low, growth-limiting concentrations of acetate. When cultured with 10 mM acetate, the wild type and mutant produced approximately the same amount of methane, close to the 1.0 mmol expected for 100-ml cultures (Table 1). However, the dry weight/methane ratio was significantly lower for the mutant versus the wild type, a result indicating that MrpA is important for efficient coupling of growth and methanogenesis with low concentrations of acetate that limit growth.
Fig 5.
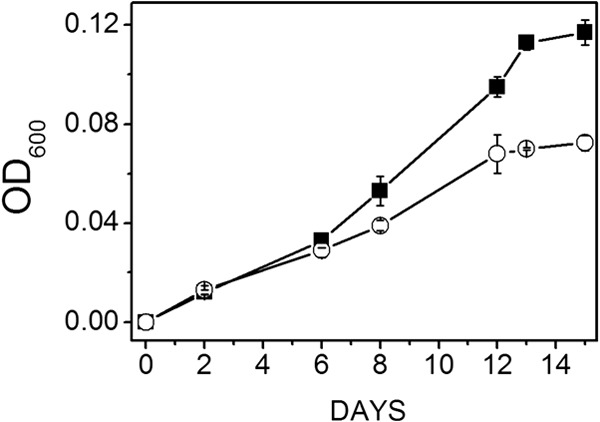
Growth profiles of wild-type versus ΔmrpA mutant strains of M. acetivorans cultured with growth-limiting 10 mM acetate. The starting pH was 6.8, and the Na+ concentration was 0.54 M. Data shown are the means ± standard deviations of three replicate experiments. Symbols: ■, wild type; ○, ΔmrpA mutant. The final optical densities for the wild type and mutant were 0.12 ± 0.01 and 0.07 ± 0.00, respectively.
Fig 6.
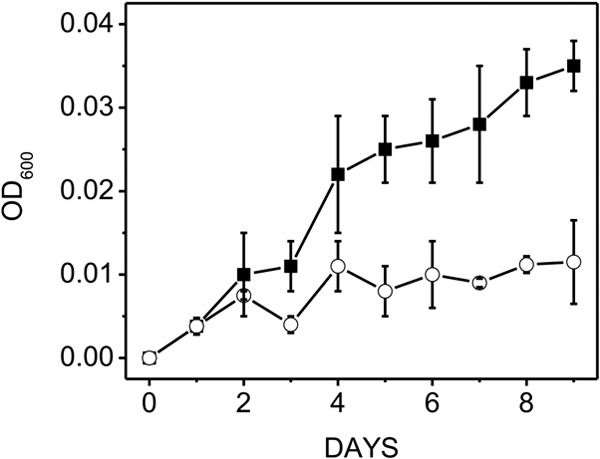
Growth profiles of wild-type versus ΔmrpA mutant strains of M. acetivorans cultured with growth-limiting 5 mM acetate. The starting pH was 6.8, and the Na+ concenration was 0.54 M. Data shown are the means ± standard deviations of four replicate experiments. Symbols: ■, wild type; ○, ΔmrpA mutant. The final optical densities for the wild type and mutant were 0.035 ± 0.003 and 0.012 ± 0.005, respectively.
Table 1.
Growth parameters for wild-type and ΔmrpA mutant strains of M. acetivorans cultured with growth-limiting 10 mM acetatea
| Parameter | Wild type | Mutant |
|---|---|---|
| Optical density (600 nm) | 0.12 ± 0.05a | 0.07 ± 0.03 |
| CH4 (mmol) | 1.09 ± 0.04 | 0.87 ± 0.10 |
| Dry wt (mg) | 2.58 ± 0.23 | 1.5 ± 0.25 |
| Dry wt/CH4 | 2.3 | 1.7 |
| Dry wt/optical density | 21.5 | 21.4 |
Values are the means ± standard deviations of three replicate experiments. Data were collected at the beginning of the stationary phase of growth (15 days). Dry weight and methane values and the dry weight/methane ratio for the mutant were significantly different from those for the wild type (P < 0.05).
