Abstract
Sixty-five CTX-M-2/15/14 extended-spectrum-β-lactamase-producing Enterobacteriaceae were isolated from 258,888 mastitic milk samples from Japanese dairy farms between 2007 and 2011. CTX-M-2-producing Klebsiella pneumoniae and CTX-M-15-producing Escherichia coli were the predominant strains isolated. There was no predominant clonal type, and clonal diversity was found even in strains isolated from a single farm.
TEXT
Since 2000, Escherichia coli and other Enterobacteriaceae species producing CTX-M-type extended-spectrum β-lactamases (ESBLs) (CTX-M) have been commonly isolated from community-acquired extraintestinal infections in humans and their companion animals (1, 2, 3, 4), from food-producing animals (3, 5, 6, 7, 8), and from retail meats, including chicken, beef, and pork (3), worldwide. The CTX-M-type genes are assumed to have been transferred separately to plasmids, including complex class 1 integrons and transposons (9), from chromosomes of different Kluyvera species (i.e., Kluyvera ascorbata, K. georgiana, and K. cryocrescens) that live in water, soil, and human and animal intestinal tracts; therefore, CTX-M has been divided into five clusters (CTX-M-1, CTX-M-2, CTX-M-8, CTX-M-9, and CTX-M-25) from base sequence homology (1, 9). CTX-M confers resistance against penicillins, oxyimino-cephalosporins, and monobactams (1, 4). Recently, the CTX-M-15-producing E. coli ST131 (O25:H4) clone has emerged as a multidrug-resistant pandemic strain affecting humans worldwide (4).
Bovine mastitis is the most common disease affecting dairy cattle (10). Both E. coli and Klebsiella pneumoniae often cause life-threatening clinical mastitis (5, 11). The incidence of bovine mastitis has been reported to be higher in Japan (30 to 35 cases per 100 cow-years at risk) (12) than in North America, Europe, and New Zealand (10 to 30 cases per 100 cow-years) (10). Only a few classes of antimicrobials are approved for the treatment of mastitis in Japan; however, large amounts of antimicrobials are used for mastitis treatment, creating selective pressure for drug-resistant organisms (13). In our previous report, we showed that Japanese dairy cattle might be a source of CTX-M-15/2/14- and CMY-2-producing Enterobacteriaceae (7). However, few studies have reported the prevalence of Enterobacteriaceae producing CTX-M in bovine mastitis (5). The aims of this study were to determine the genetic characteristics, antimicrobial susceptibility, and genetic relatedness of ESBL- and plasmid-mediated AmpC β-lactamase-producing Enterobacteriaceae isolated from bovine mastitis cases.
Screening of ESBLs.
Bacterial cultures were carried out using standard procedures on a total of milk samples from 258,888 quarters obtained from 176,808 cows affected by (mainly clinical) mastitis on 1,000 dairy farms in the Nemuro Subprefecture of Hokkaido Prefecture, Japan, between February 2007 and April 2011 (14). Streptococcus spp. (mainly Streptococcus uberis) and Enterococcus spp., coagulase-negative staphylococci, Staphylococcus aureus, E. coli, and Klebsiella spp. were the organisms most commonly isolated from culture-positive samples.
Of the isolates, 28,900 were identified as Gram-negative bacilli and were submitted for susceptibility testing by disc diffusion according to the Clinical and Laboratory Standards Institute (CLSI) guidelines (15); 419 isolates were identified as being cefazolin resistant and oxidase negative. These strains were then submitted for CLSI combination disc ESBL confirmatory tests (15) and a chromogenic oxyimino-cephalosporins hydrolysis test (Cica-β-test I; Kanto Chemical, Tokyo, Japan) to detect ESBLs, plasmidic AmpC β-lactamases, and metallo-β-lactamases. Isolates with a positive ESBL confirmatory test and/or Cica-β-test were screened for metallo-β-lactamases using the sodium mercaptoacetic acid (SMA) double-disc synergy test (SMA test) using two Kirby-Bauer discs containing ceftazidime and one disc containing SMA (Eiken Chemical, Tokyo, Japan). The ESBL test-positive Enterobacteriaceae isolates were identified using the ID 32 E API system (Sysmex bioMérieux, Tokyo, Japan).
