Abstract
The widespread use of antifungal agents, which is likely to expand with their enhanced availability, has promoted the emergence of drug-resistant strains. Antifungal susceptibility testing (AFST) is now an essential procedure for guiding appropriate antifungal therapy. Recently, we developed a matrix-assisted laser desorption ionization–time of flight mass spectrometry (MALDI-TOF MS)-based method that enables the detection of fungal isolates with reduced echinocandin susceptibility, relying on the proteome changes that are detectable after a 15-h exposure of fungal cells to serial drug concentrations. Here, we describe a simplified version of this approach that facilitates discrimination of the susceptible and resistant isolates of Candida albicans after a 3-h incubation in the presence of “breakpoint” level drug concentrations of the echinocandin caspofungin (CSF). Spectra at concentrations of 0 (null), 0.03 (intermediate), and 32 (maximal) μg/ml of CSF were used to create individual composite correlation index (CCI) matrices for 65 C. albicans isolates, including 13 fks1 mutants. Isolates are then classified as susceptible or resistant to CSF if the CCI values of spectra at 0.03 and 32 μg/ml are higher or lower, respectively, than the CCI values of spectra at 0.03 and 0 μg/ml. In this way, the drug resistance of C. albicans isolates to echinocandin antifungals can be quickly assessed. Furthermore, the isolate categorizations determined using MALDI-TOF MS-based AFST (ms-AFST) were consistent with the wild-type and mutant FKS1 genotypes and the AFST reference methodology. The ms-AFST approach may provide a rapid and reliable means of detecting emerging antifungal resistance and accelerating the initiation of appropriate antifungal treatment.
INTRODUCTION
Antifungal resistance has emerged prominently over the past few decades and continues to increase (1), posing a growing concern for the management of patients with invasive fungal infections, especially those caused by Candida species (1). Despite the advent of antifungal agents, such as the highly active triazoles (2) and the newest drugs, the echinocandins (3), invasive candidiasis is associated with excessive morbidity, mortality, and costs (4), particularly for critically ill patients (5). The general consensus is that clinical outcomes are improved when treatments are started early (6). Yet, concomitant with more antifungal drugs being available in clinical practice (7), the use of empirical or preemptive antifungal therapy leads undesirably to higher selective pressure toward species developing secondary resistance or intrinsically resistant species replacing susceptible ones (8). While Candida albicans is still the major infecting species (9, 10), decreased antifungal susceptibility has been increasingly observed in Candida species which are normally drug susceptible (11), thus reinforcing the importance of performing routine Candida susceptibility tests (12). Antifungal susceptibility testing (AFST) is important not only for optimal selection of the most appropriate antifungal agents, i.e., “likely to be active for a given infection” (1), but also, and perhaps more importantly, for the detection of resistance, i.e., “to determine which agents will not work” (1).
Although various standardized microdilution-based procedures are currently approved for AFST of fungal species by the Clinical and Laboratory Standards Institute (CLSI) and the European Committee on Antibiotic Susceptibility Testing (EUCAST) (13, 14), the role of these procedures is often restricted to specialized mycology laboratories which contribute to the setting of interpretive AFST breakpoints (14). To facilitate standardized detection of antifungal drug resistance, commercial MIC methods (e.g., the Sensititre YeastOne colorimetric plate [TREK Diagnostic Systems, Cleveland, OH] and the Etest [bioMérieux, Marcy l'Étoile, France]) for in vitro Candida AFST have been developed as modifications of broth or agar microdilution methods that conform to the CLSI standards (15) and validated by comparison with the CLSI reference method using the interpretive CLSI breakpoints (1). In spite of their ease of use, standardization, and flexibility (1), some of these methods, which are approved by the U.S. Food and Drug Administration for clinical testing, are generally limited by visual (and subjective) MIC endpoint determination and slow turnaround times (the endpoint is achieved at 24 to 48 h).
As a simple and fast technique for the analysis of large biomolecules, matrix-assisted laser desorption ionization–time of flight mass spectrometry (MALDI-TOF MS) of whole microbial cells or their extracts recently evolved from a niche research procedure to a widely used practical application that has revolutionized the manner of classifying and identifying microbial species (16) and now represents an excellent alternative to traditional clinical laboratory procedures (17, 18). Recently, we developed a MALDI-TOF MS-based assay for testing antifungal susceptibilities of clinically relevant Candida and Aspergillus species to the echinocandin caspofungin (CSF) (19) by means of a composite correlation index (CCI)-based approach (20). The method reliably and accurately allows a determination of the minimal profile change concentration (MPCC) (21), an endpoint value that is an alternative to the classical MIC, for CSF by relying on the proteome changes which are detectable after a 15-h exposure of fungal cells to serial drug concentrations (19). Endpoint readings by MALDI-TOF MS provide a clear time savings (15 h versus 24 h) over the CLSI- or EUCAST-based methods, but implementing MALDI-TOF MS into the daily workflow appears to offer only a subtle advantage to clinicians compared to that for recent strategies aimed at speeding up AFST (22, 23).
