Abstract
We evaluated the FDA-cleared InBios dengue virus (DENV) IgM capture enzyme-linked immunosorbent assay (ELISA) for qualitative detection of anti-DENV IgM antibodies from 79 serum samples obtained from dengue virus-infected patients or suspected dengue cases. The agreement, sensitivity, and specificity of the InBios assay compared to the gold standard in-house DENV IgM capture ELISA were 94, 92, and 94%, respectively. We conclude that the InBios DENV IgM capture ELISA can be effectively used for rapid diagnosis of acute or recent DENV infection.
TEXT
Dengue virus (DENV) a mosquito-borne flavivirus, is a significant human pathogen of global importance (1). Dengue, an acute viral disease, is caused by any one of the four DENV serotypes, DENV-1, -2, -3, and -4 (2). Although most of the reported dengue cases in the United States are acquired by travelers or immigrants (3), autochthonous dengue fever outbreaks have occurred in Brownsville, TX (2005), and southern Florida (2009 to 2011) and Hawaii (2011) (4). To date, there is no vaccine or specific antiviral treatment for dengue virus infection in humans, and effective management of severe dengue virus disease can be augmented by rapid diagnosis during the acute stage of infection (5, 6).
In the majority of DENV infections, immunoglobulin M (IgM) antibodies can be detected within 3 to 5 days following the onset of fever (7). In secondary DENV infection, IgM antibody titers are usually lower than those in primary DENV infection but follow similar kinetics (8). An ideal IgM serologic test should have sufficient sensitivity to detect low DENV IgM antibody titers and be specific enough to discriminate DENV infection in areas where multiple flaviviruses and other pathogens cocirculate (9). Several rapid diagnostic tests are commercially available for detection of anti-DENV IgM antibodies (9). Therefore, it is important to evaluate the performance characteristics of these kits in terms of sensitivity and specificity in order to ensure accurate and rapid diagnosis of dengue virus infection (5). Recently, the U.S. Food and Drug Administration (FDA) cleared the InBios DENV Detect IgM capture enzyme-linked immunosorbent assay (ELISA) (InBios International, Inc., Seattle, WA) for qualitative detection of anti-DENV IgM antibodies (4). This test can detect acute or recent DENV infections and can be used by public health laboratories for rapid confirmation of dengue cases during dengue outbreaks (4).
(These research data are part of the master's thesis of M.N. submitted to the University of Hawaii.)
In this study, we evaluated the InBios DENV IgM capture ELISA in comparison with the in-house DENV IgM antibody capture (MAC) ELISA using 79 well-characterized clinical serum samples collected from Hawaii, Vietnam, Niue, Singapore, and American Samoa, where dengue outbreaks have occurred in the past. Samples were coded and collected in compliance with the University of Hawaii Institutional Review Board guidelines (CHS 16857 and 16873). All serum samples were frozen at −70°C prior to assay.
The InBios DENV IgM capture ELISA was conducted according to the manufacturer's instructions. Briefly, serum samples were diluted 1:100, using DENV sample dilution buffer, and were incubated in microtiter wells coated with anti-human IgM antibodies for 1 h at 37°C followed by separate incubation with either dengue virus-derived recombinant antigens (DENRA) or normal cell antigen (NCA). NCA was derived from culture supernatant of the COS-1 cell line. After incubation and washing, the wells were treated with a DENV-specific monoclonal antibody labeled with the enzyme horseradish peroxidase (HRP). After a second incubation of 1 h at 37°C and a washing step, the wells were incubated with tetramethylbenzidine (TMB) substrate. After addition of stopping solution, absorbance was read at 450 nm. The ratio of the DENRA and the control antigen wells (NCA), designated immune status ratio (ISR), was used to determine the presence of DENV antibodies in the serum sample. All serum samples with an ISR below 1.65 were considered negative for anti-DENV IgM antibodies, whereas samples with an ISR above 2.84 were considered positive for anti-DENV IgM antibodies. Serum samples with ISRs between 1.65 and 2.84 were considered equivocal. Equivocal serum samples were retested in duplicate. Serum samples that remained equivocal after repeat testing were tested using the plaque reduction neutralization test (PRNT), if sufficient serum was available.
