Abstract
Carbapenemase-producing Enterobacteriaceae (CPE) are rapidly spreading worldwide. Early detection of fecal CPE carriers is essential for effective infection control. Here, we evaluated the performance of a meropenem combined disk test (CDT) for rapidly differentiating CPE isolates directly from rectal swabs. The screening method was applied for 189 rectal swabs from hospitalized patients at high risk for CPE carriage. Swabs were suspended in 1 ml saline and cultured for confluent growth onto a MacConkey agar plate with a meropenem (MER) disk alone, a MER disk plus phenyl boronic acid (PBA), a MER disk plus EDTA, and a MER disk plus PBA and EDTA. An inhibition zone of ≤25 mm around the MER disk alone indicated carriage of carbapenem-resistant organisms. Furthermore, ≥5-mm differences in the inhibition zone between MER disks without and with the inhibitors (PBA, EDTA, or both) were considered positive results for detecting Klebsiella pneumoniae carbapenemase (KPC), metallo-β-lactamase (MBL), or both carbapenemases, respectively. For comparison, rectal suspensions were tested using MacConkey plates with ertapenem (MacERT) disks and PCR (PCR-S) for carbapenemase genes. Of the 189 samples, 97 were genotypically confirmed as CPE positive by one of the three protocols tested. The CDT, MacERT disks, and PCR-S assays exhibited sensitivities of 94.8%, 96.9%, and 94.8% and specificities of 100%, 98.9%, and 100%, respectively, for detecting CPE-positive swabs. Moreover, the CDT correctly differentiated the production of KPC, MBL, or both carbapenemases in 78 of the 97 (80.4%) CPE-positive rectal swabs. Our results demonstrate that the CDT may provide a simple and inexpensive method for detecting and differentiating the carbapenemase type within a single day without requiring further testing and additional delay, supporting the timely implementation of infection control measures.
INTRODUCTION
The ongoing dissemination of carbapenemase-producing Enterobacteriaceae (CPE) represents a significant public health issue in many regions of the world. Early detection and infection control strategies are key factors for successfully restricting the further spread of CPE (1–3). Toward this goal, surveillance cultures have been applied effectively as part of the multifaceted nationwide interventional strategies contributing to a decrease in the CPE prevalence in countries where it is highly endemic (4). Furthermore, the prompt identification of CPE carriers at the hospital level may guide the recognition of cases brought into the hospital and may allow the early cohorting of colonized patients, thus assisting in the control of hospital epidemics (5–8). In addition, the identification of an individual patient's carriage status provides information about patients at risk for CPE infections who should be monitored closely, and it facilitates the administration of appropriate empirical treatments (9, 10).
Several approaches have been developed for rectal screening. In-house-prepared protocols include selective agar plates with impregnated carbapenems or with carbapenem disks (11–13). These approaches have been applied with or without initial broth enrichment, which causes an additional delay, while its performance still remains controversial (12, 13). Several chromogenic media, such as CHROMagar KPC and Colorex KPC (CHROMagar, Paris, France) (11, 14–16), Brilliance CRE (Oxoid, Basingstoke, United Kingdom) (14, 17), and chromID Carba (bioMérieux, Marcy l'Etoile, France) (16, 18), have also been used for the active screening of carbapenem-resistant Enterobacteriaceae (CRE) or CPE. In addition, the specific culture medium Supercarba was developed and evaluated for the detection of CPE (14, 19), while the nonspecific chromogenic medium chromID extended-spectrum β-lactamase (ESBL) (bioMérieux) has been applied for CRE detection (16, 20). These culture-based techniques exhibited variable performance characteristics; while they detect carbapenem-resistant organisms, they cannot differentiate the carbapenemase type present in the same day, requiring subsequent phenotypic or molecular testing of isolated colonies, with a further delay of at least 1 day (21). Rapid and specific PCR techniques have also been used to directly identify the carbapenemase type (15, 22–25); however, they do not yield isolates for susceptibility testing, they are not widely applicable, and they are relatively costly.
We have previously shown that a meropenem combined disk test (CDT) provides a simple and accurate phenotypic method for the specific differentiation of Enterobacteriaceae isolates possessing Klebsiella pneumoniae carbapenemases (KPCs), metallo-β-lactamases (MBLs), or both carbapenemase types (26). The aim of the present study was to evaluate this method as an easy and low-cost approach for the rapid detection of CPE and the differentiation of carbapenemase types directly from rectal swabs.