In this proof-of-concept study, we used a MALDI-TOF MS-based assay (ms-AFST) to assess the susceptibility of C. albicans isolates to CSF after a short time of exposure (3 h) to the drug. ms-AFST was modified using “breakpoint” drug concentrations in order to allow easy and labor-saving discrimination between CSF-susceptible and CSF-resistant C. albicans isolates, categorized according to the absence or presence, respectively, of hot spot mutations in the echinocandin target FKS1 gene.
(This study was presented in part as a poster at the 18th Congress of the International Society for Human and Animal Mycology 2012, Berlin, Germany, 11 to 15 June 2012 [24].)
MATERIALS AND METHODS
Fungal culture and sample preparation.
A panel of 65 C. albicans isolates with and without FKS1 gene (which encodes β-1,3-glucan synthase [GS], the echinocandin target) mutations that are known to be associated with resistance to CSF (25, 26) were included in the study (see Table S1 in the supplemental material). Ten fks1 mutant isolates were previously investigated (19). Before testing, each isolate was subcultured onto Sabouraud dextrose agar to ensure its purity and viability and reidentified by standard methods. For all isolates, FKS1 gene sequencing was performed as described elsewhere (27). AFST was performed by the broth microdilution method following the CLSI guidelines, and the MICs were determined using prominent growth inhibition as an endpoint (15). The CLSI-recommended quality control strains (Candida parapsilosis ATCC 22019 and Candida krusei ATCC 6258) were included in each set of experiments (15). Isolates were categorized as susceptible, intermediate, or resistant on the basis of the new CLSI breakpoints for CSF and C. albicans that were set at ≤0.25 μg/ml (susceptible), 0.5 μg/ml (intermediate), and ≥1 μg/ml (resistant) (26). AFST by MALDI-TOF MS (ms-AFST) was performed according to our published method (19). Briefly, protein extracts were prepared from fungal isolates (starting from an inoculum of 1 × 107 CFU/ml) grown in RPMI broth containing serial dilutions (ranging from 0.5 to 0.004 μg/ml) or a breakpoint concentration (intermediate, 0.03 μg/ml) of CSF (pure substance provided by Merck, Milan, Italy) at 37°C under agitation for 15 or 3 h, respectively. Growth in RPMI broth with 32 μg/ml CSF (maximal concentration) or 0 μg/ml CSF (null concentration) was included in the two experimental runs. For the MALDI-TOF sample preparation, fungal cells were washed twice with sterile water and resuspended in 10% (vol/vol) formic acid. One microliter of each cell suspension was directly spotted in triplicate onto a polished steel target plate (Bruker Daltonik, Bremen, Germany), covered with 1 μl of absolute ethanol and 1 μl of a saturated solution of α-cyano-4-hydroxycinnamic acid in 50% acetonitrile–2.5% trifluoroacetic acid (Bruker Daltonik), and air dried completely before the MALDI-TOF MS measurement.
MALDI-TOF MS measurements.
The MALDI-TOF MS was carried out with a microflex LT mass spectrometer (Bruker Daltonik). A Bruker Daltonik bacterial test standard (BTS255343) was used for calibration of the instrument. The protein mass spectra were acquired in the positive linear mode at a laser frequency of 20 Hz and analyzed in the mass range of 3,000 to 8,000 m/z, starting from measuring a larger mass range (2,000 to 20,000 m/z). The main instrument parameter settings were as follows: ion source 1 at 20 kV; ion source 2, 16.7 kV; and lens, 8.5 kV. For each spectrum, 240 laser shots (40 laser shots at 6 different spot positions) were automatically acquired with Bruker Daltonik MALDI Biotyper 3.0 software. For the experimental condition testing, 9 drug concentrations and 1 no-drug (null concentration) control, spectra were collected from 3 biological replicates (prepared from repeated cultivations on different days) and were automatically imported as raw data.
MALDI-TOF MS data analysis.
Since visual inspection of the mass spectra revealed qualitative peak differences not easily appreciable, as previously noted (19), the raw spectra generated as described above were divided into intervals of the same size, and the compositions of cross-correlations and autocorrelations of all intervals were used to obtain the CCI values through a specific tool of the MALDI Biotyper software, which is a modification of the mathematical algorithm for analyzing the relationships between the spectra of whole microbial cells (20). CCI values around 1 represent high conformance of spectra, while CCI values near 0 indicate clear diversity of the spectra. By comparing the spectra with one another at the drug concentrations tested, numerical correlation indices were obtained and automatically visualized in a CCI matrix view, which was translated into a heat map where closely related spectra are marked in “hot” (yellow to red) colors and unrelated ones in “cold” (green to blue) colors (19). In addition, spectra from the aforementioned biological replicates were related through cluster analysis by applying the hierarchical algorithm of the MALDI Biotyper software to assess the reproducibility of spectral changes.
ms-AFST performance calculation.