An in-house MAC-ELISA for detection of anti-DENV IgM antibodies was conducted based on a Centers for Disease Control and Prevention (CDC) protocol as described previously (10, 11). Briefly, the inner 60 wells of Immulon II plates (Dynatech Laboratories, Inc., Alexandria, VA) were coated with goat anti-human IgM antibodies (Kirkegaard & Perry Laboratories Inc., Gaithersburg, MD) diluted 1:2,000 in carbonate-bicarbonate buffer (pH 9.6), and the plate was incubated overnight at 4°C. The fluid in plate wells was aspirated and blocked with 200 μl/well of 1× phosphate-buffered saline (PBS), 0.05% Tween 20, and 5% milk for 30 min at room temperature. Further, plates were then washed five times with PBS containing 0.05% Tween 20 using an automated plate washer. Fifty microliters of 1:40-diluted serum samples was added in triplicate for the virus antigen wells and for the normal antigen wells, and the plates were incubated for 1 h at 37°C. Each plate included one positive- and one negative-control human serum. Plates were washed five times as described above. DENV antigens used were sucrose-acetone extracts from DENV-infected suckling mouse brain, obtained from the Division of Vector-Borne Diseases, CDC. DENV-1, -2, -3, and -4 antigens were diluted 1:80, 1:160, 1:80, and 1:80, respectively, using PBS. One hundred microliters of DENV antigen dilutions was added to each respective virus well, and 50 μl of 1:20 normal antigen was added to each control well. Plates were incubated overnight at 4°C and then washed as described above. Fifty microliters of flavivirus group-reactive monoclonal antibody 6B6C-1 conjugated to HRP (Hennessy Research, Shawnee, KS) diluted 1:2,500 in blocking buffer was added to each well. Plates were then incubated for 1 h at 37°C, washed five times as described above, rotated 180 degrees, and washed an additional five times. After washing, the wells were incubated with tetramethylbenzidine (TMB) substrate at room temperature for 10 min. The reaction was then stopped by the addition of 50 μl of TMB stop solution (Kirkegaard & Perry Laboratories Inc., Gaithersburg, MD), and absorbance values were read at 450 nm using a Biotek fluorescence plate reader (Biotek, Winooski, VT). Mean absorbance values for triplicate wells of each test serum (P) were divided by the mean values of the corresponding negative-control serum (N). A P/N ratio of ≥3.0 was considered positive whereas a P/N ratio of ≥2 and <3 was considered presumptive positive and a P/N ratio of <2 was considered negative. One of the limitations of the MAC-ELISA is cross-reactivity between circulating flaviviruses. This limitation must be considered when working in geographic regions where multiple flaviviruses cocirculate.
PRNT was conducted to detect anti-DENV neutralizing antibodies using 90% plaque reduction for all four DENV serotypes, according to the protocol recommended by the CDC (12). DENV-1 (1943), DENV-2 (Dakara), DENV-3 (H87), and DENV-4 (H241) grown in Vero cells were employed for conducting PRNT.
In this study, for evaluation of the sensitivity and specificity of the InBios DENV IgM capture ELISA, the in-house DENV MAC-ELISA was used as a gold standard. In addition to sensitivity and specificity, percent agreement, 95% confidence intervals (95% CI), and kappa coefficients were also determined as measures of agreement. Levels of agreement as defined by kappa values were categorized as nearly perfect (0.81 to 1.0), substantial (0.61 to 0.8), moderate (0.41 to 0.6), fair (0.21 to 0.4), slight (0 to 0.2), or poor (<0) (13). Equivocal results by the InBios DENV IgM capture ELISA were considered negative for calculating percent sensitivity and positive for calculating percent specificity. Data analysis was conducted using Microsoft Excel and SAS software.
Out of 79 serum samples tested using the InBios assay and MAC-ELISA, 22 samples were positive and 50 samples were negative by using both methods (Table 1). PRNT endpoint dilution was conducted for only six positive samples due to limited sample volume. PRNT results based on endpoint serum dilution coupled with patient clinical history indicated that five out of the six (83%) serum samples were from patients with recent primary DENV infection and one sample (17%) was from a patient with recent secondary DENV infection. Out of the 50 dengue virus IgM-negative serum samples by both assays, PRNT was conducted at 1:20 and 1:40 serum dilutions on 39 samples to confirm the presence or absence of DENV infection, out of which 18 samples (46%) were PRNT positive and thus indicated the presence of anti-DENV neutralizing antibodies or past DENV infection. The PRNT assay does not differentiate between anti-DENV IgM and IgG antibodies.