MATERIALS AND METHODS
Setting, patients, and surveillance samples.
Rectal swab specimens were collected from patients hospitalized between September 2010 and February 2012 at Tzaneio General Hospital, a 480-bed tertiary hospital, as part of an active surveillance program for CPE. This program included rectal screening of patients at high risk for CPE carriage: (i) intensive care unit (ICU) patients at admission and weekly, (ii) patients admitted from other institutions or hospitalized during the previous year, (iii) patients with CPE-positive clinical samples, and (iv) contacts of known CPE carriers, defined as patients hospitalized in the same ward or having received care from the same medical or nursing staff. Only the first positive sample per patient was included in the analysis. The rectal sample was collected with a nylon-flocked swab with 5 ml of Amies gel transport medium. The swab was inserted 2 to 3 cm into the rectum, rotated several times, and placed in the tube with the gel transport medium. The tube was directly transferred to the laboratory and then processed.
Rectal screening protocols.
Each swab containing a sample was suspended in 1 ml sterile saline by rotating and agitating it several times to release the microorganisms. Each suspension was subsequently cultured onto two MacConkey agar plates using separate swabs, and bacterial growth was evaluated after an overnight (18- to 24-h) incubation at 37°C in ambient air. The first plate was streaked to four quadrants for colony isolation, and two 10-μg ertapenem (MacERT) disks were added at the junction of the first/second and second/third quadrants; an inhibition zone of ≤27 mm was considered indicative of carbapenem resistance, as suggested previously (13). The second plate was streaked for confluent growth, and the CDT was applied using four 10-μg meropenem (MER) disks (Oxoid, Basingstoke, United Kingdom), including a MER disk alone, a MER disk plus 10 μl of 40 mg/ml phenyl boronic acid (PBA) (Sigma-Aldrich, Steinheim, Germany) for KPC inhibition, a MER disk plus 10 μl of 0.1 M EDTA (Sigma-Aldrich) for MBL inhibition, and a MER disk plus both PBA and EDTA for simultaneous inhibition of KPC (by PBA) and MBL (by EDTA) (26). MER disks were set about 30 mm apart. The inhibition zone of ≤25 mm around the MER disk alone was the optimal breakpoint, exhibiting the best performance for CPE detection. A difference of ≥5 mm in the inhibition zone between the MER disk without and with the inhibitors (PBA, EDTA, or both), taking also into account scattered colonies within the inhibition halo, was considered a positive result for the detection of KPC, MBL, or both carbapenemases, respectively (Fig. 1). Only lactose-fermenting colonies were evaluated. Several enterobacterial colonies with different morphotypes grown around the MER and ERT disks were picked up, subcultured, identified to the species level, and subjected to susceptibility testing by the Vitek 2 automated system (bioMérieux, Marcy l'Étoile, France). These colonies were also tested phenotypically for carbapenemase production (26) that was further confirmed by PCRs for genes encoding carbapenemases (VIM, KPC, IMP, NDM, and OXA-48) (27). PCR-negative isolates were further tested by the modified Hodge test (MHT) according to CLSI guidelines in order to exclude the presence of other carbapenemases not amplified by the PCR primers used.
Fig 1.
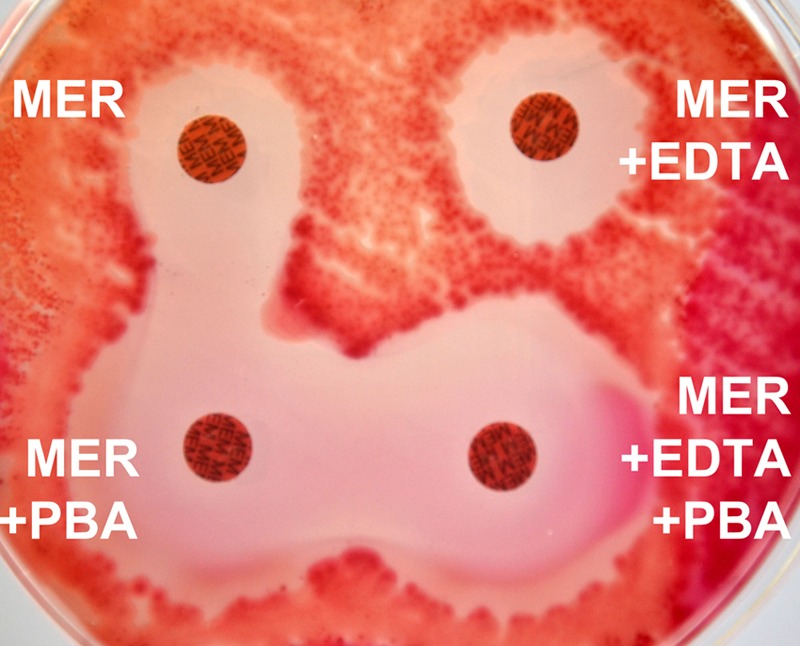
Appearance of a CDT positive for KPC-producing CPE. Inhibition zone diameters were 19 mm for the meropenem (MER) disk, 20 mm for the MER disk + EDTA, 29 mm for the MER disk + phenyl boronic acid (PBA), and 31 mm for the MER disk + PBA + EDTA. MEM, meropenem 10 mg.