Sodium/proton antiporter activity in wild-type versus ΔmrpA mutant strains of M. acetivorans cultured with either acetate or methanol.
An investigation of Na+/H+ antiporter activity for the Mrp complex of M. acetivorans was prompted by the results that indicated that MrpA is important for efficient coupling of growth with methanogenesis when cultured with acetate. Antiporter activity in cell suspensions was determined by monitoring the rate of Na+-dependent alkalinization following an acid pulse (Table 2). Wild-type acetate-grown cells showed 5-fold greater activity than wild-type methanol-grown cells, independent of which salt was preloaded. However, acetate-grown cells preloaded with NaCl showed greater activity than with KCl. Furthermore, Li+ replaced Na+ as the preferred counterion. These results are characteristic of Na+/H+ antiporter activity (29, 36–38) and confirm that acetate-grown cells have a greater capacity for this activity versus methanol-grown cells. The previously reported 5-fold-greater levels of mrpA transcript and Mrp subunits in acetate-grown versus methanol-grown M. acetivorans (11, 17) reflect the relative Na+/H+ exchange rates (Table 2), a result indicating that the majority of exchange activity in acetate-grown cells is attributable to MrpA and the Mrp complex. This role was further supported by the greater exchange rates observed between wild-type and ΔmrpA mutant strains grown with acetate (Table 2). The coordinated increase in mrpA transcript and Mrp subunits indicates that the regulation of protein abundance drives the observed antiporter activity, and not a change in the activity of a fixed level of MrpA and the Mrp complex.
Table 2.
Antiporter activity of wild-type versus ΔmrpA mutant strains of M. acetivoransa
| Preloaded salt | Antiporter activity (mean ± SD) when grown on: |
|||
|---|---|---|---|---|
| Acetate |
Methanol |
|||
| Wild type | Mutant | Wild type | Mutant | |
| NaCl | 1.01 ± 0.09 | 0.39 ± 0.03 | 0.21 ± 0.02 | 0.17 ± 0.01 |
| LiCl | 1.04 ± 0.13 | 0.37 ± 0.04 | 0.18 ± 0.02 | 0.19 ± 0.02 |
| KCl | 0.56 ± 0.04 | 0.38 ± 0.05 | 0.13 ± 0.01 | 0.2 ± 0.03 |
The units for antiporter activity are μmole per second × pH units × grams (dry weight). Values are from three replicate experiments. In all comparisons to acetate-grown wild-type cells preloaded with NaCl, except for acetate-grown wild-type cells preloaded with LiCl, values were significantly different (P < 0.05).
ATP synthesis.
The results reported here indicate that MrpA is important for the efficient coupling of growth with methanogenesis. This role for the Mrp complex was further investigated with resting cell suspensions of M. acetivorans synthesizing ATP driven by an artificial ΔΨ generated by valinomycin-induced potassium efflux. ATP synthesis increased with time in cell suspensions of the wild type and ΔmrpA mutant grown with either acetate or methanol (data not shown). ATP synthesis was dependent on increasing concentrations of NaCl for all cell suspensions, with a maximum achieved at 25 mM (data not shown). Table 3 shows the mean ATP levels obtained for five replicate experiments in which cell suspensions contained 25 mM NaCl. The results indicate wild-type acetate-grown cells have approximately 2-fold higher activity than wild-type methanol-grown cells, which is consistent with the 3-fold increase of ATP synthase in acetate-grown versus methanol-grown M. acetivorans (11). ATP synthesis was stimulated by the addition of 25 mM NaCl in cells grown with either substrate. Additions of either the protonophore CCCP or the Na+ ionophore ETH125 reduced ATP synthesis for the wild type grown with either substrate, a result consistent with the M. acetivorans ATP synthase translocating both Na+ and H+ (15). Most importantly, ATP synthesized in the acetate-grown ΔmrpA mutant was significantly lower than for the acetate-grown wild type. This trend held for acetate-grown cell suspensions containing CCCP or ETH157. These results, together with results indicating MrpA is important for coupling growth and methanogenesis, indicate MrpA plays a role in the synthesis of ATP by acetate-grown M. acetivorans. The difference in ATP synthesized for methanol-grown wild-type and ΔmrpA mutant strains was substantially less than for the acetate-grown strains, a result supporting the proposed role for MrpA, considering that methanol-grown cells of M. acetivorans contain 5-fold less mrpA transcript levels than acetate-grown cells (17). Furthermore, the coordinated increase in mrpA transcript and Mrp subunits indicates that the regulation of abundance of the Mrp complex is responsible for the difference in ATP synthesis and not a change in the activity of a fixed level of the complex.