CTX-M genes and antimicrobial susceptibility.
The ESBL- and/or Cica-β-test-positive and the SMA-test-negative isolates (n = 65) were analyzed by multiplex PCR for the presence of blaCTX-M genes (16) and plasmid-mediated AmpC β-lactamase genes (i.e., CMY, ACC, FOX, MOX, DHA, CIT, and EBC groups) (17). The CTX-M types of the CTX-M-positive isolates were identified by bidirectional sequencing using group-specific PCR primers for blaCTX-M-1 group (18), blaCTX-M-2 group and blaCTX-M-9 group (19). AmpC-positive isolates were analyzed using type-specific PCR primers (e.g., blaCMY-1 and blaCMY-2 genes), and blaTEM and blaSHV genes were analyzed and bidirectionally sequenced using previously described primers (19) (see Table S1 in the supplemental material). Comparison of nucleotide sequences and identification of each CTX-M, TEM, and SHV type were carried out using BLAST (http://blast.ncbi.nlm.nih.gov/Blast.cgi).
For these 65 isolates, the MICs of 23 antimicrobials were determined by the CLSI broth microdilution method (15, 20) using a custom-designed microtiter panel (Opt Panel MP; Kyokuto Pharmaceutical, Tokyo, Japan). The 23 drugs were ampicillin, cefazolin, cefuroxime, ceftazidime, ceftazidime/clavulanic acid, cefotaxime, cefotaxime/clavulanic acid, ceftriaxone, cefpodoxime, ceftiofur, cefquinome, cefepime, cefmetazole, moxalactam, imipenem, meropenem, aztreonam, gentamicin, amikacin, oxytetracycline, trimethoprim-sulfamethoxazole (SXT), enrofloxacin, and ciprofloxacin. Additional susceptibility tests for cefoxitin, kanamycin, chloramphenicol, and levofloxacin were performed by the CLSI disc diffusion method (15, 20). The breakpoints for veterinary pathogens were used for 12 antimicrobial agents (15), and the breakpoints for human Enterobacteriaceae isolates were used for the other 12 antimicrobial agents (20). E. coli ATCC 25922 and Pseudomonas aeruginosa ATCC 27853 were used as quality control strains.
Sixty-five of the 419 cefazolin-resistant isolates were identified as CTX-M-producing strains. Fifty-one isolates (78.5%), which included 41 K. pneumoniae, 6 Klebsiella oxytoca, 2 Citrobacter koseri, 1 E. coli, and 1 Enterobacter aerogenes isolate, harbored blaCTX-M-2; 10 E. coli isolates (15.4%) harbored blaCTX-M-15; and 4 isolates (6.2%; 2 K. pneumoniae and 2 E. coli isolates) harbored blaCTX-M-14. No isolates contained the plasmidic AmpC gene (Table 1; Fig. 1 and 2).
Table 1.