In addition, DNA was extracted directly from the same rectal suspensions using a QIAamp DNA mini kit (Qiagen, Inc., Valencia, CA) and was subjected to PCR for the carbapenemase genes (PCR-S) mentioned above.
Limits of CPE differentiation by the CDT screening protocol.
The limits of differentiation (LODs) were determined by spiking CPE-negative rectal swabs, as determined by PCR-S. Nine previously well-characterized CPE strains from our collections that consisted of four KPC-positive K. pneumoniae clinical strains (meropenem MICs, 2, 8, 16, and >32 μg/ml), three VIM-positive K. pneumoniae clinical strains (meropenem MICs, 2, 4, and 16 μg/ml), one KPC- and VIM-positive K. pneumoniae clinical strain (meropenem MIC, 16 μg/ml), and one OXA-48-positive K. pneumoniae clinical strain (meropenem MIC, 1 μg/ml) were used to assess the LOD of the carbapenemase present by the CDT protocol. These strains were thawed, subcultured onto MacConkey plates before use, and suspended in saline to the density of a 0.5 McFarland standard, followed by serial 10-fold dilutions. An aliquot of 100 μl from each dilution (≈107 CFU and lower) was suspended in 900 μl of a CPE-negative rectal suspension and then swabbed onto the CDT plates and also onto the MacConkey plates without MER disks to estimate viable colony counts. The experiments were performed in triplicate. The LOD was the lowest concentration of each strain that resulted in the differentiation of the carbapenemase present.
Sensitivity and specificity.
The sensitivity, specificity, positive predictive value (PPV), negative predictive value (NPV), and overall accuracy were calculated for each of the screening methods. True-positive results for carbapenemase production were considered to be all samples yielding presumptive positive CPE colonies by any of the two MacConkey culture methods described above that were confirmed by PCR testing on isolated colonies or positive for carbapenemase genes by PCR-S. False-positive results were considered to be all presumptive CPE colonies by the CDT and MacERT disks that were CPE negative by confirmatory PCR assays. False-negative results for each individual culture-based protocol were considered to be those not yielding presumptive CPE colonies but being CPE positive by PCR-S or the other culture-based method. False-negative samples by PCR-S were considered to be those having negative PCR-S results but yielding genotypically confirmed CPE colonies by any of the two culture-based protocols. Furthermore, the results of the CDT were evaluated for the accurate identification of the specific carbapenemase type present, as defined by the PCR-S and/or PCR confirmatory assays on isolated colonies.
RESULTS
Detection of CPE isolates in surveillance rectal swabs.
A total of 189 rectal swabs, collected from 165 patients during the study period, were included in the analysis of the present study. CPE isolates were identified in 97 of the 189 (51.3%) swabs by at least one of the tested protocols; they were subsequently confirmed by phenotypic and molecular assays to be 60 KPC positive, 9 VIM positive, 25 KPC and VIM positive, and 3 OXA-48 positive. In particular, the CDT detected as CPE positive 92 of the 97 samples (sensitivity, 94.8%), of which 57 were shown to be KPC positive, 9 to be MBL positive, 25 to be KPC and MBL positive, and 1 OXA-48 to be positive; no false-positive results were produced (specificity, 100%). By the MacERT protocol, 94 CPE-positive samples were identified (sensitivity, 96.9%; 58 KPC positive, 9 MBL positive, 25 KPC and MBL positive, and 2 OXA-48 positive), and one false-positive result was noted (specificity, 98.9%). Lastly, the PCR-S assays yielded 92 CPE-positive samples (sensitivity, 94.8%; 56 KPC positive, 9 MBL positive, 24 KPC and MBL positive, and 3 OXA-48 positive) and no false-positive results. The performance characteristics of the protocols tested are presented in Table 1. Of the 97 CPE-positive samples, 87 (89.7%) were detected by all three methods tested, while 8 (8.2%) and 2 (2.1%) were detected by two methods and one method, respectively.