By ms-AFST analysis, each C. albicans isolate was arbitrarily classified as susceptible or resistant to CSF if the mean CCI value derived from the correlation of replicate spectra at 0.03 and 32 μg/ml was, respectively, higher or lower than the mean CCI value derived from the correlation of replicate spectra at 0.03 and 0 μg/ml (unpaired t test; P < 0.05), as exemplified in Fig. 1. The FKS1 genotype was used as a gold standard to evaluate ms-AFST performance in discriminating between fks1 mutant and wild-type (WT) isolates in order to obtain the percent categorical agreement. Very major errors were recorded when fks1 mutant isolates were classified as susceptible by ms-AFST, and major errors were recorded when WT isolates were classified as resistant by ms-AFST. As our method did not allow isolates to be categorized as intermediate, minor errors were identified only when the CLSI MIC indicated an intermediate result for either fks1 mutant or WT isolates (Table 1).
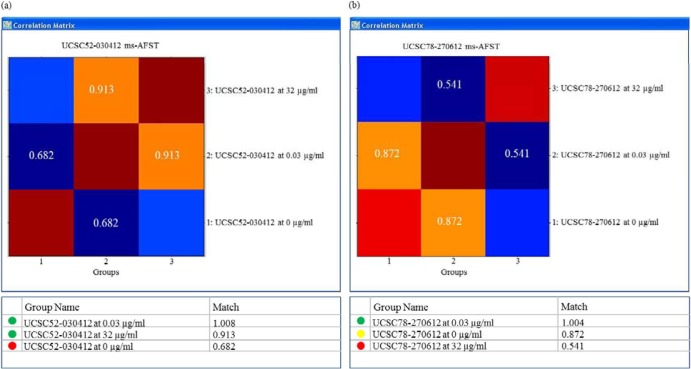
Fig 1.
Representative examples of susceptibility categorization by MALDI-TOF MS-based analysis for two C. albicans isolates, according to the FKS1 genotype. The composite correlation index (CCI)-derived matrices from each isolate's MALDI-TOF mass spectra and the corresponding CCI values are depicted in the top and bottom panels, respectively. (a) The isolate UCSC52-030412 (WT) was defined as susceptible, as the CCI values of the spectra at 0.03 and 32 μg/ml were higher than the CCI values of the spectra at 0.03 and 0 μg/ml (i.e., CCI for 0.03 versus 32 μg/ml > CCI for 0.03 versus 0 μg/ml). (b) The isolate UCSC78-270612 (fks1 mutant) was defined as resistant, as the CCI values of the spectra at 0.03 and 32 μg/ml were lower than the CCI values of the spectra at 0.03 and 0 μg/ml (i.e., CCI for 0.03 versus 32 μg/ml < CCI for 0.03 versus 0 μg/ml).
Table 1.
Performances of ms-AFST and the CLSI method for 62 clinical C. albicans isolates according to the presence or absence of FKS1 hot spot mutationsa
| Method | Total no. of isolates (fks1/WT) | No. (%) of misclassified isolates |
|||
|---|---|---|---|---|---|
| CA | VMEs | MEs | mEs | ||
| ms-AFST | 62 (11/51) | 61/62 (98.4) | 1/62 (1.6) | 0/62 | NAb |
| CLSI | 62 (11/51) | 61/62 (98.4) | 0/62 | 0/62 | 1/62 (1.6) |
Very major errors (VMEs) were identified when fks1 mutant isolates were classified as absent by ms-AFST or the CLSI method. Major errors (MEs) were identified when wild-type (WT) isolates were classified as present by ms-AFST or the CLSI method. Minor errors (mEs) were identified when fks1 mutant and WT isolates were classified as intermediate by the CLSI method only. CA, categorical agreement.
NA, not applicable.
RESULTS
As reported in our previous work (19), the MALDI-TOF MS-based approach assesses the minimal profile change concentration (MPCC) for CSF, which is defined as the CCI value at which a fungal spectrum is more similar to the one observed at the maximal effective drug concentration (32 μg/ml) than to the fungal spectrum observed at the null drug concentration (0 μg/ml). To determine this, for each concentration tested (32 to 0.004 μg/ml), the correlation of the corresponding mass spectrum with the spectra at the two extreme concentrations (null or maximal) is calculated after the spectra are obtained following a 15-h incubation of a C. albicans isolate with CSF (19). Here, our intention was to simplify and then convert the aforementioned MALDI-TOF MS assay into a rapid drug susceptibility test, namely, ms-AFST.