Origins of CTX-M-2- and CTX-M-14 ESBL-producing Enterobacteriaceae isolates other than CTX-M-2-producing K. pneumoniae and E. coli
| Isolate | Isolation date, farm, and cow | Bacterial species | CTX-M genotype | TEM or SHV genotype |
|---|---|---|---|---|
| MCKo1 | July 2008, F, 20 | K. oxytoca | CTX-M-2 | OKP-A |
| MCKo2 | Sept. 2008, H, 22 | K. oxytoca | CTX-M-2 | TEM-1 |
| MCKo3 | May 2009, O, 30 | K. oxytoca | CTX-M-2 | TEM-1 |
| MCEa1 | Sep. 2009, H, 36 | E. aerogenes | CTX-M-2 | Negative |
| MCKo4 | Jan. 2010, M, 42 | K. oxytoca | CTX-M-2 | Negative |
| MCCk1 | Apr. 2010, U, 47 | C. koseri | CTX-M-2 | Negative |
| MCCk2 | Apr. 2010, V, 48 | C. koseri | CTX-M-2 | Negative |
| MCKo5 | Aug. 2010, F, 50 | K. oxytoca | CTX-M-2 | SHV-1 |
| MCKo6 | Sept. 2010, F, 53 | K. oxytoca | CTX-M-2 | Negative |
| MCK45 | Nov. 2010, Y, 57 | K. pneumoniae | CTX-M-14 | Negative |
| MCK46 | Nov. 2010, Y, 57 | K. pneumoniae | CTX-M-14 | Negative |
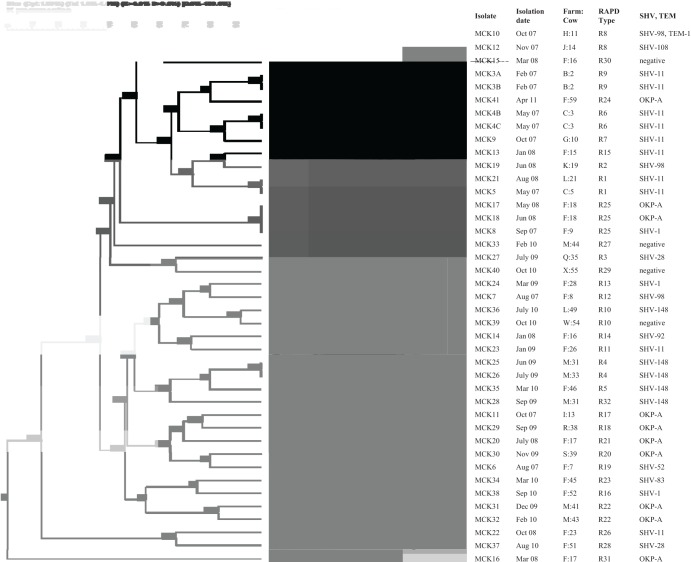
Fig 1.
RAPD-PCR of 41 K. pneumoniae isolates producing CTX-M-2. Cluster analysis was performed by the unweighted pair group method using arithmetic averages with a 1.0% band position tolerance window and 1.0% optimization. DNA relatedness was calculated based on the Dice coefficient. Thirty-two band patterns were typed using similarity cutoff values of approximately ≥85%.
Fig 2.
PFGE patterns and cluster analysis of 13 CTX-M-producing E. coli isolates (MCE1 to 13) from mastitis cases, obtained using XbaI. Cluster analysis was performed by the unweighted pair group method using arithmetic averages with a 1.0% band position tolerance window and 1.0% optimization. DNA relatedness was calculated based on the Dice coefficient. Twelve band patterns were typed using similarity cutoff values of ≥90%. The Salmonella strain Braenderup CCUG50923 was used as a marker for assessing PFGE banding patterns. FCE3, -4, -5, and -16 were isolated from feces from cattle on farm M in our previous study (7).
Thirty-seven (90.2%) of 41 CTX-M-2-producing K. pneumoniae isolates also harbored genes encoding SHV-1/11/28/52/83/92/98/108/148, OKP-A, or TEM-1. Four (66.6%) of 6 CTX-M-2-producing K. oxytoca isolates also harbored blaTEM-1, blaSHV-1, or blaOKP-A. Four (40.0%) of 10 CTX-M-15-producing E. coli isolates also harbored blaTEM-1, but no E. coli isolates harbored blaSHV (Table 1; Fig. 1 and 2). The gene sequences of the CTX-M-2/1/9, TEM, and SHV groups were 99 to 100% homologous with those of the CTX-M-2/15/14, TEM-1, and SHV subtypes which are available on GenBank.
The 65 CTX-M-producing Enterobacteriaceae were isolated from 61 quarters of 58 mastitis cases on 25 dairy farms in the Nemuro Subprefecture. Each of the 25 farms fed between 180 and 500 Holstein cattle with total mixed ration in free-stall barns or with grass-silage and concentrates fed separately in tie-stall barns; almost all used sawdust bedding. Their rolling yearly herd averages for milk production were 7,800 to 9,500 kg. The 58 affected cows had either subclinical or local to systemic clinical mastitis. Despite antimicrobial treatment, six cows were culled; and the remaining cows' clinical signs resolved 3 to 10 weeks after onset.