Table 1.
Performance of the three screening methods for detection of CPE in surveillance swabs
| Methoda | Performance (% [95% CI])b |
||||
|---|---|---|---|---|---|
| Sensitivity | Specificity | PPV | NPV | Accuracy | |
| CDT | 94.8 (87.8–98.0) | 100 (95–100) | 100 (95–100) | 94.8 (87.8–98.0) | 97.4 (95.1–99.6) |
| MacERT | 96.9 (90.5–99.1) | 98.9 (93.2–99.9) | 98.9 (93.4–99.9) | 96.8 (90.2–99.1) | 97.9 (95.8–99.9) |
| PCR-S | 94.8 (87.8–98.0) | 100 (95–100) | 100 (95–100) | 94.8 (87.8–98.0) | 97.4 (95.1–99.6) |
CDT, combined disk test; MacERT, MacConkey agar plate with an ertapenem disk; PCR-S, PCR from a rectal swab.
CI, confidence interval; PPV, positive predictive value; NPV, negative predictive value.
The 94 samples that were CPE positive by culture methods yielded a total of 105 distinct CPE isolates (104 from K. pneumoniae and 1 from Escherichia coli). Confirmatory PCR assays revealed that 79 of these isolates carried the blaKPC gene, 19 carried the blaVIM gene, 5 carried the blaKPC and blaVIM genes, and 2 carried the blaOXA-48 gene.
Limits of differentiation by the CDT protocol.
The LODs, determined by spiking carbapenemase-negative rectal swabs, ranged from 4.1 × 104 to 1.8 × 107 CFU/ml of rectal suspension, depending on the carbapenem MICs of the suspended strains. The MBL-producing organisms had LODs ranging from 4.1 × 104 to 6 × 107 CFU/ml rectal suspension, the KPC-producing organisms had LODs ranging from 5.6 × 104 to 8.4 × 106 CFU/ml, the KPC- and MBL-producing organisms had an LOD of 3.8 × 105 CFU/ml, and the OXA-48-producing organisms had an LOD of 1.8 × 107 CFU/ml.
Differentiation of carbapenemase type in surveillance rectal swabs.
The performances of the CDT and PCR-S assays for direct differentiation of carbapenemase types in the 97 CPE-positive swabs are presented in Table 2. The MacERT method was excluded from this analysis, as this method does not directly differentiate the carbapenemase type present. Overall, the CDT correctly differentiated the carbapenemase type in 78/97 (80.4%) CPE-positive rectal swabs: 57/60 samples with KPC only, 9/9 samples with MBL only, and 11/25 samples with KPC and MBL. On the other hand, the PCR-S assays correctly identified the carbapenemase genes present in 87 of the 97 (89.7%) CPE-positive rectal swabs. It should be noted that all 97 CPE-positive samples were detected by either the CDT or the PCR-S assays.
Table 2.
Ability of the CDT and PCR-S assays to directly differentiate carbapenemase types present in 97 CPE-positive surveillance swabs
| Carbapenemase type (n) | No. (%) positive bya: |
|
|---|---|---|
| CDT | PCR-S | |
| KPC (60) | 57 (95) | 56 (93.3) |
| VIM (9) | 9 (100) | 9 (100) |
| KPC + VIM (25) | 11 (44) | 19 (76) |
| OXA-48 (3) | 1 (33.3) | 3 (100) |
| Total (97) | 78 (80.4) | 87 (89.7) |
CDT, combined disk test; PCR-S, PCR from a rectal swab
The CDT and PCR-S assays produced concordant results for the presence of CPE isolates and differentiation of carbapenemase types in 160 of the 189 (84.7%) surveillance swabs. The analysis of the 29 discrepant results between the CDT and PCR-S assays is presented in Fig. 2. The CDT correctly differentiated KPC producers from producers of MBL or both enzymes in 78 of the 92 (84.8%) CDT-positive samples. Of the remaining 14 samples, the CDT identified only KPC in 12 cases and only MBL in 2 cases. These samples were found by the PCR-S or by PCR confirmatory assays to be positive for the blaKPC and blaVIM genes. The PCR-S assays produced five false-negative results; in four of them, genotypically confirmed KPC-producing K. pneumoniae isolates were recovered by the CDT, and one sample was positive for two different CPE strains carrying the blaKPC and blaVIM genes, respectively. In addition, the PCR-S assays detected only the blaKPC or blaVIM gene in five samples positive by the CDT for both KPC and VIM.