First, we conducted pilot experiments in which 11 randomly selected C. albicans isolates from a well-characterized set of susceptible WT isolates and fks1 hot spot mutant isolates (see Table S1 in the supplemental material) were exposed for 3 h to CSF concentrations ranging from 0.5 to 0.004 μg/ml (samples from three repeated independent fungal cultures were evaluated). This interval was chosen with the aim of identifying a breakpoint concentration at just below 0.25 μg/ml—the newly revisited CLSI clinical breakpoint (CBP) for C. albicans—to be utilized as an intermediate point and the corresponding spectrum to be compared with the spectra obtained at 0 and 32 μg/ml. Thus, by generating individual correlation matrices using the spectra from all 11 isolates, we found that the CCI values obtained by matching each breakpoint spectrum with the spectrum at 32 μg/ml (maximal concentration) were, respectively, higher (for WT isolates) or lower (for fks1 mutant isolates) than the CCI values obtained when the same breakpoint spectrum was matched with the spectrum at 0 μg/ml (null concentration) (see Table S2 in the supplemental material). Overall, the CCI values from the biological replicates, prepared from three repeated cultivations of the 11 isolates, differ uniformly (P < 0.05, unpaired t test), in spite of the unexplained variations in CCI values that apparently did not follow a linear trend in decreases or increases toward the null or maximal drug concentrations (data not shown). Although all the CSF concentrations within the 0.5- to 0.04-μg/ml range can be regarded as evaluable, we selected the 0.03-μg/ml concentration as the breakpoint to be used in a “three-point” AFST assay. In support of that, this concentration represents the upper limit of our 24-h WT MIC distribution, and notably, it is lower than the CLSI-established epidemiological cutoff value (ECV) for C. albicans and CSF (13). The MIC distribution of the WT fungal populations provides a measure of the ECVs (14). By differentiating the WT strains (those without mutational or acquired resistance mechanisms) from the non-WT strains (those having mutational or acquired resistance mechanisms), ECVs may effectively serve to monitor the emergence of resistant isolates harboring fks mutations, which are associated with reduced therapeutic responses. Likewise, the revisited species-specific CBPs for Candida and the echinocandins more efficiently detect resistance associated with fks mutations and optimize the clinical usefulness of in vitro AFST for the echinocandins (26), although CBPs often divide the WT distributions of the important target species (28, 29), running the risk of artificial categorizations of isolates with identical susceptibilities that lead to very major (false-susceptible) and major (false-resistant) reporting errors (1).
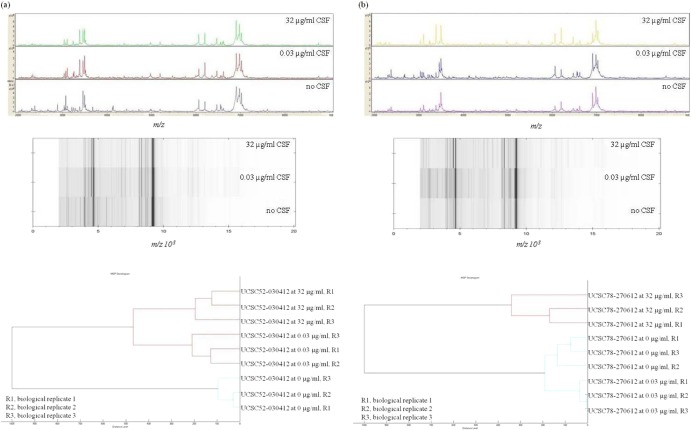
All 65 C. albicans study isolates were challenged with the established CSF concentrations (0, 0.03, and 32 μg/ml) and were processed for the MALDI-TOF MS analysis, as described above. Figure 2 shows the MALDI-TOF MS signatures for two representative WT and fks1 mutant isolates, illustrating alterations in the mass spectra according to the different drug conditions employed. Cluster analysis of the MALDI-TOF MS spectra derived from biological replicates of the two isolates exposed at the three CSF concentrations showed that replicate spectra at 0, 0.03, and 32 μg/ml were grouped, for each isolate, in separate clusters, thus confirming the reproducibility and reliability of the spectral changes obtained (Fig. 2). As shown in Table S1 in the supplemental material, the overall mean CCI values obtained from the matching of spectra at 0.03 μg/ml with those at 0 μg/ml were significantly higher or lower than the mean CCI values obtained from the matching of spectra at 0.03 μg/ml with those at 32 μg/ml. Differences between the mean CCI values did not reach statistical significance for only three isolates, UCSC64-150512, DSP1040, and DSP1014, and these isolates were then excluded from the analysis. Table 1 shows the results obtained with ms-AFST and CLSI reference methods for 62 of the 65 isolates tested. Using the FKS1 genotype as a gold standard, we correctly classified 51 of 51 (100%) isolates as CSF susceptible, and we correctly classified 10 of 11 (90.9%) as CSF resistant. Overall, excellent agreement (98.4%) was observed with one very major categorical error. This error related to the isolate DSP1012 (see Table S1 in the supplemental material) that, as it harbors a D648Y FKS1 genotype, belongs to those isolates with lower levels of echinocandin resistance and then to isolates most challenging to identify with conventional microbial susceptibility testing (30). Also in this evaluation, the CLSI reference method results were comparable to those by ms-AFST (Table 1).