The isolation rate of strains producing CTX-M-2/15/14 in bovine mastitis was 0.22% of the 28,900 Gram-negative bacillus isolates from the milk samples from 258,888 quarters of 176,808 cows. The CTX-M-2/15-producing K. pneumoniae and E. coli strains were the most common ESBL producers causing bovine mastitis in this study. In France, CTX-M-1/14-producing E. coli and K. pneumoniae strains had an isolation rate of 0.4% (6 of 1,427 E. coli and K. pneumoniae isolates) from bovine mastitis cases (5). There were no significant (P > 0.05) differences between the isolation rates found in our study and the French study (5) by the chi-square test using StatFlex version 6.0 (Artech Co., Ltd., Osaka, Japan). Among human and animal isolates in Western European countries and Japan, the most common CTX-M types were the CTX-M-1 cluster (CTX-M-1/15/55) and CTX-M-9 cluster (CTX-M-9/14/27) (2, 3, 4, 5, 6). Except for the dominance of CTX-M-2, our results are similar to these previous reports. The blaTEM-1, blaSHV-1, and blaSHV-11 genes detected in the present study encode non-ESBL enzymes (21); however, it is not clear whether the SHV-28/52/83/92/98/108/148 and OKP-A are ESBLs, because the kinetic parameters of their purified enzymes were not determined.
Isolates producing CTX-M exhibited high resistance to oxyimino-cephalosporins; however, they exhibited high rates of susceptibility to cefmetazole, moxalactam, imipenem, meropenem, gentamicin, and amikacin. The isolates producing CTX-M-2 and CTX-M-14 showed high rates of susceptibility to ceftazidime and fluoroquinolones. In contrast, the CTX-M-15-producing E. coli strains showed significantly higher rates of resistance to ceftazidime, aztreonam, cefepime, SXT, oxytetracycline, ciprofloxacin, levofloxacin, cefoxitin, and kanamycin than CTX-M-2/14-producing Klebsiella spp. and/or other CTX-M-2/14-producing Enterobacteriaceae (P < 0.05) by the chi-square tests (Table 2). Our results are consistent with a previous study (1), and the CTX-M types other than CTX-M-15, CTX-M-16, and CTX-M-27 efficiently hydrolyze cefotaxime and ceftriaxone but not ceftazidime (1).
Table 2.
Antimicrobial susceptibilities of isolates producing CTX-M-type ESBL from mastitic cows
| Antimicrobial agenta | Group 1 |
Group 2 |
Group 3 |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| MIC range (μg/ml) | MIC50 (μg/ml) | MIC90 (μg/ml) | Breakpoint (μg/ml; mm)b | No. of isolates (%) |
MIC range (μg/ml) | No. of resistant isolates (%) (B)h | MIC range (μg/ml) | No. of resistant isolates (%) (C)h | |||
| Susceptible | Intermediate | Resistant (A)h | |||||||||
| Ampicilline | >512 | >512 | >512 | ≥32 | 0 | 0 | 49 (100) | >512 | 10 (100) | >512 | 6 (100) |
| Cefazoline | >256 | >256 | >256 | ≥32 | 0 | 0 | 49 (100) | >256 | 10 (100) | >256 | 6 (100) |
| Cefuroximee | >256 | >256 | >256 | ≥32c | 0 | 0 | 49 (100) | >256 | 10 (100) | >256 | 6 (100) |