Table 3.
ATP synthesis in resting cell suspensions driven by a potassium diffusion potentiala
| Amendment | ATP synthesis on growth substrate |
|||
|---|---|---|---|---|
| Acetate |
Methanol |
|||
| Wild type | ΔmrpA mutant | Wild type | ΔmrpA mutant | |
| None | 3.7 ± 0.5 | 1.63 ± 0.28 | 2.4 ± 0.18 | 2.44 ± 0.3 |
| 25 mM NaCl | 6.7 ± 0.32 | 1.93 ± 0.2 | 3.57 ± 0.22 | 2.82 ± 0.1 |
| 25 mM NaCl + 25 μM CCCP | 3.06 ± 0.05 | 1.22 ± 0.19 | 2.1 ± 0.13 | 1.7 ± 0.13 |
| 25 mM NaCl + 60 μM ETH157 | 3.7 ± 0.22 | 1.7 ± 0.1 | 2.41 ± 0.33 | 2.39 ± 0.3 |
Values are nmoles of ATP produced per milligram of protein, determined 2 min after the addition of 75 μM valinomycin to buffered cell suspensions containing the indicated amendments. Values are the means ± standard errors of five replicate experiments.
DISCUSSION
This work represents the first investigation of an Mrp complex from the domain Archaea, and the first from several species of methanogens for which the genomes have been annotated for Mrp complexes (39). As expected, MrpA from M. acetivorans was shown to be required for Na+/H+ antiporter activity. However, it is not known if MrpA is both necessary and sufficient for activity. Based on previous investigations of Mrp complexes from the domain Bacteria, it is likely that other subunits of the 7-subunit M. acetivorans Mrp complex are necessary to catalyze Na+/H+ antiporter activity, and loss of MrpA results in incomplete assembly of the Mrp complex necessary for activity, as proposed for the homologous Mrp complex from B. subtilis (35, 40). Indeed, it has been proposed that MrpA channels Na+, whereas MrpD channels H+ in the complex from B. subtilis (21).
The results obtained with methanol-grown M. acetivorans indicated that the Mrp complex is not required for pH homoeostasis or tolerance to Na+ stress. However, the results indicated that MrpA is essential for optimal ATP synthesis and growth of M. acetivorans with acetate. A physiological role is envisioned for the Mrp complex of M. acetivorans in adjusting the Na+/H+ ratio optimally for ATP synthesis by the ATP synthase and thereby increasing the thermodynamic efficiency of acetate conversion to methane. The proposed role is based on results presented here, recent genetic evidence that the Rnf complex generates a high external Na+ gradient (14), and that activity of the ATP synthase is concurrently coupled to both Na+ and H+ (15). In the pathway of acetate conversion to methane (Fig. 7), Na+ gradients are generated in the Mtr- and Rnf-catalyzed reactions, yielding the previously proposed theoretical stoichiometry of 2 Na+ and 4 Na+, respectively, for a total of 6 Na+ (16). Additionally, the Hdr-catalyzed oxidation of reduced MP generates a gradient with a stoichiometry of 2 H+. We propose that the Mrp complex functions to adjust the 6 Na+/2 H+ ratio optimally for ATP synthesis by the ATP synthase. A mechanism by which the Mrp complex could sense the Na+/H+ ratio is obscure, although a role for subunits other than MrpA and MrpD that channel Na+ and H+ is a possibility. Although the optimal Na+/H+ ratio is yet to be determined, the finding that both CCCP and ETH125 inhibit ATP synthesis is at least consistent with a requirement for both a Na+ and H+ gradient to drive ATP synthesis. The genome of M. acetivorans is annotated with only one other Na+/H+ antiporter (MA2633), for which neither the protein mor gene expression is detectable in either acetate- or methanol-grown cells (17, 41, 42). Clearly, the results presented here warrant a comprehensive characterization of the Mrp complex to determine the Na+/H+ exchange role.
