Abstract
Two unusual occurrences of pleural trichomonosis due to a new Tetratrichomonas species previously reported but not named were confirmed. In one patient, Trichomonas tenax and a Tetratrichomonas species were also detected in the oral cavity by molecular methods. We suggest that this new Tetratrichomonas species be named Tetratrichomonas empyemagena.
CASE REPORTS
Case 1 is that of a 54-year-old male from Mexico City with uncontrolled type 2 diabetes who was admitted to the hospital with a history of 1 week of malaise, poor food intake, chills, and fever after developing right thoracic pain, dyspnea, and fatigue; he denied any history of choking or trauma in recent days. Dullness on percussion and decreased breathing sounds were noted over his right chest. His blood pressure was 120/50 mmHg, his heart rate was 120 beats/min, his respiratory rate was 30 breaths/min, his white blood cell count was 45,000/mm3, his platelet count was 565,000/mm3, his glucose level was 503 mg/dl, his albumin level was 1.6 g/dl, and his arterial HCO3 level was 5.6 meq/liter. An initial chest X-ray and computed tomography (CT) scan showed a right pleural effusion that was drained (700 ml), and the pleural fluid was sent for microbiological analysis. Its macroscopic appearance was that of a brownish purulent fluid. Microscopic examination revealed Gram-positive cocci and Gram-negative bacilli, as well as numerous motile and flagellated protozoa that were initially identified as trichomonads because of their morphological characteristics (form, size, and motility). These flagellates were not cultured, and their identification was confirmed by microscopic examination of an empyema fluid smear. Samples were mixed with whole blood at 1:3, spread onto glass slides, air dried, methanol fixed, and stained with Giemsa stain (Fig. 1A and B). Some aliquots of empyema fluid were stored at −20°C for molecular analysis. Bacteriological cultures of the fluid were performed for aerobic and anaerobic microorganisms, yielding Staphylococcus auricularis, but no bacilli were identified until after PCR sequencing of an empyema fluid sample. A Prevotella species was demonstrated by using universal primers for the 16S rRNA gene (1). Antibiotic therapy with metronidazole and piperacillin-tazobactam was administered.
Fig 1.
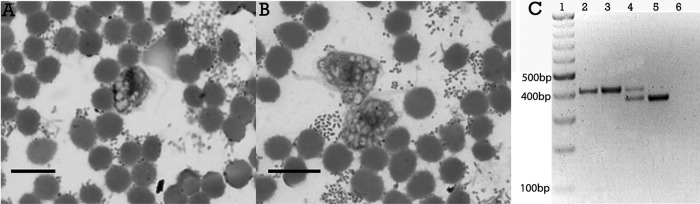
Giemsa-stained smears of empyema fluid. (A) Trichomonads showing piriform and ellipsoidal shapes. (B) Amoeboid forms with some pseudopods. Internal structures such as the axostyle-pelta complex and flagella are not clearly visible; only food vacuoles are evident. In both cases, several bacteria are present. Bars, 14 μm. (C) Ethidium bromide-stained agarose gel showing PCR amplicons of the ITS1-5.8S-rRNA-ITS2 gene of Trichomonadinae. Lanes: 1, 100-bp DNA molecular size marker; 2 and 3, total DNA obtained from empyema fluid samples from cases 1 and 2, respectively; 4, DNA from an oral sample from case 2; 5, DNA from a vaginal specimen with T. vaginalis as a positive control; 6, negative control.