Fig 2.
MALDI-TOF mass spectra and relative gel MALDI views of the mass profiles obtained from a WT (UCSC52-030412) C. albicans isolate (a) and an fks1 mutant (UCSC78-270612) C. albicans isolate (b), under different caspofungin (CSF) treatment conditions (0, 0.03, or 32 μg/ml); minimal alterations following exposure to CSF are visible for both isolates. The bottom panels show hierarchical cluster analyses of spectra derived from biological replicates of each isolate. Replicate spectra of the WT and fks1 mutant isolates form separate clusters according to the three CSF concentrations used, thus indicating the reproducibility of ms-AFST. Distance is displayed in relative units.
DISCUSSION
As a member of the echinocandin class of antifungal agents, CSF is presumed to bind and inhibit the catalytic subunit of the GS enzyme complex, Fksp, encoded by three related genes, FKS1, FKS2, and FKS3 (25). Resistance in susceptible species such as C. albicans is uncommon (26) but has been attributed to mutations in the FKS1 gene within two hot spot regions (25). These mutations result in elevated MICs, although the most significant MIC increases have been shown to be related to amino acid changes at Ser-645 (S645P, S645F, and S645Y) within hot spot region 1, which are consistent with the most prominent decreases in the drug sensitivity of the GS enzyme (27). The inhibition of GS by echinocandins blocks the biosynthesis of β-1,3-glucan, a critical cell wall component of many pathogenic fungi, which compromises the integrity of the growing cell wall, leading to osmotic instability and the death of susceptible Candida cells (31). Accordingly, exposure of C. albicans to CSF (0.0075 μg/ml for 6 h) was observed to induce an increase in the abundance of several proteins associated with cell wall biosynthesis and integrity, including Rho1p, which is a low-molecular-weight GTPase that regulates the activity of the GS enzyme, in addition to the proteins involved in oxidative and osmotic stress (32).
Although resistance to CSF in C. albicans may be monitored by MALDI-TOF MS through Fks1p and/or Fks2p enzyme modification (33), the specific adaptive proteomic changes in response to drug exposure have not been well characterized. In particular, the nature of biomarkers utilized for the differentiation of truly resistant (fks1-containing) isolates from those of the WT genotype has not been adequately evaluated. To this point, our proof-of-concept study provides strong evidence that minimal drug-induced alterations in protein expression and thus of the mass spectrum of C. albicans can be exploited as rapid and sensitive markers for the emergence of decreased CSF susceptibility in this species. Analogously, in the bacterial field, MALDI-TOF MS has been used for fast and accurate identification of methicillin-resistant and vancomycin-intermediate Staphylococcus aureus strains (34) and has been shown to discriminate between cfiA-negative and cfiA-positive Bacteroides fragilis isolates and then predict carbapenem resistance in a routine laboratory setting (35).
Not surprisingly, the accuracy of ms-AFST for identifying C. albicans isolates with an Fks1-mediated resistance mechanism parallels that displayed by the new species-specific CBPs or, for those species where clinical data are lacking, the efficacy of ECVs in separating non-WT from WT strains, as recently documented (13). Only one isolate was misclassified by ms-AFST, but interestingly, it possesses an atypical fks1 mutation. However, despite the progress in the CLSI AFST of Candida species (13), including validation of 24-h reading times for all antifungal agents (15), MIC determination using the CLSI microdilution reference methodology or its surrogates (36) remains a labor-intensive procedure that requires expertise. This poses a compelling necessity for new, simpler approaches as valid alternatives to the currently available AFST methods. The MALDI-TOF MS-based assay described here may provide such an approach, as it complements the growing use of MS technology for microbial identification in clinical laboratories and has the potential for automated high-throughout use in the near future.
Nonetheless, further experiments are required to assess the scale and range of ms-AFST to facilitate routine adoption by clinical laboratories. First, our method was evaluated with only one Candida species and one drug, and it will be necessary to extend its application to species other than C. albicans, such as Candida glabrata (the other major clinically important Candida species), and to anidulafungin, micafungin, and other classes of antifungal drugs. In this regard, our preliminary results indicate that ms-AFST works well also with C. glabrata (i.e., WT and fks1 mutant isolates) and CSF (data not shown). Second, until it is possible to test several drug classes simultaneously and with all Candida species, ms-AFST in its present state requires laboratories to utilize two AFST methods (i.e., MALDI-TOF MS and the CLSI method), and personnel require training on software algorithms such as CCI-based matching. Finally, the cost benefit of saving 21 h of a patient's initial therapy with CSF, which may not be appropriate if the ms-AFST result indicates that the infecting Candida isolate is resistant, would be counterbalanced only partly by the additional expenses when MALDI-TOF MS, as currently described, is used instead of the CLSI method by which AFST results are available 21 h later.
