Abstract
Among 36 Mycobacterium masilliense and 22 M. abscessus isolates identified by erm(41) PCR and sequencing analysis of rpoB and 23S rRNA genes, the rate of accurate differentiation between these two subspecies was 100% by cluster analysis of spectra generated by Bruker Biotyper matrix-assisted laser desorption ionization–time of flight mass spectrometry.
TEXT
Mycobacterium abscessus complex, a rapidly growing mycobacterium, is the cause of an increasing number of community- and health care-associated infections in humans (1). The isolation of M. abscessus complex from patients with various clinical infections has been reported in many countries, including Taiwan (2–7). The M. abscessus complex comprises three closely related subspecies, namely, M. massiliense, M. bolletii, and M. abscessus (sensu stricto) (4, 5, 7–9). Identification of M. abscessus complex members to the species level depends on sequencing analysis of several genes, including the erm(41) gene, the 23S rRNA gene, and several housekeeping genes (e.g., rpoB and hsp65) (7, 9–12). A previous report found that erm(41) PCR can differentiate M. massiliense from M. abscessus and M. bolletii but that sequencing analysis of rpoB and hsp65 was less reliable at differentiating between the two (9). M. abscessus subsp. bolletti is now the recommended taxonomic name for M. massiliense (4, 12). Differences of in vitro susceptibilities to clarithromycin between M. massiliense (M. abscessus subsp. bolletti) and M. abscessus (sensu stricto) isolates and of treatment response rates of lung diseases caused by these two species with clarithromycin-based antibiotic therapy have been reported (7–12). However, the use of molecular methods to differentiate among these subspecies is not possible in many routine microbiology laboratories.
The use of matrix-assisted laser desorption ionization–time of flight mass spectrometry (MALDI-TOF MS) is relatively new in the field of microbiology for species identification of yeasts and bacteria, including mycobacteria (13–17). Although MALDI-TOF MS has been shown to be a highly accurate method for identifying M. abscessus complex isolates to the species level, this method has not been shown to be able to differentiate between subspecies of the M. abscessus complex, i.e., M. massiliense (M. abscessus subsp. bolletti) and M. abscessus (sensu stricto) (15, 16).
In this study, we evaluated a total of 58 isolates of the M. abscessus complex obtained from various clinical specimens from patients treated during the period from January 2011 to December 2012 at the National Taiwan University Hospital, a 2,900-bed tertiary-care medical center in northern Taiwan. The isolates were presumptively identified as M. abscessus complex based on conventional biochemical methods as previously described (4–6). These isolates were further identified to the subspecies level by sequencing the erm(41) gene and by performing sequence analysis techniques targeting the rpoB and 23S rRNA genes. The following primer pairs were used: ermF (5′-GAC CGG GGC CTT CTT CGT GAT-3′) and ermR1 (5′-GAC TTC CCC GCA CCG ATT CC-3′) for the whole erm(41) gene; rpoB F (5′-GGCAAGGTCACCCCGAAGGG-3′) and rpoB R (5′-AGCGGCTGCTGGGTGATCATC-3′) for the rpoB gene; and 19 (5′-GTAGCGAAATTCCTTGTCGG-3′) and 21 (5′-TTCCCGCTTAGATGCTTTCAG-3′) for the 23S rRNA gene (4, 5). The sequences obtained were compared with published sequences in the GenBank database by using the BLASTN algorithm (http://www.ncbi.nlm.nih.gov/blast).
Among the 58 M. abscessus complex isolates, 22 were confirmed to be M. abscessus (sensu stricto) [smaller DNA of 673 bp amplified by erm(41) PCR; GenBank accession numbers JF346872.1 for the rpoB gene and EU590128.1 for 23S rRNA] and 36 were M. massiliense [smaller DNA of 397 bp amplified by erm(41) PCR; GenBank accession numbers CP003699.1 for the rpoB gene and GU143887.1 for 23S rRNA].