Table 1.
Sensitivity, specificity, percent agreement, and kappa value for InBios DENV detect IgM capture ELISA compared to the in-house MAC-ELISA
| Result by InBios DENV Detect IgM capture ELISA (n = 79)b | No. of samples with result by in-house MAC-ELISAc |
% (95% CI) |
Kappa value | ||||
|---|---|---|---|---|---|---|---|
| Positive | Negative | Presumptive positive | Sensitivity | Specificity | Agreement | ||
| Positive | 22 | 2 | 0 | ||||
| Negative | 0 | 50 | 0 | ||||
| Equivocala | 2 | 1 | 2 | ||||
| Total | 24 | 53 | 2 | 92 (73–99) | 94 (84–99) | 94 (86–98) | 0.87 |
Equivocal samples were considered false negative for sensitivity and false positive for specificity.
InBios IgM positive, ISR of >2.84; InBios IgM equivocal, ISR of 1.65 to 2.84; InBios IgM negative, ISR of <1.65.
MAC-ELISA positive, P/N of ≥3; MAC-ELISA presumptive positive, P/N of ≥2 and <3; MAC-ELISA negative, P/N of <2.
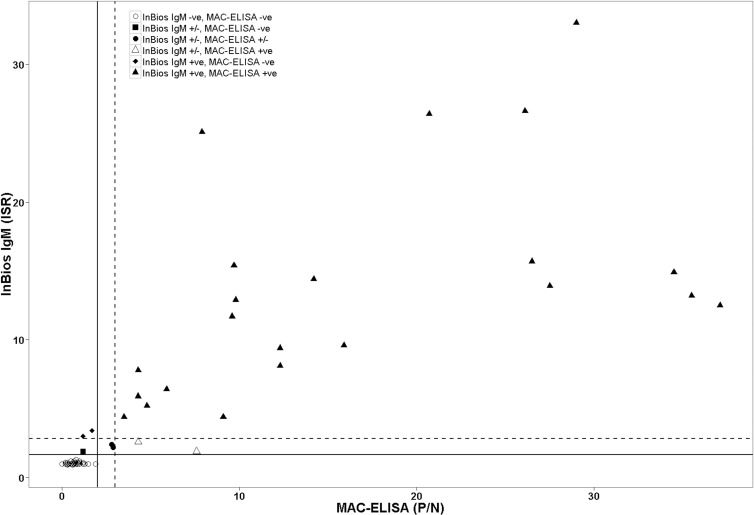
We observed a significant correlation in detecting anti-DENV IgM antibodies between the InBios IgM capture ELISA and the in-house MAC-ELISA (Pearson's r = 0.80, P < 0.0001) (Fig. 1). The agreement, sensitivity, and specificity of InBios IgM capture ELISA were 94% (95% CI, 86 to 98%), 92% (95% CI, 73 to 99%), and 94% (95% CI, 84 to 99%), respectively (Table 1). In addition, the InBios assay showed near-perfect agreement (κ = 0.87) (13) with the in-house MAC-ELISA (Table 1). Out of 79 serum samples, two serum samples (samples 3 and 4) were MAC-ELISA presumptive positive and InBios equivocal (Table 2). Serum samples 3 and 4 were obtained from DENV-infected patients in Singapore. These two serum samples were DENV-NS1 positive as well as anti-DENV IgM positive using an independent IgM assay (data not shown). In addition, serum sample 2, obtained from a DENV-infected patient in Singapore, was MAC-ELISA positive and InBios equivocal; however, it was anti-DENV IgM positive by independent IgM assay. Although it is not certain, these three samples may have been collected from patients with secondary DENV infection or during the early phase of primary DENV infection, as the levels of IgM antibodies were low.
Fig 1.
Correlation between the in-house DENV MAC-ELISA and the InBios DENV IgM capture ELISA. Scatter plot depicting in-house DENV MAC-ELISA versus InBios DENV IgM capture ELISA determined using 79 human serum samples. Left of the vertical solid line are MAC-ELISA negatives, P/N of <2; right of the vertical dashed line are MAC-ELISA positives, P/N of ≥3; and all values between the vertical solid and dashed lines are MAC-ELISA presumptive positive samples, P/N of ≥2 and <3. Below the horizontal solid line are InBios IgM ELISA negatives, ISR of <1.65; above the dotted horizontal line are InBios IgM ELISA positives, ISR of >2.84; all samples between these two lines are InBios IgM capture ELISA equivocal (ISR of 1.65 to 2.84).