Case 2 is that of a 33-year-old man from Mexico City with a history of smoking and alcohol consumption who was admitted to the hospital with a 2-week nonproductive cough and chills, dyspnea, and left pleural chest pain. His blood pressure was 70/40 mmHg, his heart rate was 150 beats/min, and his respiratory rate was 40 breaths/min. During subsequent days, fever and diaphoresis were added to the clinical course, as well as worsening of dyspnea until he developed acute respiratory failure and septic shock. Thorax auscultation revealed bilateral crackles and diminished breath sounds on the left side; he was placed on mechanical ventilation. A chest X-ray and a thorax CT scan showed a right pneumothorax and left pleural effusion. He underwent bilateral chest tube insertion, and a purulent, brownish, and fetid fluid containing a large number of cells, Gram-positive cocci and bacilli, and Gram-negative rods was drained from the left side. In addition, numerous motile flagellates with the typical appearance of trichomonads were visible in fresh preparations. Culture yielded only Streptococcus sanguinis. A Prevotella species was demonstrated by PCR sequencing as described above. As in case 1, empyema fluid smears were stained with Giemsa stain and some fluid samples were stored at −20°C for molecular analysis. In addition, an oral sample was taken by brushing the teeth and gums with a cytology brush and saline solution. Fresh preparations showed trichomonads with high motility, and remnant samples were frozen for molecular analysis. Treatment with metronidazole and piperacillin-tazobactam was given. In both cases, the trichomonads in fixed samples had an average size of 14.1 ± 2.1 μm long and 9.4 ± 1.1 μm wide.
Molecular analysis.
In order to identify the trichomonad species involved in both cases, DNA was extracted from empyema fluid and from oral lavage saline solution with the Puregene DNA Purification System (Gentra Systems) according to the manufacturer's protocol. The internal transcribed spacer 1 (ITS1)-5.8S rRNA-ITS2 region was amplified with primers described previously (2–4). Negative (human DNA) and positive (vaginal specimen with Trichomonas vaginalis) controls were included. PCR products were separated by agarose gel electrophoresis (Fig. 1C). Unexpectedly, single amplicons of ∼400 bp were observed in both empyema fluids, and two bands of ∼370 and ∼400 bp were seen in the oral sample. The T. vaginalis sample has a size of ∼370 bp. All amplicons were purified with the QIAquick Gel Extraction kit (Qiagen, Hilden, Germany), and both strands were sequenced by a commercial supplier. These sequences were compared with 181 trichomonad ITS1-5.8S rRNA-ITS2 sequences available in the National Center for Biotechnology Information database. Multiple alignments were performed with the CLUSTAL W program v1.8 (5) and manually adjusted with the MEGA program v4 (6, 7), while the ModelTest 3.7 program (8) was used to determine the appropriate model of molecular evolution. The sequences were analyzed with the general time-reversible model with gamma distribution. A phylogenetic reconstruction using Bayesian inference was performed with the program Mr. Bayes 3.1.2 (9–11). The analysis was executed for 1 million generations. Sampling trees were built every 100 generations, and those with scores lower than the stationary phase were discarded, while those that reached the stationary phase were collected and used to build a consensus tree. The phylogenetic inference (Fig. 2) showed that both of the trichomonads found in the lungs of our patients were grouped in a separate clade with high levels of statistical support (a posterior probability of 0.96) with sequences of the new Tetratrichomonas species previously reported by Mantini et al. (4). They were 99% identical, and they were distant from Tetratrichomonas gallinarum and other species of the Tetratrichomonas genus. The sequence of one of the amplicons obtained from the oral sample of case 2 (∼370 bp) clustered with a sequence of Trichomonas tenax, while the other (∼400 bp) was identical to the empyema fluid isolate sequence.