For MALDI-TOF MS analysis, a 10-μl inoculation loop was used to obtain M. abscessus complex colonies that had been grown in Middlebrook 7H11 agar plates (Becton, Dickinson-Diagnostic Systems, Sparks, MD) for 3 days. The MALDI-TOF MS protein extraction protocol for mycobacteria was performed as described previously, with modifications (16). A full loop of colonies was suspended in 500 μl of distilled water in a 1.5-ml screw-cap Eppendorf tube and was inactivated at 100°C for 30 min to kill the mycobacteria. The microtube was centrifuged at 13,000 rpm for 2 min, and the supernatant was discarded. The pellet was washed with 70% ethanol, vortexed briefly, and centrifuged at 13,000 rpm for 2 min. The supernatant was decanted, the residual fluid was removed, and the pellet was dried at room temperature for 2 min. Twenty microliters of 0.5-mm-diameter silica beads and 20 μl of pure acetonitrile were added, and the mixture vortexed for 1 min. Twenty microliters of 70% formic acid was added, and the mixture vortexed for 10 s. One microliter of the supernatant was used for analysis by MALDI TOF MS.
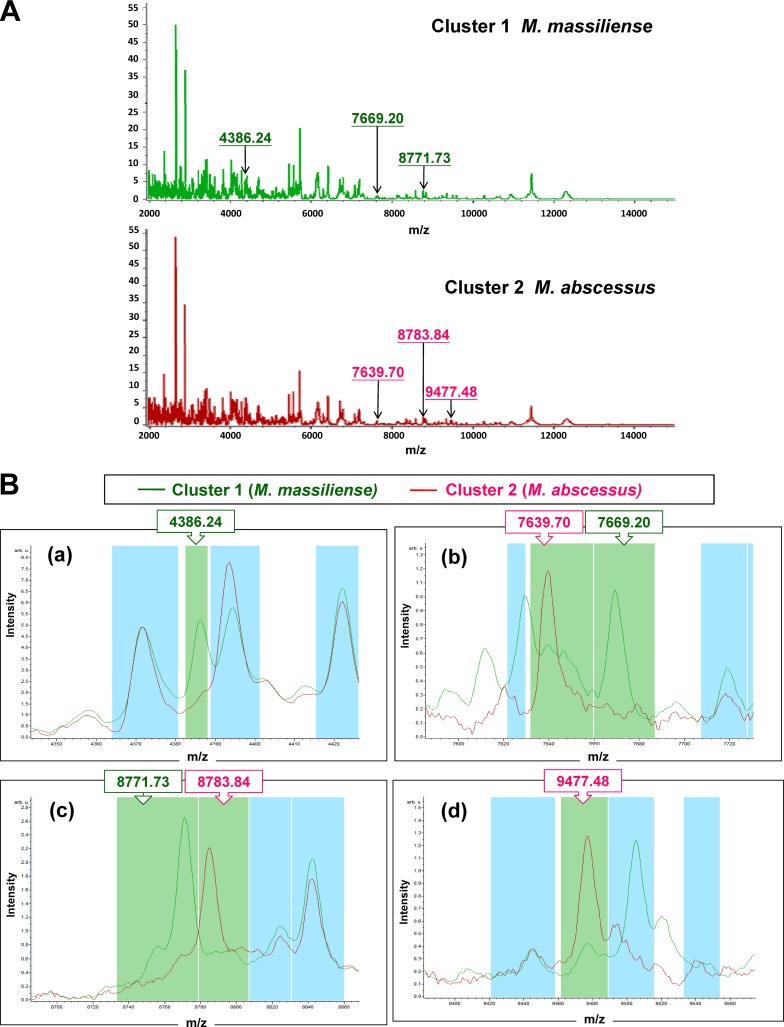
MALDI-TOF MS measurement of the isolates was performed using a MALDI Biotyper system (Microflex LT; Bruker Daltonik GmbH, Bremen, Germany) with Compass Flex Series version 1.3 software and a 60-Hz nitrogen laser (337-nm wavelength). Spectra were collected in the linear positive mode in a mass range covering 1,960 to 20,132 m/z. ClinProTools (version 3.0.22; Bruker Daltoniks GmbH, Bremen, Germany) was used to generate classification models to differentiate among M. massiliense and M. abscessus (sensu stricto). All 58 isolates were initially identified by Bruker Biotyper MALDI-TOF MS as M. abscessus complex because of the lack of M. massiliense and M. abscessus (sensu stricto) in the database (MaldiBiotyperDB version 3.3.1.0 and Mycolib version 1.0.3). About 70 to 100 prominent ion peaks were noted in the m/z 2,000 to 12,000 range. Of these isolates, 50 (86.2%) had identification scores of >2.0 (range, 2.033 to 2.312) and 8 (13.8%) had identification scores of <2.0 (range, 1.74 to 1.911). Clustering analysis of the spectra from the 58 isolates of the M. abscessus complex, including 36 M. massiliense and 22 M. abscessus (sensu stricto) isolates, revealed two characteristic clusters (clusters 1 and 2) (Fig. 1A). Six peaks in the spectra of the isolates, i.e., 4,386.24 m/z, 7,639.70 m/z, 7,669.20 m/z, 8,771.73 m/z, 8,783.84 m/z, and 9,477.48 m/z, which were generated by ClinProTools with the genetic algorithm, were used to define clusters 1 and 2. The signals 4,386.24 m/z, 7,669.2 m/z, and 8,771.73 m/z were observed in the spectrum for cluster 1 (all were M. massiliense) but not in the spectrum for cluster 2 (all were M. abscessus [sensu stricto]), and those of 7,639.7 m/z, 8,783.84 m/z, and 9,477.48 m/z were observed in the cluster 2 spectrum but not in the cluster 1 spectrum (Fig. 1B).