| Ceftazidime | ≤2 to 64 | ≤2 | ≤2 | ≥16c | 45 (91.8) | 3 (6.1) | 1 (2.0)**B | 16–64 | 10 (100)**A,C | ≤2 | 0**B |
| CAZ/CLA | ≤0.25 to >8 | ≤0.25 | 0.5 | 0.5 to >8 | ≤0.25 to 0.5 | ||||||
| Cefotaxime | ≤16 to 256 | 64 | 128 | ≥4c | -g | 4 (8.2)g | 45 (91.8) | 512 to >512 | 10 (100) | 128 to >512 | 6 (100) |
| CTX/CLA | ≤1 to >8 | ≤1 | ≤1 | ≤1 to 8 | ≤1 | ||||||
| Cefpodoxime | ≤8 to >64 | >64 | >64 | ≥16 | 1 (2.0) | 0 | 48 (98.0) | >64 | 10 (100) | >64 | 6 (100) |
| Ceftriaxone | 64 to >512 | 512 | >512 | ≥4c | 0 | 0 | 49 (100) | >512 | 10 (100) | 512 to >512 | 6 (100) |
| Ceftiofure | 64 to >512 | >512 | >512 | ≥8 | 0 | 0 | 49 (100) | >512 | 10 (100) | >512 | 6 (100) |
| Cefquinomee | 16 to >128 | 128 | >128 | —d | >128 | >128 | |||||
| Cefepime | ≤8 to 64 | ≤8 | ≤8 | ≥32c | 43 (87.8) | 3 (6.1) | 3 (6.1)**B,C | >64 | 10 (100)**A | ≤8 to >64 | 4 (66.6)**A |
| Cefmetazole | ≤4 to >32 | ≤4 | ≤4 | ≥64c | 47 (95.9) | 0 | 2 (4.1) | ≤4 to 32 | 0 | ≤4 to >32 | 1 (16.6) |
| Moxalactam | ≤8 to >32 | ≤8 | ≤8 | ≥64c | 48 (98.0) | 0 | 1 (2.0) | ≤8 | 0 | ≤8 | 0 |
| Imipenem | ≤1 | ≤1 | ≤1 | ≥16 | 49 (100) | 0 | 0 | ≤1 | 0 | ≤1 | 0 |
| Meropenem | ≤2 to 4 | ≤2 | ≤2 | ≥4c | 48 (98.0) | 0 | 1 (2) | ≤2 | 0 | ≤2 | 0 |
| Aztreonam | ≤8 to >64 | ≤8 | 16 | ≥16c | —g | 43 (87.8)g | 6 (12.2)**B, *C | 64 to >64 | 10 (100)**A,*C | ≤8 to 32 | 3 (50)*A,B |
| Gentamicine | ≤2 to >16 | ≤2 | ≤2 | ≥16 | 46 (93.9) | —d | 3 (6.1) | ≤2 to >16 | 1 (10.0) | ≤2 | 0 |
| Amikacin | ≤4 to >16 | ≤4 | ≤4 | ≥64 | 48 (98.0) | 1 (2.0)g | ≤4 to 8 | 0 | ≤4 | 0 | |
| OTETe | ≤4 to >16 | ≤4 | >16 | ≥16 | 24 (49.0) | 11 (22.4) | 14 (28.6)*B | ≤4 to >16 | 7 (70.0)*A | ≥16 | 6 (100) |
| SXTe | ≤0.5/9.5 to >4/76 | ≤0.5/9.5 | 4/76 | ≥4/76 | 42 (85.7) | —d | 7 (14.3)**B,*C | 4/76 to >4/76 | 7 (70.0)**A | ≤0.5/9.5 to >4/76 | 3 (50)*A |
| Enrofloxacine | ≤0.25 to 2 | ≤0.25 | 2 | ≥2 | 45 (91.8) | 2 (4.1) | 2 (4.1) | ≤0.25 to >2 | 1 (10.0) | ≤0.25 to 0.5 | 0 |
| Ciprofloxacin | ≤0.5 to 2 | ≤0.5 | ≤0.5 | ≥4c | 46 (93.9) | 3 (6.1) | 0*B | ≤0.5 to >2 | 1 (10.0)*A | ≤0.5 to 1 | 0 |
| Cefoxitinf | ≤14 mmc | 48 (98.0) | 1 (2.0) | 0*B,**C | S to R | 1 (10.0)*A | S to R | 1 (16.6)**A | |||
| Kanamycine,f | ≤13 mm | 38 (77.6) | 1 (2.0) | 10 (20.4)*B, C | S to R | 5(50.0)*A | S to R | 4 (66.6)*A | |||
| CHLf | ≤13 mm | 41 (83.7) | 0 | 8 (16.3) | S to R | 4 (40.0) | S to R | 3 (50) | |||
| Levofloxacinf | ≤13 mmc | 49 (100) | 0 | 0*B | S to R | 1(10.0)*A | S | 0 | |||
Assessed by the broth microdilution or disk diffusion method for 41 K. pneumoniae and 6 K. oxytoca isolates producing CTX-M-2 and 2 K. pneumoniae isolates producing CTX-M-14 (group 1), 10 E. coli isolates producing CTX-M-15 (group 2), and other Enterobacteriaceae isolates producing CTX-M-2 or CTX-M-14 (group 3; CTX-M-2: E. coli, n = 1; C. koseri, n = 2; E. aerogenes, n = 1; E. coli, CTX-M-14, n = 2). Abbreviations: CAZ/CLA, ceftazidime/clavlanic acid; CTX, cefotaxime; OTET, oxytetracycline; SXT, trimethoprim/sulfamethoxazol; CHL, chloramphenicol.