However, the key to successful therapies and outcomes of patients with invasive Candida infections is prompt initiation of an appropriate antifungal treatment, as antifungal resistance is associated with poorer outcomes (37, 38). In the case of CSF resistance, the occurrence of mutations in the FKS gene is an established important event for both increases in MICs and clinical failures, i.e., breakthrough infections (39). Given that all three of the echinocandins are considered the agents of first choice for the initial treatment of most episodes of invasive candidiasis (40), it is clear that routine AFST can aid not only in the selection of agents for primary therapy but also in a deescalating strategy in order to optimize the effectiveness of antifungal therapy (13).
AFST continues to be a very dynamic field of medical mycology (41), since modifications of the available methods as well as other methodologies are increasingly being investigated (29, 30, 36). In this context, the recent appearance of biophysical methods, such as MALDI-TOF MS, in routine diagnostic microbiology laboratories holds the promise of significantly accelerating the detection of infections caused by fungal pathogens that are resistant to one or more antifungals (19, 21). We have already witnessed the use of MALDI-TOF MS being expanded from fungal identification to antifungal susceptibility testing (19, 42–44). Thus, the present study extends our previous observations by demonstrating that the proposed simplified procedure, i.e., ms-AFST, can be used reliably for the accurate detection of CSF-resistant and CSF-susceptible C. albicans.
In conclusion, as ms-AFST provides useful information sooner than conventional AFST methods, it may be successfully adapted to clinical mycology, especially in those institutions that have acquired MALDI-TOF MS technology for species-level identification purposes. In that case, when applied to the direct analysis of positive blood cultures (43), our approach can provide clinicians with “real-time” identification and AFST results simultaneously to help them in the care of patients with invasive candidiasis. Despite the great potential of ms-AFST, further study is required to better define its reproducibility and standardization.
Supplementary Material
ACKNOWLEDGMENTS
We thank Leopoldo Dimiziani for technical advice on MALDI-TOF MS.
This work was supported by a UCSC-Linea D1 grant to M.S., by NIH grant AI069397 to D.S.P., and a grant from Pfizer to D.S.P. for support for the Echinocandin Resistance Reference Center.
M.K. is employed by the mass spectrometry company Bruker Daltonik. The other authors declare no conflicts of interest.
Footnotes
Published ahead of print 3 July 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JCM.00903-13.
REFERENCES
- 1.Pfaller MA. 2012. Antifungal drug resistance: mechanisms, epidemiology, and consequences for treatment. Am. J. Med. 125:S3–S13 [DOI] [PubMed] [Google Scholar]
- 2.Pasqualotto AC, Denning DW. 2008. New and emerging treatments for fungal infections. J. Antimicrob. Chemother. 61:i19–i30 [DOI] [PubMed] [Google Scholar]
- 3.Sucher AJ, Chahine EB, Balcer HE. 2009. Echinocandins: the newest class of antifungals. Ann. Pharmacother. 43:1647–1657 [DOI] [PubMed] [Google Scholar]
- 4.Arendrup MC. 2010. Epidemiology of invasive candidiasis. Curr. Opin. Crit. Care 16:445–452 [DOI] [PubMed] [Google Scholar]
- 5.Marchetti O, Eggimann P, Calandra T. 2010. Invasive candidiasis in critically ill patients: does progressing knowledge improve clinical management and outcome? Curr. Opin. Crit. Care 16:442–444 [DOI] [PubMed] [Google Scholar]
- 6.Karthaus M, Rüping MJ, Cornely OA, Steinbach A, Groll AH, Lass-Flörl C, Ostermann H, Ruhnke M, Vehreschild JJ. 2011. Current issues in the clinical management of invasive Candida infections—the AGIHO, DMykG, ÖGMM and PEG web-based survey and expert consensus conference 2009. Mycoses 54:e546–e556 [DOI] [PubMed] [Google Scholar]
- 7.Bal AM. 2010. The echinocandins: three useful choices or three too many? Int. J. Antimicrob. Agents 35:13–18 [DOI] [PubMed] [Google Scholar]
- 8.Kanafani ZA, Perfect JR. 2008. Antimicrobial resistance: resistance to antifungal agents: mechanisms and clinical impact. Clin. Infect. Dis. 46:120–128 [DOI] [PubMed] [Google Scholar]
- 9.Castón-Osorio JJ, Rivero A, Torre-Cisneros J. 2008. Epidemiology of invasive fungal infection. Int. J. Antimicrob. Agents 32:S103–S109 [DOI] [PubMed] [Google Scholar]