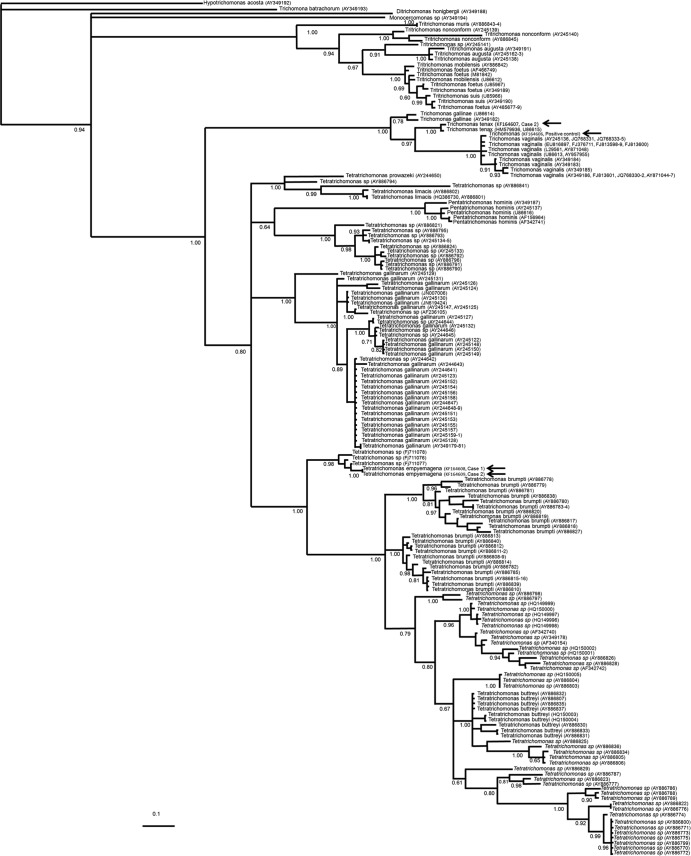
Fig 2.
Bayesian phylogenetic tree of several members of the family Trichomonadinae based on ITS1-5.8S-rRNA-ITS2 sequences. The values at the nodes are posterior percentage probabilities obtained by using 106 generations. GenBank accession numbers are shown in parentheses. The arrows in the center of the tree indicate the new species, placed as an independent clade. T. tenax from the oral sample and T. vaginalis, used as the positive control, were grouped in their respective species clades.
Pulmonary trichomonosis is a rare event; however, some studies published over the last few years indicate that its occurrence is underestimated (12, 13). Indeed, although precise morphological identification guidelines for this parasite are available and generally the typical flagellate form is easily recognized by microbiologists, trichomonads may not be identified even by expert biologists because the parasite is able to present amoeboid forms during adhesion to host cells (14) and sometimes its internal structures are not visible (14, 15).
According to Mallat et al. (3) and Leterrier et al. (13), T. tenax is often suspected to be responsible for pulmonary trichomonosis even though it is a commensal of the human oral cavity, particularly in people with poor oral hygiene and advanced periodontal disease. It has been proposed that trichomonad parasites probably enter the respiratory tract by aspiration from the oropharynx. In addition, the parasite is known to feed on the aspirated bacteria and therefore it does not cause pulmonary disease by itself (3, 16). In case 2, we confirmed the simultaneous presence of T. tenax and a new Tetratrichomonas species in the mouth of the patient and only the presence of the latter in the empyema fluid. Besides, as reported in other cases (2–4, 13), coinfection with trichomonads and bacteria from the oropharyngeal microbiota supports the hypothesis that the source of this parasite was most likely the oral cavity. However, it is not clear why only the new Tetratrichomonas species was identified in the lung; we suggest that T. tenax did not develop in the lungs because of a competitive phenomenon and/or an adverse microenvironment during pleural invasion. Two other interesting features in both cases were the seasonality and the presence of some risk factors in the patients. Our hospital is the only public general concentration hospital in the south of Mexico City, pulmonary trichomonosis was never before identified, and the presence of two similar cases in the same winter season may be a relevant finding about the biology of Trichomonadinae; therefore, epidemiological studies should be undertaken. Proliferation of trichomonads seems to depend on the presence of bacteria such as oral streptococci or anaerobes. Here we confirmed a Prevotella species and cocci in both patients, similar to a previously reported empyema case due to a new Tetratrichomonas species (4); thus, it is probably that these bacteria promote a microenvironment adequate for the establishment and growth of Tetratrichomonas. In addition, both patients had underlying diseases (uncontrolled type 2 diabetes and alcohol abuse, respectively), as generally reported in other cases of pulmonary trichomonosis (13).