Breakpoint for resistance in accordance with CLSI document M31-A3 (2008) (15) for veterinary pathogens.
Breakpoint for resistance in accordance with CLSI document M100-S21 (2011) (20) for isolates from human infections with Enterobacteriaceae.
Breakpoint for resistance or intermediate is not defined in CLSI documents M31-A3 (2008) (15) and M100-S21 (2011) (20).
Antimicrobial agent approved for cattle in Japan; the other 17 antimicrobials in this table are unapproved for cattle.
Susceptibilities were tested by disc diffusion method according to CLSI documents M31-A3 (2008) (15) and M100-S21 (2011) (20).
Isolates in the susceptible, intermediate, and resistant categories could not be differentiated.
Significant differences were determined by the χ2 test for comparison with the group indicated by the capital letter (A, B, or C): *, P < 0.05; **, P < 0.01.
Molecular subtyping profiles.
Random amplified polymorphic DNA (RAPD)-PCR analysis of the 41 CTX-M-2-producing K. pneumoniae isolates was performed using the oligonucleotide RAPD7 as previously described (22). Pulsed-field gel electrophoresis (PFGE) of a total of 13 CTX-M-producing E. coli isolates was conducted according to the PulseNet standardized laboratory protocol (23) using XbaI (Roche Applied Science, Mannheim, Germany) and the CHEF-DR III electrophoresis systems (Bio-Rad, Hercules, CA). Dendrograms of RAPD patterns and PFGE patterns were analyzed using BioNumerics software, version 5.1 (Applied Maths, Austin, TX). Four CTX-M-15-producing E. coli strains isolated from bovine feces on farm M in our previous study (7) were used for comparison with the E. coli isolates from mastitis cases.
Multilocus sequence typing (MLST) of the 13 CTX-M-producing E. coli isolates was conducted according to standard protocols using the E. coli database on the MLST website. (http://mlst.ucc.ie/mlst/dbs/Ecoli). The 13 E. coli isolates were serotyped according to O and H antigens using the pathogenic E. coli Seiken set 1 and set 2 antisera, respectively (Denka Seiken, Tokyo, Japan).
The 41 CTX-M-2-producing K. pneumoniae isolates from 15 farms revealed 32 RAPD types. More than half of the strains were isolated from 2 farms (F and M). The 18 isolates from farm F revealed 16 RAPD types. There was not a predominant RAPD type among the 41 isolates. However, two or three isolates each of K. pneumoniae (MCK17/18/8, MCK25/26, and MCK31/32), which were isolated from 2 different cows on same farm (F or M), showed closely related RAPD types (R25, R4, and R22, respectively) (Fig. 1).
The 13 E. coli isolates from 7 farms belonged to 10 STs and showed 12 PFGE types. Two isolates each of E. coli (MCE1/3, MCE4/6, and MCE9/10), which were isolated from 2 different cows on same farm (D, M, or P), had the same ST and closely related PFGE types (ST23/W, ST58/V1 and V2, and ST10/Z1 and Z2, respectively). There were not any closely related strains between the 5 mastitis and the 4 fecal E. coli CTX-M-15-producing isolates from farm M (Fig. 2). Most of the E. coli isolates had untypeable O and H antigens (OUT, HUT). Neither E. coli clone ST131 (O25:H4) nor enterohemorrhagic E. coli O157, O26, or O111 or other serotypes commonly isolated from human infections (8) were detected from the 13 isolates.