- 10.Falagas ME, Roussos N, Vardakas KZ. 2010. Relative frequency of albicans and the various non-albicans Candida spp. among candidemia isolates from inpatients in various parts of the world: a systematic review. Int. J. Infect. Dis. 14:e954–e966 [DOI] [PubMed] [Google Scholar]
- 11.Oxman DA, Chow JK, Frendl G, Hadley S, Hershkovitz S, Ireland P, McDermott LA, Tsai K, Marty FM, Kontoyiannis DP, Golan Y. 2010. Candidaemia associated with decreased in vitro fluconazole susceptibility: is Candida speciation predictive of the susceptibility pattern? J. Antimicrob. Chemother. 65:1460–1465 [DOI] [PubMed] [Google Scholar]
- 12.Lass-Flörl C, Perkhofer S, Mayr A. 2010. In vitro susceptibility testing in fungi: a global perspective on a variety of methods. Mycoses 53:1–11 [DOI] [PubMed] [Google Scholar]
- 13.Pfaller MA, Diekema DJ. 2012. Progress in antifungal susceptibility testing of Candida spp. by use of Clinical and Laboratory Standards Institute broth microdilution methods, 2010 to 2012. J. Clin. Microbiol. 50:2846–2856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Alastruey-Izquierdo A, Cuenca-Estrella M. 2012. EUCAST and CLSI: how to assess in vitro susceptibility and clinical resistance. Curr. Fungal Infect. Rep. 6:229–234 [Google Scholar]
- 15.Clinical and Laboratory Standards Institute 2012. Reference method for broth dilution antifungal susceptibility testing of yeasts; 4th informational supplement. CSLI M27-S4. Clinical and Laboratory Standards Institute, Wayne, PA [Google Scholar]
- 16.Freiwald A, Sauer S. 2009. Phylogenetic classification and identification of bacteria by mass spectrometry. Nat. Protoc. 4:732–742 [DOI] [PubMed] [Google Scholar]
- 17.Bizzini A, Greub G. 2010. Matrix-assisted laser desorption ionization time-of-flight mass spectrometry, a revolution in clinical microbial identification. Clin. Microbiol. Infect. 16:1614–1619 [DOI] [PubMed] [Google Scholar]
- 18.Murray PR. 2010. Matrix-assisted laser desorption ionization time-of-flight mass spectrometry: usefulness for taxonomy and epidemiology. Clin. Microbiol. Infect. 16:1626–1630 [DOI] [PubMed] [Google Scholar]
- 19.De Carolis E, Vella A, Florio AR, Posteraro P, Perlin DS, Sanguinetti M, Posteraro B. 2012. Use of matrix-assisted laser desorption ionization–time of flight mass spectrometry (MALDI-TOF MS) for caspofungin susceptibility testing of Candida and Aspergillus species. J. Clin. Microbiol. 50:2479–2483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Arnold RJ, Reilly JP. 1998. Fingerprint matching of E. coli strains with matrix-assisted laser desorption/ionization time-of-flight mass spectrometry of whole cells using a modified correlation approach. Rapid Commun. Mass Spectrom. 12:630–636 [DOI] [PubMed] [Google Scholar]
- 21.Marinach C, Alanio A, Palous M, Kwasek S, Fekkar A, Brossas JY, Brun S, Snounou G, Hennequin C, Sanglard D, Datry A, Golmard JL, Mazier D. 2009. MALDI-TOF MS-based drug susceptibility testing of pathogens: the example of Candida albicans and fluconazole. Proteomics 9:4627–4631 [DOI] [PubMed] [Google Scholar]
- 22.Rudensky B, Broide E, Berko N, Wiener-Well Y, Yinnon AM, Raveh D. 2008. Direct fluconazole susceptibility testing of positive Candida blood cultures by flow cytometry. Mycoses 51:200–204 [DOI] [PubMed] [Google Scholar]
- 23.Ingham CJ, Boonstra S, Levels S, de Lange M, Meis JF, Schneeberger PM. 2012. Rapid susceptibility testing and microcolony analysis of Candida spp. cultured and imaged on porous aluminum oxide. PLoS One 7:e33818. 10.1371/journal.pone.0033818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.De Carolis E, Vella A, Sanguinetti M, Posteraro B. 2012. Abstr. 18th Congr. Int. Soc. Hum. Anim. Mycol., poster P591 [Google Scholar]
- 25.Perlin DS. 2007. Resistance to echinocandin-class antifungal drugs. Drug Resist. Updat. 10:121–130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pfaller MA, Diekema DJ, Andes D, Arendrup MC, Brown SD, Lockhart SR, Motyl M, Perlin DS, CLSI Subcommittee for Antifungal Testing 2011. Clinical breakpoints for the echinocandins and Candida revisited: integration of molecular, clinical, and microbiological data to arrive at species-specific interpretive criteria. Drug Resist. Updat. 14:164–176 [DOI] [PubMed] [Google Scholar]