Fig 7.
Pathway for conversion of acetate to methane in M. acetivorans. Ack, acetate kinase; Pta, phosphotransacetylase; CoA-SH, coenzyme A; H4SPT, tetrahydrosarcinapterin; Fdr, reduced ferredoxin; Fdo, oxidized ferredoxin; Cdh, CO dehydrogenase/acetyl-CoA synthase; CoM-SH, coenzyme M; Mtr, methyl-H4SPT:CoM-SH methyltransferase; CoB-SH, coenzyme B; MP, methanophenazine; Hdr-DE, heterodisulfide reductase; Rnf, Rnf complex; Mrp, Mrp complex; Atp, ATP synthase.
The proposed role of the Mrp complex is supported by the finding that growth of the ΔmrpA mutant relative to wild type is impeded to a greater extent as the concentration of acetate is lowered and the free energy available for ATP synthesis is decreased. The standard free energy available via conversion of acetate to methane (ΔG°′ = −36 kJ mol of CH4−1) allows for the synthesis of approximately 1 ATP (ΔG°′ = −31.8 kJ mol−1) (43). Thus, the amount of energy available for acetotrophic growth is marginal at best, especially when considering that 1 ATP is required for activation of acetate to acetyl-CoA in the pathway (Fig. 7). Moreover, it is highly unlikely that standard conditions of equimolar reactants and products prevail in the marine sediment from which M. acetivorans was isolated. Indeed, it has been reported that pore water in a variety of marine sediments contain micromolar amounts of acetate (44), concentrations that severely restrict the amount of energy available for ATP synthesis, which argues in favor of an essential requirement for the Mrp complex during acetotrophic growth of M. acetivorans in its native environment. M. acetivorans was isolated from a submarine canyon containing sediments with a high fraction of organic material that continually accumulated from the littoral zone (23), which results in an estimated fraction of methanogenesis from acetate of between 50 and 70% based on isotopic analysis (2). Since the pH and salinity were not recorded for the site and the time of isolation, the pH (6.8) and salinity level (0.54 M) tested were based on the published values for optimal growth of M. acetivorans (23).
The expression of genes encoding the Mrp complex are downregulated nearly 10-fold in methanol- versus acetate-grown M. acetivorans (17), and growth of M. acetivorans with methanol was unaltered by loss of mrpA, indicating that the Mrp complex has a less important role when grown with methanol than during acetotrophic growth. One explanation is the much greater energy available via methanol conversion to methane (ΔG°′ = −106.5 kJ mol CH4−1) than with acetate (ΔG°′ = −36 kJ mol CH4−1) (45), obviating a strict requirement for Mrp to optimize the thermodynamic efficiency of the ATP synthase. The ΔmrpA mutant was found to be less tolerant to Na+ stress conditions than the wild type, which may be the consequence of energy diverted away from ATP synthesis toward lowering the intracellular concentrations of Na+ exacerbated by a decrease in the thermodynamic efficiency of ATP synthesis in the absence of Mrp.
A joint function for Mrp and Rnf complexes in Na+-dependent chemiosmotic mechanisms of ATP synthesis is supported by strict mutual encoding of the complexes in genomes of Methanosarcinaceae members isolated from marine environments, where Na+-dependent ATP synthesis is a dominant characteristic (46). Genomes of obligate H2-oxidizing CO2-reducing methanogens isolated from either marine or freshwater environments, incapable of acetotrophic or methylotrophic growth, do not encode both Rnf and Mrp complexes (18, 39). These species are incapable of electron transport coupled to generation of ion gradients, negating the requirement for Rnf and Mrp complexes (48).