The genetic diversity in the 18 K. pneumoniae isolates obtained from bovine mastitis cases on farm F suggests that these were opportunistic infections originating from a wide variety of environmental sources (11). However, the presence of some strains of K. pneumoniae and E. coli showing closely related genotypes, which were isolated from the different cows on the same farm, suggests a contagious infection or an infection from an environmental point source (11). Similar to our results, E. coli clones ST10/23/58 producing CTX-M-14/1 have also been isolated from bovine mastitis in France (5). Consistent with this French study, we detected no E. coli clone ST131 (O25:H4) producing CTX-M-15/27. Thus, these results suggest that cattle, unlike humans, canines and felines, have little significance as a source of this clone (2, 4). In contrast, recently, two enterohemorrhagic E. coli strains (with serotypes O111:H8 and O26:H11) belonging to the B1 phylogenetic group and carrying blaCTX-M-15/9 were isolated from diarrheic cattle in France (8).
In conclusion, the genes encoding CTX-M2/15/14 are present at a low frequency in Enterobacteriaceae isolates causing bovine mastitis on Japanese dairy farms. The ESBL producers were dominated by CTX-M-2-producing K. pneumoniae and CTX-M-15-producing E. coli strains which showed multidrug resistance to ceftazidime, aztreonam, and cefepime. There was not a predominant clonal type, and even the 18 K. pneumoniae strains isolated from a single farm showed clonal diversity by molecular subtyping.
Supplementary Material
Footnotes
Published ahead of print 10 July 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JCM.00920-13.
REFERENCES
- 1.Bonnet R. 2004. Growing group of extended-spectrum β-lactamases: the CTX-M enzymes. Antimicrob. Agents Chemother. 48:1–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Harada K, Nakai Y, Kataoka Y. 2012. Mechanisms of resistance to cephalosporin and emergence of O25b-ST131 clone harboring CTX-M-27 β-lactamase in extraintestinal pathogenic Escherichia coli from dogs and cats in Japan. Microbiol. Immunol. 56:480–485 [DOI] [PubMed] [Google Scholar]
- 3.Overdevest I, Willemsen I, Rijnsburger M, Eustace A, Xu L, Hawkey P, Heck M, Savelkoul P, Vandenbroucke-Grauls C, van der Zwaluw K, Huijsdens X, Kluytmans J. 2011. Extended-spectrum β-lactamase genes of Escherichia coli in chicken meat and humans, the Netherlands. Emerg. Infect. Dis. 17:1216–1222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Peiranoa G, Pitou JDD. 2010. Molecular epidemiology of Escherichia coli producing CTX-M β-lactamases: the worldwide emergence of clone ST131 O25:H4. Int. J. Antimicrob. Agents 35:316–321 [DOI] [PubMed] [Google Scholar]
- 5.Dahmen S, Métayer V, Gay E, Madec JY, Haenni M. 2013. Characterization of extended-spectrum beta-lactamase (ESBL)-carrying plasmids and clones of Enterobacteriaceae causing cattle mastitis in France. Vet. Microbiol. 162:793–799 [DOI] [PubMed] [Google Scholar]
- 6.Madec JY, Poirel L, Saras E, Gourguechon A, Girlich D, Nordmann P, Haenni M. 2012. Non-ST131 Escherichia coli from cattle harbouring human-like bla (CTX-M-15)-carrying plasmids. J. Antimicrob. Chemother. 67:578–581 [DOI] [PubMed] [Google Scholar]
- 7.Ohnishi M, Okatani AT, Esaki H, Harada K, Sawada T, Murakami M, Marumo K, Kato Y, Sato R, Shimura K, Hatanaka N, Takahashi T. 2013. Herd Prevalence of Enterobacteriaceae producing CTX-M-type and CMY-2 β-lactamases among Japanese dairy farms. J. Appl. Microbiol. 115:282–289 [DOI] [PubMed] [Google Scholar]