Fig 1.
(A) Two clusters of M. abscessus complex spectra, i.e., cluster 1 (M. massiliense) and cluster 2 (M. abscessus [sensu stricto]), analyzed by clustering analysis of MALDI-TOF MS results. (B) The six peaks used to define cluster 1 (M. massiliense) and cluster 2 (M. abscessus [sensu stricto]), which were generated by ClinProTools with the genetic algorithm, are 4,386.24 m/z, 7,639.70 m/z, 7,669.20 m/z, 8,771.73 m/z, 8,783.84 m/z, and 9,477.48 m/z. The signals of 4,386.24 m/z, 7,669.2 m/z, and 8,771.73 m/z were observed in cluster 1 spectra but not in cluster 2 spectra, and those of 7,639.7 m/z, 8,783.84 m/z, and 9,477.48 m/z (class 2) were observed in cluster 2 spectra but not in cluster 1 spectra. The absolute intensities of the ions are shown on the y axis, and the masses (m/z) of the ions are shown on the x axis. The m/z values represent the mass-to-charge ratio.
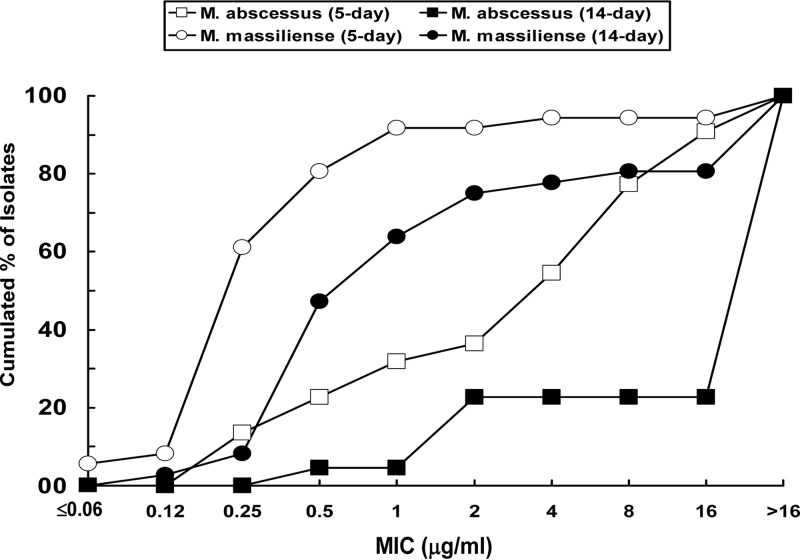
The MICs of 15 antimicrobial agents against the 58 M. abscessus complex isolates were determined using the Sensititre RAPMYCO panel test (TREK Diagnostic Systems, West Sussex, United Kingdom) (5). The MICs of all agents tested were read on the fifth day (day 5) after incubation, and those of clarithromycin were also read on extended incubation (14 days). All the antimicrobial agents tested exhibited poor in vitro activities against both M. abscessus (sensu stricto) and M. massiliense isolates (Table 1). The MIC distribution of clarithromycin against M. massiliense and M. abscessus (sensu stricto) is shown in Fig. 2. The MIC90 of clarithromycin was 16 μg/ml for M. abscessus (sensu stricto) and 1 μg/ml for M. massiliense when the susceptibility assays were read on day 5 and >16 μg/ml when the susceptibility assays were read on day 14 for both species isolates (Table 1). Seven of the M. abscessus (sensu stricto) isolates had clarithromycin MICs of ≤2 μg/ml when the broth susceptibility test results were read on day 5. However, these seven M. abscessus (sensu stricto) isolates all showed clarithromycin MIC values of >16 (≥32) mg/ml on day 14, indicating that these isolates exhibited inducible resistance to clarithromycin (7). In contrast, the clarithromycin MIC values of 33 of 36 M. massiliense isolates were ≤2 μg/ml when the broth susceptibility test results were read on day 5; however, 11 (33.3%) of the 33 isolates exhibited clarithromycin MICs of >16 (≥32) μg/ml during the 14-day observation.