Fig 2.
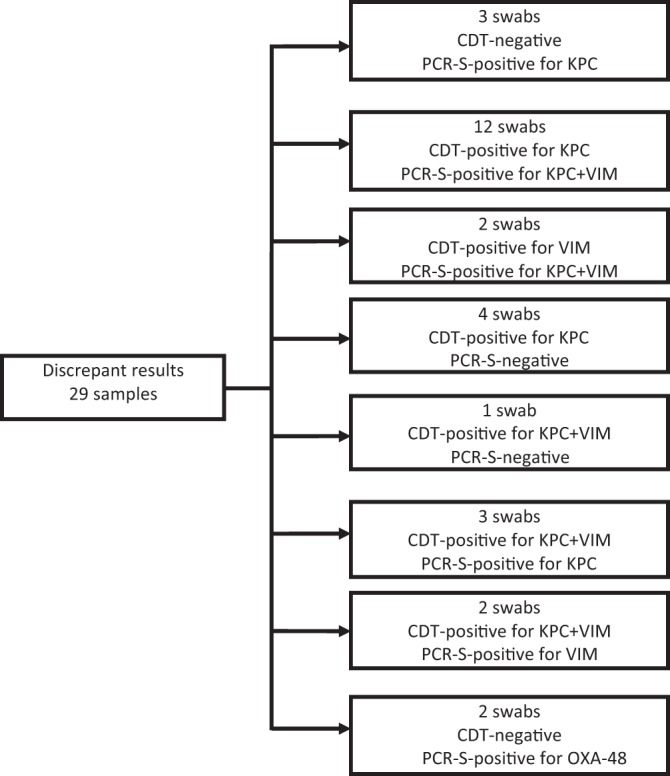
Analysis of discrepant results for CPE isolates and type of carbapenemase present for the methods that differentiate carbapenemase types. CDT, combined disk test; PCR-S, PCR from a rectal swab.
The turnaround time (TAT) of the CDT for specific results for the carbapenemase type was 1 day, and no subsequent testing was necessary. By the PCR-S assay, a specific result was available on the same day of the sampling. On the other hand, the MacERT disk required 1 or 2 extra days compared with the time for the CDT to phenotypically differentiate the carbapenemase type.
It should be noted that 8 samples tested positive by PCR-S for the blaVIM genes, which proved to be due to the presence of VIM-producing Pseudomonas aeruginosa. These results were excluded from our analysis, because the proposed protocol is intended for the identification of CPE. Coincidentally, these samples were also positive for KPC-producing CPE isolates that were correctly identified by all three methods studied.
DISCUSSION
The prompt identification of fecal CPE carriers is an integral part of the interventional strategies required to constrain the global threat of hospital infections due to CPE (1, 2). Several protocols, including culture-based and molecular techniques, have been developed and evaluated in this context. A reference method for screening for carriers has not been proposed, and the performance of each method varies considerably between studies, depending on the comparator method used (11, 18, 21). Thus, there is no consensus on the optimal method for rectal screening. The culture-based methods that have been evaluated to date used either in-house-prepared or commercially available chromogenic media (11–16, 18–20). These methods are intended to detect the presence of carbapenem-resistant and/or carbapenemase-producing isolates but do not provide information about the carbapenemase type present; subsequent testing is necessary to identify the type of carbapenemase, increasing the workload, the cost, and the turnaround time for results.
The simultaneous occurrence or spread of different carbapenemase types in several countries (3, 28) necessitates the identification of the specific enzyme to allow appropriate implementation of patient cohorting. PCR-based assays performed directly on rectal swabs exhibit high performance (15, 22–25); however, they may not be available for routine testing in clinical laboratories, especially in regions with low resources. To facilitate the specific identification of CPE carriers in a routine laboratory practice, we evaluated a simple and inexpensive in-house approach for the rectal detection of CPE and simultaneous differentiation of KPC and MBL production.