Nature has evolved diverse Fd:CoM-S-S-CoB oxidoreductase systems in acetotrophic methanogens that generate ion gradients driving ATP synthesis which are different from M. acetivorans and do not require Rnf and Mrp complexes for growth. Although M. barkeri and M. mazei utilize the growth substrate acetate, the genomes of these freshwater species do not encode Rnf and Mrp complexes (49). Instead, unlike M acetivorans and other marine-dwelling members of the Methanosarcinaceae, M. barkeri and M. mazei are capable of metabolizing H2, which is both produced and consumed in the oxidation of Fd and reduction of CoM-S-S-CoB during growth with acetate, yielding a theoretical 5 H+ (16). The H+ gradient coupled with 2 Na+ translocated by Mtr yields a theoretical ratio of 2 Na+/5 H+, compared to 6 Na+/2 H+ for M. acetivorans. Unfortunately, specificity for translocation of Na+ and H+ by the ATP synthase of M. barkeri and M. mazei has not been investigated. Methanosaeta spp. only utilize acetate as a growth substrate and do not metabolize H2 or encode Rnf and Mrp complexes, suggesting yet another mechanism for generating ion gradients that drive ATP synthesis (50). Indeed, it is proposed that Methanosaeta thermophila utilizes a Fd:CoM-S-S-CoB oxidoreductase system comprised of a truncated form of F420H2 dehydrogenase and MP and that generates a H+ gradient driving ATP synthesis (51).
ACKNOWLEDGMENTS
The work was supported by National Science Foundation grant number 0820734 to J.G.F., by the Person Endowment from The Pennsylvania State University to J.G.F., and in part by grants from the U.S. Department of Energy, Office of Science, numbers DE-FG02-93ER20106 and DE-FG02-07ER64502, to K.R.S.
Footnotes
Published ahead of print 8 July 2013
REFERENCES
- 1.Whiticar MJ, Faber E, Schoell M. 1986. Biogenic methane formation in marine and freshwater environments: CO2 reduction vs. acetate fermentation-isotope evidence. Geochim. Cosmochim. Acta 50:693–709 [Google Scholar]
- 2.Jenden PD, Kaplan IR. 1986. Comparison of microbial gases from the Middle America Trench and Scripps Submarine Canyon: implications for the origin of natural gas. Appl. Geochem. 1:631–646 [Google Scholar]
- 3.Guss AM, Kulkarni G, Metcalf WW. 2009. Differences in hydrogenase gene expression between Methanosarcina acetivorans and Methanosarcina barkeri. J. Bacteriol. 191:2826–2833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ferry JG. 2010. How to make a living exhaling methane. Annu. Rev. Microbiol. 64:453–473 [DOI] [PubMed] [Google Scholar]
- 5.Meuer J, Bartoschek S, Koch J, Kunkel A, Hedderich R. 1999. Purification and catalytic properties of Ech hydrogenase from Methanosarcina barkeri. Eur. J. Biochem. 265:325–335 [DOI] [PubMed] [Google Scholar]
- 6.Welte C, Kratzer C, Deppenmeier U. 2010. Involvement of Ech hydrogenase in energy conservation of Methanosarcina mazei. FEBS J. 277:3396–3403 [DOI] [PubMed] [Google Scholar]
- 7.Welte C, Kallnik V, Grapp M, Bender G, Ragsdale S, Deppenmeier U. 2010. Function of Ech hydrogenase in ferredoxin-dependent, membrane-bound electron transport in Methanosarcina mazei. J. Bacteriol. 192:674–678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Meuer J, Kuettner HC, Zhang JK, Hedderich R, Metcalf WW. 2002. Genetic analysis of the archaeon Methanosarcina barkeri Fusaro reveals a central role for Ech hydrogenase and ferredoxin in methanogenesis and carbon fixation. Proc. Natl. Acad. Sci. U. S. A. 99:5632–5637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Welte C, Deppenmeier U. 2011. Proton translocation in methanogens. Methods Enzymol. 494:257–280 [DOI] [PubMed] [Google Scholar]