Table 1.
In vitro susceptibilities of M. massiliense and M. abscessus (sensu stricto) to 15 antimicrobial agents
| Antimicrobial agent | MIC (μg/ml) for: |
|||||
|---|---|---|---|---|---|---|
|
M. massiliense (n = 36) |
M. abscessus (sensu stricto) (n = 22) |
|||||
| Range | 50% | 90% | Range | 50% | 90% | |
| Amoxicillin-clavulanate | 16/8–>64/32 | >64/32 | >64/32 | 64/32–>64/32 | >64/32 | >64/32 |
| Cefoxitin | 32–>128 | 64 | 128 | 32–>128 | 64 | 128 |
| Ceftriaxone | 64–>64 | >64 | >64 | 64–>64 | >64 | >64 |
| Cefepime | >32 | >32 | >32 | >32 | >32 | >32 |
| Imipenem | 8–>64 | 32 | >64 | 16–>64 | 32 | 64 |
| Clarithromycin (day 5) | 0.06–>16 | 0.25 | 1 | 0.25–>16 | 4 | 16 |
| Clarithromycin (day 14) | 0.12–>16 | 1 | >16 | 0.5–>16 | >16 | >16 |
| Ciprofloxacin | >4 | >4 | >4 | 2–>4 | >4 | >4 |
| Moxifloxacin | 8–>8 | >8 | >8 | 1–>8 | >8 | >8 |
| Tobramycin | 8–>16 | >16 | >16 | 16–>16 | 16 | >16 |
| Amikacin | 8–>64 | 16 | >64 | 8–>64 | 16 | 64 |
| Trimethoprim-sulfamethoxazole | 1/19–>8/152 | >8/152 | >8/152 | 1/19–>8/152 | 8/152 | >8/152 |
| Doxycycline | 16–>16 | >16 | >16 | 16–>16 | >16 | >16 |
| Minocycline | 8–>8 | >8 | >8 | >8 | >8 | >8 |
| Tigecycline | 0.06–>4 | 2 | >4 | 0.5–>4 | 2 | 4 |
| Linezolid | 16–>32 | >32 | >32 | 16–>32 | >32 | >32 |
Fig 2.
Distribution of MICs of clarithromycin against M. massiliense (n = 36) and M. abscessus (sensu stricto) (n = 22) determined by the Sensititre RAPMYCOI panel test (TREK Diagnostic Systems). Results were read on the 5th day of incubation and on day 14 (extended incubation).
Previous studies have shown that rapidly growing mycobacteria, especially those of the M. abscessus complex, are the most common clinical isolates of mycobacteria and are a major cause of pulmonary disease due to nontuberculous mycobacteria (NTM) in Asia (2, 3, 18–20). Saleeb et al. reported that MALDI-TOF MS can be incorporated into the work flow of the microbiology laboratory for rapid and accurate identification of most species of mycobacteria (16). However, they also indicated that three pairs of closely related strains, namely, M. abscessus and M. massiliense, M. mucogenicum and M. phocaicum, and M. chimaera and M. intracellulare, could not be differentiated from each other by MALDI-TOF MS (16). Differentiation between two subspecies among M. abscessus complex isolates is important clinically because of the differences in their susceptibility profiles and clinical relevance (4, 5, 7, 9, 21). M. massiliense isolates tend to be more susceptible to clarithromycin than M. abscessus (sensu stricto) isolates (5, 7, 9, 21). In this study, we found that M. abscessus (sensu stricto) isolates were less susceptible to clarithromycin and had higher rates of resistance to that antibiotic than isolates of M. massiliense. Furthermore, a surprisingly high proportion of M. massiliense has been reported among M. abscessus complex central nervous system infections, and a higher proportion of M. abscessus (sensu stricto) among M. abscessus complex otologic infections (4, 5, 18).