The bacterial loads needed to allow the differentiation of the carbapenemase present by the CDTs, as estimated by the spiking of CPE-negative rectal samples, were rather high, as was also reported previously for disk-based screening tests (11), which are considered somewhat less sensitive than chromogenic media for the detection of CPE (21). The relatively high LOD of the CDT could have been anticipated, as this protocol was originally designed to be applied in isolated colonies (26), where heavy inocula of a 0.5 McFarland standard (108 CFU/ml) are swabbed. Despite this LOD, the CDT in our study correctly detected most CPE-positive swabs. This might be attributed to the predictably high bacterial loads of CPE-colonized ICU patients, whose normal flora are significantly affected due to exposure to large doses of antibiotics.
The proposed CDT protocol exhibited high sensitivity (94.8%) and excellent specificity for the detection of CPE while effectively differentiating KPC and/or MBL production. Compared with the alternative approaches tested, the CDT had performance characteristics comparable to those of the MacERT disk, and considering that the CDT saves time and labor, it can be deemed clearly superior. Furthermore, the CDT applied in this study exhibited sensitivity and specificity equal to those of the PCR-S assay, in contrast with a recent report of an investigation of a local outbreak that used a two-disk combined test and showed a lower sensitivity than PCR directly from the rectal swabs (29). The CDTs exhibited five false-negative results, which might be due either to low meropenem MICs, producing large inhibition zones around all meropenem disks, or to low-level CPE carriage. In fact, these false-negative swabs contained three KPC-producing isolates (two had meropenem MICs of 2 μg/ml, and one had a meropenem MIC of 4 μg/ml) and two OXA-48 producers, each having meropenem MICs of 1 μg/ml. The PCR-S assays also exhibited five false-negative results that might be attributed to low bacterial inocula, yielding insufficient DNA templates. It should be noted that the CDTs overall correctly differentiated the carbapenemase type in a relatively lower proportion of CPE-positive swabs (80.4%) than the PCR-S assays (89.7%). However, the PCR-S assay has significant disadvantages: it targets only specific carbapenemase genes (25) (potentially missing genes not amplified by the primers used), it does not reveal enzyme production, it does not yield viable organisms for identification, susceptibility testing, and typing, and it is not suitable for application as a daily routine.
The described CDT protocol cannot differentiate other carbapenemase types apart from KPC and MBLs, such as OXA-48. However, non-MBL- and non-KPC-producing CPE isolates can be detected as colonies grown in the proximity of all disks if they exhibit reduced susceptibility to meropenem. The performance of our protocol for the detection of OXA-48 producers cannot be evaluated due to the low number of positive samples. The detection of OXA-48 producers by culture-based methods is difficult (18) due to their usually low MICs for carbapenems and expanded-spectrum cephalosporins and mainly requires PCR (21). Finally, we did not evaluate the performance of CDT for the differentiation of CRE isolates due to ESBL or AmpC production combined with porin loss, as we did not detect such isolates in surveillance samples from the survey period.
In conclusion, this simple combined disk algorithm enables simultaneous detection and differentiation of the carbapenemase type within 1 day of rectal screening without requiring subsequent testing, delay, and cost, thus supporting the timely implementation of infection control measures. We believe that it is clearly cost-effective for implementation in active CPE surveillance programs. Depending on the local epidemiology of circulating carbapenemase genes, this test can be applied in a simpler way by using only 2 disks (a MER disk only and a MER disk plus PBA or a MER disk plus EDTA).
ACKNOWLEDGMENT
This study was internally funded.