- 27.Garcia-Effron G, Park S, Perlin DS. 2009. Correlating echinocandin MIC and kinetic inhibition of fks1 mutant glucan synthases for Candida albicans: implications for interpretive breakpoints. Antimicrob. Agents Chemother. 53:112–122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Arendrup MC, Kahlmeter G, Rodriguez-Tudela JL, Donnelly JP. 2009. Breakpoints for susceptibility testing should not divide wild-type distributions of important target species. Antimicrob. Agents Chemother. 53:1628–1629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Arendrup MC, Park S, Brown S, Pfaller M, Perlin DS. 2011. Evaluation of CLSI M44-A2 disk diffusion and associated breakpoint testing of caspofungin and micafungin using a well-characterized panel of wild-type and fks hot spot mutant Candida isolates. Antimicrob. Agents Chemother. 55:1891–1895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Astvad KM, Perlin DS, Johansen HK, Jensen RH, Arendrup MC. 2013. Evaluation of caspofungin susceptibility testing by the new Vitek 2 AST-YS06 yeast card using a unique collection of FKS wild-type and hot spot mutant isolates, including the five most common Candida species. Antimicrob. Agents Chemother. 57:177–182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Walker LA, Gow NA, Munro CA. 2010. Fungal echinocandin resistance. Fungal Genet. Biol. 47:117–126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hoehamer CF, Cummings ED, Hilliard GM, Rogers PD. 2010. Changes in the proteome of Candida albicans in response to azole, polyene, and echinocandin antifungal agents. Antimicrob. Agents Chemother. 54:1655–1664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Havlicek V, Lemr K, Schug KA. 2013. Current trends in microbial diagnostics based on mass spectrometry. Anal. Chem. 85:790–797 [DOI] [PubMed] [Google Scholar]
- 34.Lu JJ, Tsai FJ, Ho CM, Liu YC, Chen CJ. 2012. Peptide biomarker discovery for identification of methicillin-resistant and vancomycin-intermediate Staphylococcus aureus strains by MALDI-TOF. Anal. Chem. 84:5685–5692 [DOI] [PubMed] [Google Scholar]
- 35.Wybo I, De Bel A, Soetens O, Echahidi F, Vandoorslaer K, Van Cauwenbergh M, Piérard D. 2011. Differentiation of cfiA-negative and cfiA-positive Bacteroides fragilis isolates by matrix-assisted laser desorption ionization–time of flight mass spectrometry. J. Clin. Microbiol. 49:1961–1964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pfaller MA, Chaturvedi V, Diekema DJ, Ghannoum MA, Holliday NM, Killian SB, Knapp CC, Messer SA, Miskou A, Ramani R. 2012. Comparison of the Sensititre YeastOne colorimetric antifungal panel with CLSI microdilution for antifungal susceptibility testing of the echinocandins against Candida spp., using new clinical breakpoints and epidemiological cutoff values. Diagn. Microbiol. Infect. Dis. 73:365–368 [DOI] [PubMed] [Google Scholar]
- 37.Parkins MD, Sabuda DM, Elsayed S, Laupland KB. 2007. Adequacy of empirical antifungal therapy and effect on outcome among patients with invasive Candida species infections. J. Antimicrob. Chemother. 60:613–618 [DOI] [PubMed] [Google Scholar]
- 38.Bassetti M, Trecarichi EM, Righi E, Sanguinetti M, Bisio F, Posteraro B, Soro O, Cauda R, Viscoli C, Tumbarello M. 2007. Incidence, risk factors, and predictors of outcome of candidemia. Survey in 2 Italian university hospitals. Diagn. Microbiol. Infect. Dis. 58:325–331 [DOI] [PubMed] [Google Scholar]
- 39.Perlin DS. 2011. Current perspectives on echinocandin class drugs. Future Microbiol. 6:441–457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chen SC, Slavin MA, Sorrell TC. 2011. Echinocandin antifungal drugs in fungal infections: a comparison. Drugs 71:11–41 [DOI] [PubMed] [Google Scholar]
- 41.Arikan S. 2007. Current status of antifungal susceptibility testing methods. Med. Mycol. 45:569–587 [DOI] [PubMed] [Google Scholar]
- 42.De Carolis E, Posteraro B, Lass-Flörl C, Vella A, Florio AR, Torelli R, Girmenia C, Colozza C, Tortorano AM, Sanguinetti M, Fadda G. 2012. Species identification of Aspergillus, Fusarium and Mucorales with direct surface analysis by matrix-assisted laser desorption ionization time-of-flight mass spectrometry. Clin. Microbiol. Infect. 18:475–484 [DOI] [PubMed] [Google Scholar]
- 43.Spanu T, Posteraro B, Fiori B, D'Inzeo T, Campoli S, Ruggeri A, Tumbarello M, Canu G, Trecarichi EM, Parisi G, Tronci M, Sanguinetti M, Fadda G. 2012. Direct MALDI-TOF mass spectrometry assay of blood culture broths for rapid identification of Candida species causing bloodstream infections: an observational study in two large microbiology laboratories. J. Clin. Microbiol. 50:176–179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Posteraro B, Vella A, Cogliati M, De Carolis E, Florio AR, Posteraro P, Sanguinetti M, Tortorano AM. 2012. Matrix-assisted laser desorption ionization–time of flight mass spectrometry-based method for discrimination between molecular types of Cryptococcus neoformans and Cryptococcus gattii. J. Clin. Microbiol. 50:2472–2476 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.