The genus Tetratrichomonas can be recognized because it has four anterior flagella, a long posterior flagellum with a free distal end, and a typically discoidal parabasal body, i.e., the Golgi complex with adjacent striated fibrils (17, 18), that can parasitize birds, reptiles, and livestock. On the other hand, ITS1-5.8S rRNA-ITS2 is a good marker for genetic diversity research (19, 20). Molecular analysis performed by Kleina et al. (21) and Cepicka et al. (17) with this molecular marker showed that the genus Tetratrichomonas is more diverse than previously known, with numerous robust host-specific and monophyletic groups that probably represent new species, and because of its wide host range and high level of species specificity, this makes the genus Tetratrichomonas a valuable model for studying parasite evolution. In addition, a DNA sequencing study of the ITS1-5.8S rRNA-ITS2 region from a collection of axenic trichomonad strains isolated from the oral cavities and bronchi of Estonian patients with pulmonary diseases revealed the presence of another novel trichomonad species that belongs to the genus Tetratrichomonas with high similarity to the avian species Tetratrichomonas gallinarum (22).
According to Duboucher et al. (12), it is relevant to alert parasitologists to the potential occurrence of trichomonads, alone or as coinfecting agents, in pulmonary disease milieus and to underline the importance of direct microscopic examination of fresh samples, as well as the usefulness of molecular methods for species identification, because we are certain that new cases of pulmonary trichomonosis caused by this new species will appear. Thus, considering the fact that a new Tetratrichomonas species was identified in two human pleural empyema patients in the present study and previously in another patient but was not named then, we suggest that this new Tetratrichomonas species be named T. empyemagena on the basis of the pathological sign empyema and the Greek suffix gena, meaning origin or production.
Nucleotide sequence accession numbers.
The nucleotide sequences of T. empyemagena, T. tenax, and T. vaginalis obtained in this study have been submitted to GenBank under accession numbers KF164606 to KF164609.
ACKNOWLEDGMENTS
We thank Rigoberto Hernandez-Castro and Silvia Villanueva-Recillas for their scientific and technical assistance, respectively.
This work was partially supported by CONACYT grant 182089.
Footnotes
Published ahead of print 19 June 2013
REFERENCES
- 1.Lane DJ. 1991. 16S/23S rRNA sequencing, p 115–175 In Stackebrandt E, Goodfellow M. (ed), Nucleic acids techniques in bacterial systematics. Wiley, Chichester, United Kingdom [Google Scholar]
- 2.Jongwutiwes S, Silachamroon U, Putaporntip C. 2000. Pentatrichomonas hominis in empyema thoracis. Trans. R. Soc. Trop. Med. Hyg. 94:185–186 [DOI] [PubMed] [Google Scholar]
- 3.Mallat H, Podglajen I, Lavarde V, Mainardi J-L, Frappier J, Cornet M. 2004. Molecular characterization of Trichomonas tenax causing pulmonary infection. J. Clin. Microbiol. 42:3886–3887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mantini C, Souppart L, Noel C, Duong TH, Mornet M, Carroger G, Dupont P, Masseret E, Goustille J, Capron M, Duboucher C, Dei-Case E, Viscogliosi E. 2009. Molecular characterization of a new Tetratrichomonas species in a patient with empyema. J. Clin. Microbiol. 47:2336–2339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Templeton AR, Sing CF. 1993. A cladistic analysis of phenotypic associations with haplotypes inferred from restriction endonuclease mapping. IV. Nested analyses with cladogram uncertainty and recombination. Genetics 134:659–669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kimura M. 1980. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J. Mol. Evol. 16:111–120 [DOI] [PubMed] [Google Scholar]