In summary, our findings show that cluster analysis of spectra generated by MALDI-TOF MS can accurately differentiate between M. massiliense and M. abscessus (sensu stricto), although the number of isolates used in the study was small. This technique could be incorporated into the work flow of the microbiology laboratory for rapid and accurate differentiation between M. massiliense and M. abscessus (sensu stricto) among members of the M. abscessus complex.
Footnotes
Published ahead of print 3 July 2013
REFERENCES
- 1.Petrini B. 2006. Mycobacterium abscessus: an emerging rapid-growing potential pathogen. APMIS 114:319–328 [DOI] [PubMed] [Google Scholar]
- 2.Simons S, van Ingen J, Hsueh PR, Van Hung N, Dekhuijzen PN, Boeree MJ, van Soolingen D. 2011. Nontuberculous mycobacteria in respiratory tract infections, eastern Asia. Emerg. Infect. Dis. 17:343–349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lai CC, Wang HC. 2011. Clinical significance of Mycobacterium abscessus isolates at a medical center in northern Taiwan. J. Microbiol. Immunol. Infect. 44:488–489 [DOI] [PubMed] [Google Scholar]
- 4.Lee MR, Tsai HY, Cheng A, Liu CY, Huang YT, Liao CH, Liang SK, Lee LN, Hsueh PR. 2012. Otitis media and otomastoiditis caused by Mycobacterium massiliense (Mycobacterium abscessus subspecies bolletii). J. Clin. Microbiol. 50:3754–3756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lee MR, Cheng A, Lee YC, Yang CY, Lai CC, Huang YT, Ho CC, Wang HC, Yu CJ, Hsueh PR. 2012. CNS infections caused by Mycobacterium abscessus complex: clinical features and antimicrobial susceptibilities of isolates. J. Antimicrob. Chemother. 67:222–225 [DOI] [PubMed] [Google Scholar]
- 6.Lai CC, Tan CK, Chou CH, Hsu HL, Liao CH, Huang YT, Yang PC, Luh KT, Hsueh PR. 2010. Increasing incidence of nontuberculous mycobacteria, Taiwan, 2000-2008. Emerg. Infect. Dis. 16:294–296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Koh WJ, Jeon K, Lee NY, Kim BJ, Kook YH, Lee SH, Park YK, Kim CK, Shin SJ, Huitt GA, Daley CL, Kwon OJ. 2011. Clinical significance of differentiation of Mycobacterium massiliense from Mycobacterium abscessus. Am. J. Respir. Crit. Care Med. 183:405–410 [DOI] [PubMed] [Google Scholar]
- 8.Harada T, Akiyama Y, Kurashima A, Nagai H, Tsuyuguchi K, Fujii T, Yano S, Shigeto E, Kuraoka T, Kajiki A, Kobashi Y, Kokubu F, Sato A, Yoshida S, Iwamoto T, Saito H. 2012. Clinical and microbiological differences between Mycobacterium abscessus and Mycobacterium massiliense lung diseases. J. Clin. Microbiol. 50:3556–3561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kim HY, Kim BJ, Kook Y, Yun YJ, Shin JH, Kim BJ, Kook YH. 2010. Mycobacterium massiliense is differentiated from Mycobacterium abscessus and Mycobacterium bolletii by erythromycin ribosome methyltransferase gene (erm) and clarithromycin susceptibility patterns. Microbiol. Immunol. 54:347–353 [DOI] [PubMed] [Google Scholar]
- 10.Nash KA, Brown-Elliott BA, Wallace RJ., Jr 2009. A novel gene, erm(41), confers inducible macrolide resistance to clinical isolates of Mycobacterium abscessus but is absent from Mycobacterium chelonae. Antimicrob. Agents Chemother. 53:1367–1376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kim HY, Kook Y, Yun YJ, Park CG, Lee NY, Shim TS, Kim BJ, Kook YH. 2008. Proportions of Mycobacterium massiliense and Mycobacterium bolletii strains among Korean Mycobacterium chelonae-Mycobacterium abscessus group isolates. J. Clin. Microbiol. 46:3384–3390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Leão SC, Tortoli E, Euzeby JP, Garcia MJ. 2011. Proposal that Mycobacterium massiliense and Mycobacterium bolletii be united and reclassified as Mycobacterium abscessus subsp. bolletii comb. nov., designation of Mycobacterium abscessus subsp. abscessus subsp. nov. and emended description of Mycobacterium abscessus. Int. J. Syst. Evol. Microbiol. 61:2311–2313 [DOI] [PubMed] [Google Scholar]