Table 2.
Results of MAC-ELISA and InBios DENV IgM capture ELISA equivocal and discordant samplesa
| Sample no. | MAC-ELISAb |
InBios DENV IgM capture ELISAc |
PRNT result | ||||
|---|---|---|---|---|---|---|---|
| P/N | Result | DENRA (OD) | NCA (OD) | ISR | Result | ||
| 1 | 7.6 | Pos | 0.27 | 0.11 | 2.4 | Equivocal | QNS |
| 2 | 4.3 | Pos | 0.22 | 0.08 | 2.6 | Equivocal | QNS |
| 3 | 2.9 | Presumptive Pos | 0.13 | 0.06 | 2.2 | Equivocal | QNS |
| 4 | 2.8 | Presumptive Pos | 0.16 | 0.07 | 2.4 | Equivocal | QNS |
| 5 | 1.2 | Neg | 0.21 | 0.11 | 1.9 | Equivocal | DENV-2 Pos |
| 6 | 1.7 | Neg | 0.21 | 0.06 | 3.4 | Pos | DENV-1 Pos |
| 7 | 1.2 | Neg | 0.19 | 0.06 | 3.0 | Pos | DENV Pos |
Abbreviations: DENRA, dengue virus-derived recombinant antigens; NCA, normal cell antigen; ISR, immune status ratio; P/N, positive-to-negative ratio; QNS, quantity not sufficient; Pos, positive; Neg, negative; OD, optical density.
MAC-ELISA positive, P/N of ≥3; MAC-ELISA presumptive positive, P/N of ≥2 and <3; MAC-ELISA negative, P/N of <2.
InBios IgM positive, ISR of >2.84; InBios IgM equivocal, ISR of 1.65 to 2.84; InBios IgM negative, ISR of <1.65.
Out of 79 serum samples, only five serum samples (samples 1, 2, 5, 6, and 7) exhibited discordant results (Table 2). Dengue antigen (DENRA) and NCA optical density (OD) values obtained for 79 serum samples are given in Table 3. The NCA used in the InBios IgM capture ELISA detects the nonspecific reactivity of test serum samples (14). Out of the five samples that exhibited discordant results, three MAC-ELISA-negative samples (samples 5, 6, and 7) were InBios equivocal or positive (Table 2). These three samples exhibited DENRA OD values in the range of 0.19 to 0.21 (Table 2), which are closer to the highest DENRA OD value (0.14) obtained for negative serum samples using the InBios assay (Table 3). These data support the true negativity of these serum samples by the MAC-ELISA. In addition, sample 1 depicted high background as indicated by an NCA OD value of 0.11 and was thus interpreted as equivocal even though the DENRA OD value was 0.27 (Table 2). Thus, inclusion of positive as well as negative antigens is critical for interpretation of the test results as the test detects background nonspecific reactivity of serum samples (14).
Table 3.
Detection of anti-DENV IgM antibodies using InBios DENV IgM capture ELISA
| InBios DENV IgM capture ELISA result typea | No. of samples | Avg (range) |
||
|---|---|---|---|---|
| ISR | DENRA (OD) | NCA (OD) | ||
| Positive | 24 | 12.1 (3.0–33.0) | 0.83 (0.19–2.11) | 0.07 (0.06–0.14) |
| Equivocal | 5 | 2.4 (1.9–2.6) | 0.21 (0.13–0.27) | 0.08 (0.06–0.11) |
| Negative | 50 | 1.0 (0.9–1.3) | 0.07 (0.06–0.14) | 0.07 (0.06–0.13) |
InBios IgM positive, ISR of >2.84; InBios IgM equivocal, ISR of 1.65 to 2.84; InBios IgM negative, ISR of <1.65.