- 10.Galagan JE, Nusbaum C, Roy A, Endrizzi MG, Macdonald P, FitzHugh W, Calvo S, Engels R, Smirnov S, Atnoor D, Brown A, Allen N, Naylor J, Stange-Thomann N, DeArellano K, Johnson R, Linton L, McEwan P, McKernan K, Talamas J, Tirrell A, Ye W, Zimmer A, Barber RD, Cann I, Graham DE, Grahame DA, Guss AM, Hedderich R, Ingram-Smith C, Kuettner HC, Krzycki JA, Leigh JA, Li W, Liu J, Mukhopadhyay B, Reeve JN, Smith K, Springer TA, Umayam LA, White O, White RH, de Macario EC, Ferry JG, Jarrell KF, Jing H, Macario AJ, Paulsen I, Pritchett M, Sowers KR, Swanson RV, Zinder SH, Lander E, Metcalf WW, Birren B. 2002. The genome of M. acetivorans reveals extensive metabolic and physiological diversity. Genome Res. 12:532–542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li Q, Li L, Rejtar T, Lessner DJ, Karger BL, Ferry JG. 2006. Electron transport in the pathway of acetate conversion to methane in the marine archaeon Methanosarcina acetivorans. J. Bacteriol. 188:702–710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang M, Tomb JF, Ferry JG. 2011. Electron transport in acetate-grown Methanosarcina acetivorans. BMC Microbiol. 11:165. 10.1186/1471-2180-11-165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Buan NR, Metcalf WW. 2010. Methanogenesis by Methanosarcina acetivorans involves two structurally and functionally distinct classes of heterodisulfide reductase. Mol. Microbiol. 75:843–853 [DOI] [PubMed] [Google Scholar]
- 14.Schlegel K, Welte C, Deppenmeier U, Muller V. 2012. Electron transport during aceticlastic methanogenesis by Methanosarcina acetivorans involves a sodium-translocating Rnf complex. FEBS J. 279:4444–4452 [DOI] [PubMed] [Google Scholar]
- 15.Schlegel K, Leone V, Faraldo-Gomez JD, Muller V. 2012. Promiscuous archaeal ATP synthase concurrently coupled to Na+ and H+ translocation. Proc. Natl. Acad. Sci. U. S. A. 109:947–952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schlegel K, Muller V. 2013. Evolution of Na+ and H+ bioenergetics in methanogenic archaea. Biochem. Soc. Trans. 41:421–426 [DOI] [PubMed] [Google Scholar]
- 17.Li L, Li Q, Rohlin L, Kim U, Salmon K, Rejtar T, Gunsalus RP, Karger BL, Ferry JG. 2007. Quantitative proteomic and microarray analysis of the archaeon Methanosarcina acetivorans grown with acetate versus methanol. J. Proteome Res. 6:759–771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Swartz TH, Ikewada S, Ishikawa O, Ito M, Krulwich TA. 2005. The Mrp system: a giant among monovalent cation/proton antiporters? Extremophiles 9:345–354 [DOI] [PubMed] [Google Scholar]
- 19.Dzioba-Winogrodzki J, Winogrodzki O, Krulwich TA, Boin MA, Hase CC, Dibrov P. 2009. The Vibrio cholerae Mrp system: cation/proton antiport properties and enhancement of bile salt resistance in a heterologous host. J. Mol. Microbiol. Biotechnol. 16:176–186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Blanco-Rivero A, Leganes F, Fernandez-Valiente E, Fernandez-Pinas F. 2009. mrpA (all1838), a gene involved in alkali and Na+ sensitivity, may also have a role in energy metabolism in the cyanobacterium Anabaena sp. strain PCC 7120. J. Plant Physiol. 166:1488–1496 [DOI] [PubMed] [Google Scholar]
- 21.Moparthi VK, Kumar B, Mathiesen C, Hagerhall C. 2011. Homologous protein subunits from Escherichia coli NADH:quinone oxidoreductase can functionally replace MrpA and MrpD in Bacillus subtilis. Biochim. Biophys. Acta 1807:427–436 [DOI] [PubMed] [Google Scholar]