Footnotes
Published ahead of print 10 July 2013
REFERENCES
- 1.Carmeli Y, Akova M, Cornaglia G, Daikos GL, Garau J, Harbarth S, Rossolini GM, Souli M, Giamarellou H. 2010. Controlling the spread of carbapenemase-producing Gram-negatives: therapeutic approach and infection control. Clin. Microbiol. Infect. 16:102–111 [DOI] [PubMed] [Google Scholar]
- 2.Centers for Disease Control and Prevention 2012. 2012 CRE Toolkit—guidance for control of carbapenem-resistant Enterobacteriaceae (CRE). CDC, Atlanta, GA: http://www.cdc.gov/hai/organisms/cre/cre-toolkit/index.html [Google Scholar]
- 3.Gupta N, Limbago BM, Patel JB, Kallen AJ. 2011. Carbapenem-resistant Enterobacteriaceae: epidemiology and prevention. Clin. Infect. Dis. 53:60–67 [DOI] [PubMed] [Google Scholar]
- 4.Schwaber MJ, Lev B, Israeli A, Solter E, Smollan G, Rubinovitch B, Shalit I, Carmeli Y, Israel Carbapenem-Resistant Enterobacteriaceae Working Group 2011. Containment of a country-wide outbreak of carbapenem-resistant Klebsiella pneumoniae in Israeli hospitals via a nationally implemented intervention. Clin. Infect. Dis. 52:848–855 [DOI] [PubMed] [Google Scholar]
- 5.Ben-David D, Maor Y, Keller N, Regev-Yochay G, Tal I, Shachar D, Zlotkin A, Smollan G, Rahav G. 2010. Potential role of active surveillance in the control of a hospital-wide outbreak of carbapenem-resistant Klebsiella pneumoniae infection. Infect. Control Hosp. Epidemiol. 31:620–626 [DOI] [PubMed] [Google Scholar]
- 6.Calfee D, Jenkins SG. 2008. Use of active surveillance cultures to detect asymptomatic colonization with carbapenem-resistant Klebsiella pneumoniae in intensive care unit patients. Infect. Control Hosp. Epidemiol. 29:966–968 [DOI] [PubMed] [Google Scholar]
- 7.Kochar S, Sheard T, Sharma R, Hui A, Tolentino E, Allen G, Landman D, Bratu S, Augenbraun M, Quale J. 2009. Success of an infection control program to reduce the spread of carbapenem-resistant Klebsiella pneumoniae. Infect. Control Hosp. Epidemiol. 30:447–452 [DOI] [PubMed] [Google Scholar]
- 8.Poulou A, Voulgari E, Vrioni G, Xidopoulos G, Pliagkos A, Chatzipantazi V, Markou F, Tsakris A. 2012. Imported Klebsiella pneumoniae carbapenemase-producing K. pneumoniae clones in a Greek hospital: impact of infection control measures for restraining their dissemination. J. Clin. Microbiol. 50:2618–2623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Borer A, Saidel-Odes L, Eskira S, Nativ R, Riesenberg K, Livshiz-Riven I, Schlaeffer F, Sherf M, Peled N. 2012. Risk factors for developing clinical infection with carbapenem-resistant Klebsiella pneumoniae in hospital patients initially only colonized with carbapenem-resistant K. pneumoniae. Am. J. Infect. Control 40:421–425 [DOI] [PubMed] [Google Scholar]
- 10.Schechner V, Kotlovsky T, Kazma M, Mishali H, Schwartz D, Navon-Venezia S, Schwaber MJ, Carmeli Y. 2013. Asymptomatic rectal carriage of blaKPC producing carbapenem-resistant Enterobacteriaceae: who is prone to become clinically infected? Clin. Microbiol. Infect. 19:451–456 [DOI] [PubMed] [Google Scholar]
- 11.Adler A, Navon-Venezia S, Moran-Gilad J, Marcos E, Schwartz D, Carmeli Y. 2011. Laboratory and clinical evaluation of screening agar plates for detection of carbapenem-resistant Enterobacteriaceae from surveillance rectal swabs. J. Clin. Microbiol. 49:2239–2242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Landman D, Salvani JK, Bratu S, Quale J. 2005. Evaluation of techniques for detection of carbapenem-resistant Klebsiella pneumoniae in stool surveillance cultures. J. Clin. Microbiol. 43:5639–5641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lolans K, Calvert K, Won S, Clark J, Hayden MK. 2010. Direct ertapenem disk screening method for identification of KPC-producing Klebsiella pneumoniae and Escherichia coli in surveillance swab specimens. J. Clin. Microbiol. 48:836–841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Girlich D, Poirel L, Nordmann P. 2013. Comparison of the Supercarba, CHROMagar KPC, and Brilliance CRE screening media for detection of Enterobacteriaceae with reduced susceptibility to carbapenems. Diagn. Microbiol. Infect. Dis. 75:214–217 [DOI] [PubMed] [Google Scholar]