Due to unavailability of sufficient sample volume, PRNT endpoint dilution was conducted on only three discordant samples, 5, 6, and 7 (Table 2), to identify the infecting DENV serotype and to confirm primary or secondary DENV infection. Out of three discordant samples, sample 5 from Vietnam was from a patient with past DENV-2 infection, sample 6 from Hawaii was collected in 2010 from a patient infected with DENV-1 in 2001, and for sample 7 from Vietnam, the DENV serotype could not be determined because of possible multiple past DENV infections (Table 2). Serum samples 6 and 7 from Hawaii and Vietnam, respectively, from patients with previous DENV infection as determined by PRNT, produced false-positive reactions using the InBios DENV IgM capture ELISA (Table 2). However, ISR values for these two samples ranged from 3.0 to 3.4 (Table 2) and were closer to the ISR cutoff of 2.84 for InBios positives. Out of these two serum samples, serum sample 6 from Hawaii (Table 2), collected in 2010 from a patient infected with DENV-1 in 2001 (15), was previously studied in our laboratory for long-term T cell memory responses (A. Gurary, unpublished data). In humans, dengue virus epitope-specific tetramer-positive CD8+ T cells can be detected ex vivo up to a year after natural primary DENV infection and even longer after secondary DENV infection (16). In sample 6, we were unable to detect dengue virus epitope-specific tetramer-positive CD8+ T cells ex vivo. In addition, even after stimulation with cognate dengue virus epitope, sample 6 demonstrated a lower dengue virus-specific CD8+ T cell proliferative response than did samples from recently DENV-infected patients (A. Gurary, unpublished data). These data combined with MAC-ELISA data confirm the absence of recent secondary DENV infection and thus anti-DENV IgM antibodies in sample 6.
The sensitivity of other commercially available anti-DENV IgM tests for detection of anti-DENV IgM antibodies varies from 21 to 99%, whereas the specificities are 77 to 98% compared with the reference solid-phase MAC-ELISA used by the CDC (9). Recently, in one study by Blacksell et al., sensitivities and specificities of the two commercial IgM antibody ELISAs for detection of acute dengue virus infection ranged from 85 to 89% and 88 to 100%, respectively (17).
This study has several limitations. First, cross-reactivity with IgM from West Nile virus and other flaviviruses may occur with the InBios dengue virus IgM assay. Second, we did not have access to detailed clinical history or vaccination records for other flaviviruses such as yellow fever virus (YFV) for the 79 serum samples tested in this study. Third, the number of samples used in this evaluation study is small, and fourth, we did not have PRNT data for all 79 serum samples due to insufficient sample volume. Despite these limitations, the InBios DENV IgM capture ELISA is advantageous compared to the MAC-ELISA since the results can be obtained in 1 day (∼5 h), whereas the latter requires 2 to 3 days. This study for the first time evaluated the only U.S. Food and Drug Administration-cleared InBios DENV IgM capture ELISA.
In summary, this study indicates that the InBios DENV Detect IgM capture ELISA is a reliable, rapid, sensitive, and specific serological test for detection of acute or recent dengue virus infections and thus can be employed by public health laboratories for rapid detection of dengue virus infections during dengue epidemics.
ACKNOWLEDGMENTS
This study was supported in part by grant P20GM103516 from the Centers of Biomedical Research Excellence, National Institute of General Medical Sciences; grant U01AI078213 from the National Institute of Allergy and Infectious Diseases, National Institutes of Health (NIH); and grant W8IXWH0720073 from the Department of Defense and by institutional funds.
We thank James Davis of the Biostatistics and Data Management Core supported by the RMATRIX grant (U54MD007584) from the National Institute on Minority Health and Health Disparities, NIH, for conducting statistical data analysis.