- 7.Kumar S, Tamura K, Nei M. 2004. MEGA3: integrated software for molecular evolutionary genetics analysis and sequence alignment. Brief. Bioinform. 5:150–163 [DOI] [PubMed] [Google Scholar]
- 8.Posada D, Crandall KA. 1998. MODELTEST: testing the model of DNA substitution. Bioinformatics 14:817–818 [DOI] [PubMed] [Google Scholar]
- 9.Holder M, Lewis PO. 2003. Phylogeny estimation: traditional and Bayesian approaches. Nat. Rev. Genet. 4:275–284 [DOI] [PubMed] [Google Scholar]
- 10.Huelsenbeck JP, Ronquist F, Nielsen R, Bollback JP. 2001. Bayesian inference of phylogeny and its impact on evolutionary biology. Science 14:2310–2314 [DOI] [PubMed] [Google Scholar]
- 11.Ronquist F, Huelsenbeck JP. 2003. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19:1572–1574 [DOI] [PubMed] [Google Scholar]
- 12.Duboucher C, Pierce RJ, Capron M, Dei-Cas E, Viscogliosi E. 2008. Recent advances in pulmonary trichomonosis. Trends Parasitol. 24:201–202 [DOI] [PubMed] [Google Scholar]
- 13.Leterrier M, Morio F, Renard BT, Poirier AS, Miegeville M, Chambreuil G. 2012. Trichomonads in pleural effusion: case report, literature review and utility of PCR for species identification. New Microbiol. 35:83–87 [PubMed] [Google Scholar]
- 14.Gilroy S, Simcaski E, Rawling R, Granato P. 2007. Trichomonas species empyema coinfection in an alcoholic female. Clin. Microbiol. Newsl. 29:69–71 [Google Scholar]
- 15.Bellanger AP, Cabaret O, Costa JM, Foulet F, Bretagne S, Botterel F. 2008. Two unusual occurrences of trichomoniasis: rapid species identification by PCR. J. Clin. Microbiol. 46:3159–3161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lewis KL, Doherty DE, Ribes J, Seabolt JP, Bensadoun ES. 2003. Empyema caused by Trichomonas. Chest 123:291–292 [DOI] [PubMed] [Google Scholar]
- 17.Cepicka I, Hampl V, Kulda J, Flegr J. 2006. New evolutionary lineages, unexpected diversity, and host specificity in the parabasalid genus Tetratrichomonas. Mol. Phylogenet. Evol. 39:542–551 [DOI] [PubMed] [Google Scholar]
- 18.Honigberg BM. 1963. Evolutionary and systematic relationships in the flagellate order Trichomonadida Kirby. J. Protozool. 10:20–63 [DOI] [PubMed] [Google Scholar]
- 19.Martinez-Hernandez F, Jimenez-Gonzalez DE, Chenillo P, Alonso-Fernandez C, Maravilla P, Flisser A. 2009. Geographical widespread of two lineages of Taenia solium due to human migrations: can population genetic analysis strengthen this hypothesis? Infect. Genet. Evol. 9:1108–1114 [DOI] [PubMed] [Google Scholar]
- 20.Pomajbíková K, Oborník M, Horák A, Petrželková KJ, Grim JN, Levecke B, Todd A, Mulama M, Kiyang J, Modrý D. 2013. Novel insights into the genetic diversity of Balantidium and Balantidium-like cyst-forming ciliates. PLoS Negl. Trop. Dis. 7:e2140. 10.1371/journal.pntd.0002140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kleina P, Bettim-Bandinelli J, Bonatto SL, Benchimol M, Bogo MR. 2004. Molecular phylogeny of Trichomonadidae family inferred from ITS-1, 5.8S rRNA and ITS-2 sequences. Int. J. Parasitol. 34:963–970 [DOI] [PubMed] [Google Scholar]
- 22.Kutisova K, Kulda J, Cepicka I, Flegr J, Koudela B, Teras J, Tachezy J. 2005. Tetratrichomonads from the oral cavity and respiratory tract of humans. Parasitology 131:309–319 [DOI] [PubMed] [Google Scholar]