- 13.Bizzini A, Greub G. 2010. Matrix-assisted laser desorption ionization time-of-flight mass spectrometry, a revolution in clinical microbial identification. Clin. Microbiol. Infect. 16:1614–1619 [DOI] [PubMed] [Google Scholar]
- 14.Wang J, Chen WF, Li QX. 2012. Rapid identification and classification of Mycobacterium spp. using whole-cell protein barcodes with matrix assisted laser desorption ionization time of flight mass spectrometry in comparison with multigene phylogenetic analysis. Anal. Chim. Acta 2716:133–137 [DOI] [PubMed] [Google Scholar]
- 15.El Khéchine A, Couderc C, Flaudrops C, Raoult D, Drancourt M. 2011. Matrix-assisted laser desorption/ionization time-of-flight mass spectrometry identification of mycobacteria in routine clinical practice. PLoS One 6:e24720. 10.1371/journal.pone.0024720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Saleeb PG, Drake SK, Murray PR, Zelazny AM. 2011. Identification of mycobacteria in solid-culture media by matrix-assisted laser desorption ionization-time of flight mass spectrometry. J. Clin. Microbiol. 49:1790–1794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lotz A, Ferroni A, Beretti JL, Dauphin B, Carbonnelle E, Guet-Revillet H, Veziris N, Heym B, Jarlier V, Gaillard JL, Pierre-Audigier C, Frapy E, Berche P, Nassif X, Bille E. 2010. Rapid identification of mycobacterial whole cells in solid and liquid culture media by matrix-assisted laser desorption ionization-time of flight mass spectrometry. J. Clin. Microbiol. 48:4481–4486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hoefsloot W, van Ingen J, Andrejak C, Angeby K, Bauriaud R, Bemer P, Beylis N, Boeree MJ, Cacho J, Chihota V, Chimara E, Churchyard G, Cias R, Dasa R, Daley CL, Dekhuijzen PN, Domingo D, Drobniewski F, Esteban J, Fauville-Dufaux M, Folkvardsen DB, Gibbons N, Gómez-Mampaso E, Gonzalez R, Hoffmann H, Hsueh PR, Indra A, Jagielski T, Jamieson F, Jankovic M, Jong E, Keane J, Koh WJ, Lange B, Leao S, Macedo R, Mannsåker T, Marras TK, Maugein J, Milburn HJ, Mlinkó T, Morcillo N, Morimoto K, Papaventsis D, Palenque E, Paez-Peña M, Piersimoni C, Polanová M, Rastogi N, Richter E. 18 April 2013. The geographic diversity of nontuberculous mycobacteria isolated from pulmonary samples: a NTM-NET collaborative study. Eur. Respir. J. [Epub ahead of print.] 10.1183/09031936.00149212 [DOI] [PubMed] [Google Scholar]
- 19.Chen CY, Chen HY, Chou CH, Huang CT, Lai CC, Hsueh PR. 2012. Pulmonary infection caused by nontuberculous mycobacteria in a medical center in Taiwan, 2005-2008. Diagn. Microbiol. Infect. Dis. 72:47–51 [DOI] [PubMed] [Google Scholar]
- 20.Chen CY, Sheng WH, Lai CC, Liao CH, Huang YT, Tsay W, Huang SY, Tang JL, Tien HF, Hsueh PR. 2012. Mycobacterial infections in adult patients with hematological malignancy. Eur. J. Clin. Microbiol. Infect. Dis. 31:1059–1066 [DOI] [PubMed] [Google Scholar]
- 21.Nessar R, Cambau E, Reyrat JM, Murray A, Gicquel B. 2012. Mycobacterium abscessus: a new antibiotic nightmare. J. Antimicrob. Chemother. 67:810–818 [DOI] [PubMed] [Google Scholar]