- 8.Valat C, Auvray F, Forest K, Metayer V, Gay E, Peytavin de Garam C, Madec JY, Haennia M. 2012. Phylogenetic grouping and virulence potential of extended-spectrum-β-lactamase-producing Escherichia coli strains in cattle. Appl. Environ. Microbiol. 78:4677–4682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cantón R, González-Alba JM, Galán JC. 2012. CTX-M enzymes: origin and diffusion. Front. Microbiol. 3:110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Contreras GA, Rodríguez JM. 2011. Mastitis: comparative etiology and epidemiology. J. Mammary Gland Biol. Neoplasia 16:339–356 [DOI] [PubMed] [Google Scholar]
- 11.Munoz MA, Welcome FL, Schukken YH, Zadoks RN. 2007. Molecular epidemiology of two Klebsiella pneumoniae mastitis outbreaks on a dairy farm in New York State. J. Clin. Microbiol. 45:3964–3971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ministry of Agriculture, Forestry, and Fisheries (MAFF) 2009. Statistical tables of livestock mutual relief, by agricultural disaster compensation system (2008). MAFF, Tokyo, Japan. (In Japanese.) [Google Scholar]
- 13.Barlow J. 2011. Mastitis therapy and antimicrobial susceptibility: a multi species review with a focus on antibiotic treatment of mastitis in dairy cattle. J. Mammary Gland Biol. Neoplasia 16:383–407 [DOI] [PubMed] [Google Scholar]
- 14.National Mastitis Council 1999. Laboratory handbook on bovine mastitis. revised edition National Mastitis Council, Madison, WI [Google Scholar]
- 15.Clinical and Laboratory Standards Institute (CLSI) 2008. Performance standards for antimicrobial disk and dilution susceptibility tests for bacteria isolated from animals; approved standard, 3rd ed, M31-A3 CLSI, Wayne, PA [Google Scholar]
- 16.Xu L, Ensor V, Gossain S, Nye K, Hawkey P. 2005. Rapid and simple detection of blaCTX-M genes by multiplex PCR assay. J. Med. Microbiol. 54:1183–1187 [DOI] [PubMed] [Google Scholar]
- 17.Pérez-Pérez FJ, Hanson ND. 2002. Detection of plasmid-mediated AmpC β-lactamase genes in clinical isolates by using multiplex PCR. J. Clin. Microbiol. 40:2153–2162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mena A, Plasencia V, Garcia L, Hidalgo O, Avestarán JI, Alberti S, Borrell N, Pérez JL, Oliver A. 2006. Characterization of a large outbreak by CTX-M-1-producing Klebsiella pneumoniae and mechanisms leading to in vivo carbapenem resistance development. J. Clin. Microbiol. 44:2831–2837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kojima A, Ishii Y, Ishihara K, Esaki H, Asai T, Oda C, Tamura Y, Takahashi T, Yamaguchi K. 2005. Extended-spectrum-β-lactamase-producing Escherichia coli strains isolated from farm animals from 1999 to 2002: Report from the Japanese Veterinary Antimicrobial Resistance Monitoring Program. Antimicrob. Agents Chemother. 49:3533–3537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Clinical and Laboratory Standards Institute (CLSI) 2011. Performance standards for antimicrobial susceptibility testing, 21st informational supplement, M100-S21 CLSI, Wayne, PA [Google Scholar]
- 21.Paterson DL, Bonomo RA. 2005. Extended-spectrum β-lactamases: a clinical update. Clin. Microbiol. Rev. 18:657–686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ben-Hamouda T, Foulon T, Ben-Cheikh-Masmoudi A, Fendri C, Belhadj O, Ben-Mahrez K. 2003. Molecular epidemiology of an outbreak of multiresistant Klebsiella pneumoniae in a Tunisian neonatal ward. J. Med. Microbiol. 52:427–433 [DOI] [PubMed] [Google Scholar]
- 23.Centers for Disease Control and Prevention (CDC) 2004. One-day (24–28 h) standardized laboratory protocol for molecular subtyping of Escherichia coli O157:H7, nontyphoidal Salmonella serotypes, and Shigella sonnei by pulsed field gel electrophoresis (PFGE). Sections 5.1, 5.2, 5.4 CDC, Atlanta, GA [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.