- 15.Samra Z, Bahar J, Madar-Shapiro L, Aziz N, Israel S, Bishara J. 2008. Evaluation of CHROMagar KPC for rapid detection of carbapenem-resistant Enterobacteriaceae. J. Clin. Microbiol. 46:3110–3111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wilkinson KM, Winstanley TG, Lanyon C, Cummings SP, Raza MW, Perry JD. 2012. Comparison of four chromogenic culture media for carbapenemase-producing Enterobacteriaceae. J. Clin. Microbiol. 50:3102–3104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Day KM, Ali S, Mirza IA, Sidjabat HE, Silvey A, Lanyon CV, Cummings SP, Abbasi SA, Raza MW, Paterson DL, Perry JD. 2013. Prevalence and molecular characterization of Enterobacteriaceae producing NDM-1 carbapenemase at a military hospital in Pakistan and evaluation of two chromogenic media. Diagn. Microbiol. Infect. Dis. 75:187–191 [DOI] [PubMed] [Google Scholar]
- 18.Vrioni G, Daniil I, Voulgari E, Ranellou K, Koumaki V, Ghirardi S, Kimouli M, Zambardi G, Tsakris A. 2012. Comparative evaluation of a prototype chromogenic medium (ChromID CARBA) for detecting carbapenemase-producing Enterobacteriaceae in surveillance rectal swabs. J. Clin. Microbiol. 50:1841–1846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nordmann P, Girlich D, Poirel L. 2012. Detection of carbapenemase producers in Enterobacteriaceae by use of a novel screening medium. J. Clin. Microbiol. 50:2761–2766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Carrër A, Fortineau N, Nordmann P. 2010. Use of ChromID extended-spectrum beta-lactamase medium for detecting carbapenemase-producing Enterobacteriaceae. J. Clin. Microbiol. 48:1913–1914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gazin M, Paasch F, Goossens H, Malhotra-Kumar S, MOSAR WP2 and SATURN WP1 Study Teams 2012. Current trends in culture-based and molecular detection of extended-spectrum-β-lactamase-harboring and carbapenem-resistant Enterobacteriaceae. J. Clin. Microbiol. 50:1140–1146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hindiyeh M, Smollen G, Grossman Z, Ram D, Davidson Y, Mileguir F, Vax M, Ben David D, Tal I, Rahav G, Shamiss A, Mendelson E, Keller N. 2008. Rapid detection of blaKPC carbapenemase genes by real-time PCR. J. Clin. Microbiol. 46:2879–2883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mangold KA, Santiano K, Broekman R, Krafft CA, Voss B, Wang V, Hacek DM, Usacheva EA, Thomson RB, Jr, Kaul KL, Peterson LR. 2011. Real-time detection of blaKPC in clinical samples and surveillance specimens. J. Clin. Microbiol. 49:3338–3339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Richter SN, Frasson I, Biasolo MA, Bartolini A, Cavallaro A, Palù G. 2012. Ultrarapid detection of blaKPC1/2–12 from perirectal and nasal swabs by use of real-time PCR. J. Clin. Microbiol. 50:1718–1720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Singh K, Mangold KA, Wyant K, Schora DM, Voss B, Kaul KL, Hayden MK, Chundi V, Peterson LR. 2012. Rectal screening for Klebsiella pneumoniae carbapenemases: comparison of real-time PCR and culture using two selective screening agar plates. J. Clin. Microbiol. 50:2596–2600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tsakris A, Poulou A, Pournaras S, Voulgari E, Vrioni G, Themeli-Digalaki K, Petropoulou D, Sofianou D. 2010. A simple phenotypic method for the differentiation of metallo-beta-lactamases and class A KPC carbapenemases in Enterobacteriaceae clinical isolates. J. Antimicrob. Chemother. 65:1664–1671 [DOI] [PubMed] [Google Scholar]
- 27.Pournaras S, Poulou A, Voulgari E, Vrioni G, Kristo I, Tsakris A. 2010. Detection of the new metallo-beta-lactamase VIM-19 along with KPC-2, CMY-2 and CTX-M-15 in Klebsiella pneumoniae. J. Antimicrob. Chemother. 65:1604–1607 [DOI] [PubMed] [Google Scholar]
- 28.Cantón R, Akóva M, Carmeli Y, Giske CG, Glupczynski Y, Gniadkowski M, Livermore DM, Miriagou V, Naas T, Rossolini GM, Samuelsen O, Seifert H, Woodford N, Nordmann P, the European Network on Carbapenemases 2012. Rapid evolution and spread of carbapenemases among Enterobacteriaceae in Europe. Clin. Microbiol. Infect. 18:413–431 [DOI] [PubMed] [Google Scholar]
- 29.Giani T, Tascini C, Arena F, Ciullo I, Conte V, Leonildi A, Menichetti F, Rossolini GM. 2012. Rapid detection of intestinal carriage of Klebsiella pneumoniae producing KPC carbapenemase during an outbreak. J. Hosp. Infect. 81:119–122 [DOI] [PubMed] [Google Scholar]