- 22.Ito M, Guffanti AA, Krulwich TA. 2001. Mrp-dependent Na+/H+ antiporters of Bacillus exhibit characteristics that are unanticipated for completely secondary active transporters. FEBS Lett. 496:117–120 [DOI] [PubMed] [Google Scholar]
- 23.Sowers KR, Baron SF, Ferry JG. 1984. Methanosarcina acetivorans sp. nov., an acetotrophic methane-producing bacterium isolated from marine sediments. Appl. Environ. Microbiol. 47:971–978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wolin EA, Wolin MJ, Wolfe RS. 1963. Formation of methane by bacterial extracts. J. Biol. Chem. 238:2882–2886 [PubMed] [Google Scholar]
- 25.Miller TL, Wolin MJ. 1974. A serum bottle modification of the Hungate technique for cultivating obligate anaerobes. Appl. Microbiol. 27:985–987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang JK, White AK, Kuettner HC, Boccazzi P, Metcalf WW. 2002. Directed mutagenesis and plasmid-based complementation in the methanogenic archaeon Methanosarcina acetivorans C2A demonstrated by genetic analysis of proline biosynthesis. J. Bacteriol. 184:1449–1454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Metcalf WW, Zhang JK, Apolinario E, Sowers KR, Wolfe RS. 1997. A genetic system for Archaea of the genus Methanosarcina: liposome-mediated transformation and construction of shuttle vectors. Proc. Natl. Acad. Sci. U. S. A. 94:2626–2631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pritchett MA, Zhang JK, Metcalf WW. 2004. Development of a markerless genetic exchange method for Methanosarcina acetivorans C2A and its use in construction of new genetic tools for methanogenic archaea. Appl. Environ. Microbiol. 70:1425–1433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Terracciano JS, Schreurs WJA, Kashket ER. 1987. Membrane H+ conductance of Clostridium thermoaceticum and Clostridium acetobutylicum: evidence for electrogenic Na+:H+ antiport in Clostridium thermoaceticum. Appl. Environ. Microbiol. 53:782–786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mountfort DO, Morschel E, Beimborn DB, Schonheit P. 1986. Methanogenesis and ATP synthesis in a protoplast system of Methanobacterium thermoautotrophicum. J. Bacteriol. 168:892–900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jasso-Chavez R, Moreno-Sanchez R. 2003. Cytosol-mitochondria transfer of reducing equivalents by a lactate shuttle in heterotrophic Euglena. Eur. J. Biochemistry 270:4942–4951 [DOI] [PubMed] [Google Scholar]
- 32.Gornall AG, Bardawill CJ, David MM. 1949. Determination of serum proteins by means of the Biuret reaction. J. Biol. Chem. 177:751–766 [PubMed] [Google Scholar]
- 33.Schauer NL, Ferry JG. 1980. Metabolism of formate in Methanobacterium formicicum. J. Bacteriol. 142:800–807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kajiyama Y, Otagiri M, Sekiguchi J, Kudo T, Kosono S. 2009. The MrpA, MrpB, and MrpD subunits of the Mrp antiporter complex in Bacillus subtilis contain membrane-embedded and essential acidic residues. Microbiology 155:2137–2147 [DOI] [PubMed] [Google Scholar]
- 35.Morino M, Natsui S, Swartz TH, Krulwich TA, Ito M. 2008. Single gene deletions of mrpA to mrpG and mrpE point mutations affect activity of the Mrp Na+/H+ antiporter of alkaliphilic Bacillus and formation of hetero-oligomeric Mrp complexes. J. Bacteriol. 190:4162–4172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hiramatsu T, Kodama K, Kuroda T, Mizushima T, Tsuchiya T. 1998. A putative multisubunit Na+/H+ antiporter from Staphylococcus aureus. J. Bacteriol. 180:6642–6648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schonheit P, Beimborn DB. 1985. Presence of a Na+:H+ antiporter in Methanobacterium thermoautotrophicum and its role in Na+ dependent methanogenesis. Arch. Microbiol. 142:354–361 [Google Scholar]