The funders had no role in the study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Footnotes
Published ahead of print 3 July 2013
REFERENCES
- 1.Guzman MG, Halstead SB, Artsob H, Buchy P, Farrar J, Gubler DJ, Hunsperger E, Kroeger A, Margolis HS, Martinez E, Nathan MB, Pelegrino JL, Simmons C, Yoksan S, Peeling RW. 2010. Dengue: a continuing global threat. Nat. Rev. Microbiol. 8:S7–S16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Solomon T, Mallewa M. 2001. Dengue and other emerging flaviviruses. J. Infect. 42:104–115 [DOI] [PubMed] [Google Scholar]
- 3.Hynes NA. 2012. Dengue: a reemerging concern for travelers. Cleve. Clin. J. Med. 79:474–482 [DOI] [PubMed] [Google Scholar]
- 4.Adalja AA, Sell TK, Bouri N, Franco C. 2012. Lessons learned during dengue outbreaks in the United States, 2001–2011. Emerg. Infect. Dis. 18:608–614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Peeling RW, Artsob H, Pelegrino JL, Buchy P, Cardosa MJ, Devi S, Enria DA, Farrar J, Gubler DJ, Guzman MG, Halstead SB, Hunsperger E, Kliks S, Margolis HS, Nathanson CM, Nguyen VC, Rizzo N, Vazquez S, Yoksan S. 2010. Evaluation of diagnostic tests: dengue. Nat. Rev. Microbiol. 8:S30–S38 [DOI] [PubMed] [Google Scholar]
- 6.Thomas SJ, Endy TP. 2011. Critical issues in dengue vaccine development. Curr. Opin. Infect. Dis. 24:442–450 [DOI] [PubMed] [Google Scholar]
- 7.Simmons CP, Farrar JJ, Nguyen VV, Wills B. 2012. Dengue. N. Engl. J. Med. 366:1423–1432 [DOI] [PubMed] [Google Scholar]
- 8.Halstead SB. 2007. Dengue. Lancet 370:1644–1652 [DOI] [PubMed] [Google Scholar]
- 9.Hunsperger EA, Yoksan S, Buchy P, Nguyen VC, Sekaran SD, Enria DA, Pelegrino JL, Vazquez S, Artsob H, Drebot M, Gubler DJ, Halstead SB, Guzman MG, Margolis HS, Nathanson CM, Lic NRR, Bessoff KE, Kliks S, Peeling RW. 2009. Evaluation of commercially available anti-dengue virus immunoglobulin M tests. Emerg. Infect. Dis. 15:436–440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Innis BL, Nisalak A, Nimmannitya S, Kusalerdchariya S, Chongswasdi V, Suntayakorn S, Puttisri P, Hoke CH. 1989. An enzyme-linked immunosorbent assay to characterize dengue infections where dengue and Japanese encephalitis co-circulate. Am. J. Trop. Med. Hyg. 40:418–427 [DOI] [PubMed] [Google Scholar]
- 11.Martin DA, Muth DA, Brown T, Johnson AJ, Karabatsos N, Roehrig JT. 2000. Standardization of immunoglobulin M capture enzyme-linked immunosorbent assays for routine diagnosis of arboviral infections. J. Clin. Microbiol. 38:1823–1826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Roehrig JT, Hombach J, Barrett AD. 2008. Guidelines for plaque-reduction neutralization testing of human antibodies to dengue viruses. Viral Immunol. 21:123–132 [DOI] [PubMed] [Google Scholar]
- 13.Landis JR, Koch GG. 1977. The measurement of observer agreement for categorical data. Biometrics 33:159–174 [PubMed] [Google Scholar]
- 14.Welch RJ, Anderson BL, Litwin CM. 2008. Evaluation of a new commercial enzyme immunoassay for the detection of IgM antibodies to West Nile virus using a ratio method to eliminate nonspecific reactivity. J. Clin. Lab. Anal. 22:362–366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Imrie A, Meeks J, Gurary A, Sukhbataar M, Kitsutani P, Effler P, Zhao Z. 2007. Differential functional avidity of dengue virus-specific T-cell clones for variant peptides representing heterologous and previously encountered serotypes. J. Virol. 81:10081–10091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Friberg H, Bashyam H, Toyosaki-Maeda T, Potts JA, Greenough T, Kalayanarooj S, Gibbons RV, Nisalak A, Srikiatkhachorn A, Green S, Stephens HA, Rothman AL, Mathew A. 2011. Cross-reactivity and expansion of dengue-specific T cells during acute primary and secondary infections in humans. Sci. Rep. 1:51. 10.1038/srep00051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Blacksell SD, Jarman RG, Gibbons RV, Tanganuchitcharnchai A, Mammen MP, Jr, Nisalak A, Kalayanarooj S, Bailey MS, Premaratna R, de Silva HJ, Day NP, Lalloo DG. 2012. Comparison of seven commercial antigen and antibody enzyme-linked immunosorbent assays for detection of acute dengue infection. Clin. Vaccine Immunol. 19:804–810 [DOI] [PMC free article] [PubMed] [Google Scholar]