- 38.Swartz TH, Ito M, Ohira T, Natsui S, Hicks DB, Krulwich TA. 2007. Catalytic properties of Staphylococcus aureus and Bacillus members of the secondary cation/proton antiporter-3 (Mrp) family are revealed by an optimized assay in an Escherichia coli host. J. Bacteriol. 189:3081–3090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Anonymous 2013. The Comprehensive Microbial Resource, data release version 22.0. J. Craig Venter Institute, Rockville, MD: http://cmr.jcvi.org/tigr-scripts/CMR/CmrHomePage.cgi [Google Scholar]
- 40.Morino M, Natsui S, Ono T, Swartz TH, Krulwich TA, Ito M. 2010. Single site mutations in the hetero-oligomeric Mrp antiporter from alkaliphilic Bacillus pseudofirmus OF4 that affect Na+/H+ antiport activity, sodium exclusion, individual Mrp protein levels, or Mrp complex formation. J. Biol. Chem. 285:30942–30950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Li Q, Li L, Rejtar T, Karger BL, Ferry JG. 2005. The proteome of Methanosarcina acetivorans. Part II, comparison of protein levels in acetate- and methanol-grown cells. J. Proteome Res. 4:129–136 [DOI] [PubMed] [Google Scholar]
- 42.Li Q, Li L, Rejtar T, Karger BL, Ferry JG. 2005. The proteome of Methanosarcina acetivorans. Part I, an expanded view of the biology of the cell. J. Proteome Res. 4:112–128 [DOI] [PubMed] [Google Scholar]
- 43.Thauer RK, Jungermann K, Decker K. 1977. Energy conservation in chemotrophic anaerobic bacteria. Bacteriol. Rev. 41:100–180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.King GM. 1991. Measurement of acetate concentrations in marine pore waters by using an enzymatic approach. Appl. Environ. Microbiol. 57:3476–3481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Thauer RK. 1998. Biochemistry of methanogenesis: a tribute to Marjory Stephenson. Microbiology 144:2377–2406 [DOI] [PubMed] [Google Scholar]
- 46.Spring S, Scheuner C, Lapidus A, Lucas S, Glavina Del Rio T, Tice H, Copeland A, Cheng JF, Chen F, Nolan M, Saunders E, Pitluck S, Liolios K, Ivanova N, Mavromatis K, Lykidis A, Pati A, Chen A, Palaniappan K, Land M, Hauser L, Chang YJ, Jeffries CD, Goodwin L, Detter JC, Brettin T, Rohde M, Goker M, Woyke T, Bristow J, Eisen JA, Markowitz V, Hugenholtz P, Kyrpides NC, Klenk HP. 2010. The genome sequence of Methanohalophilus mahii SLP(T) reveals differences in the energy metabolism among members of the Methanosarcinaceae inhabiting freshwater and saline environments. Archaea 2010:690737. 10.1155/2010/690737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Reference deleted.
- 48.Thauer RK, Kaster AK, Seedorf H, Buckel W, Hedderich R. 2008. Methanogenic archaea: ecologically relevant differences in energy conservation. Nat. Rev. Microbiol. 6:579–591 [DOI] [PubMed] [Google Scholar]
- 49.Maeder DL, Anderson I, Brettin TS, Bruce DC, Gilna P, Han CS, Lapidus A, Metcalf WW, Saunders E, Tapia R, Sowers KR. 2006. The Methanosarcina barkeri genome: comparative analysis with Methanosarcina acetivorans and Methanosarcina mazei reveals extensive rearrangement within methanosarcinal genomes. J. Bacteriol. 188:7922–7931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Smith KS, Ingram-Smith C. 2007. Methanosaeta, the forgotten methanogen? Trends Microbiol. 7:150–155 [DOI] [PubMed] [Google Scholar]
- 51.Welte C, Deppenmeier U. 2011. Membrane-bound electron transport in Methanosaeta thermophila. J. Bacteriol. 193:2868–2870 [DOI] [PMC free article] [PubMed] [Google Scholar]