Abstract
Recombination events were found in two human coxsackievirus B3 strains, Beijing0811 and SD2012CHN. The strains were isolated separately from five newborns diagnosed with severe hospital-acquired acute myocarditis in Beijing in 2008 and from two children diagnosed with hand, foot, and mouth disease with concurrent acute myocarditis in Shandong in 2012.
TEXT
Human enteroviruses (HEVs) belonging to the genus Enterovirus under the family Picornaviridae are the causes of various diseases, including hand, foot, and mouth disease (HFMD), myocarditis, meningitis, herpangina, and nonspecific febrile illness (1). HEVs are classified into four species (A to D) based on their molecular and antigenic properties (The Pirbright Institute). HEV-B is comprised of 61 conventional serotypes, including coxsackieviruses (CV) B1 to B6 and A9, echoviruses (ECoV) 1 to 7, 9, 11 to 21, 24 to 27, and 29 to 33, human enteroviruses 69, 73 to 75, 77 to 88, 93, 97, 98, 100, 101, 106, 107, 110, and 111, and simian enterovirus 5 (SA5) (2). As a member of the HEV-B species, the human coxsackievirus B3 serogroup (CVB3) is one of the most common human pathogens associated with clinical acute and chronic viral myocarditis. Some acute CVB3 infections are severe and lethal. Previous studies have indicated that CVB3 is also associated with severe cases of HFMD (3–5).
In the city of Beijing in 2008, five newborns with fever, shortness of breath, and coagulation disorders in the same hospital were clinically diagnosed with hospital-acquired acute myocarditis. Using pharyngeal swabs from the five newborns (each sampling was done with the informed consent of the parents or guardian of the patient), one CVB3 strain was confirmed by using reverse transcription and seminested PCR, as described previously (6). In the city of Pingdu in Shandong Province in 2012, we identified one CVB3 strain in pharyngeal swabs from two children clinically diagnosed with HFMD, and concurrent acute myocarditis sampling was performed (the sampling was done with the informed consent of the parents of the patients). These two CVB3 strains produced typical cytopathic effects (CPEs) in human rhabdomyosarcoma (RD) cells. After propagation and purification of the plaque by cell culture, overlapping fragments of the two viruses were obtained by using degenerate primers based on the genome sequences of existing CVB3 strains available in GenBank, and the complete genome sequences of the two viruses were obtained by assembling overlapping fragments using the SeqMan program available in the Lasergene 7 package. The two CVB3 strains were designated Beijing0811 and SD2012CHN (7). The complete genomes of these two isolates were submitted to GenBank (accession numbers GQ141875 and JX976770).
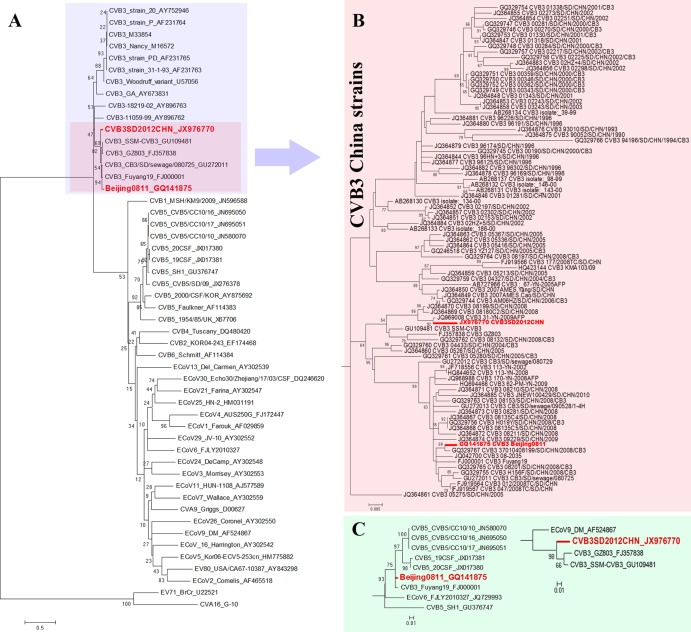
The genomic RNA is 7,402 nucleotides (nt) for both Beijing0811 and SD2012CHN. The characteristic order of Beijing0811 is 5′ (743 nt)-P1 (capsid region, 2,553 nt)-P2 (noncapsid region, 1,734 nt)-P3 (noncapsid region, 2,271 nt)-3′ (101 nt). The characteristic order of SD2012CHN is 5′ (744 nt)-P1 (2,562 nt)-P2 (1,725 nt)-P3 (2,271 nt)-3′ (100 nt). Based on the ClustalW algorithm of MegAlign in the Lasergene 7 package, pairwise similarity analysis revealed 88% nucleotide sequence identity among the genome sequences, 92% nucleotide identity among the P1 regions, 86% nucleotide identity among the P2 regions, and 83% nucleotide identity among the P3 regions of Beijing0811 and SD2012CHN. The VP1 of these two viruses shared 93% nucleotide identity. Among CVB3 strains, the VP1 of Beijing0811 and SD2012CHN exhibited the highest nucleotide identity (92% to 98%) with CVB3 strains in mainland China and less than 83% nucleotide identity with CVB3 strains in other regions. The maximum-likelihood phylogenetic tree obtained using MEGA5.0 (8) based on the complete VP1 nucleotide sequences of HEV-B members (Fig. 1A) indicated that the two new CVB3 strains clustered with other CVB3 strains and were closely related to CVB3 China strains. The phylogenetic tree based on the partial VP1 sequences of 89 CVB3 China strains (Fig. 1B) indicated that Chinese CVB3 strains divided into a distinct lineage from other CVB3 strains in other regions, and Beijing0811 and SD2012CHN were separately divided into different sublineages. This finding reveals that CVB3 has been circulating in China for many years and represents an independent evolution propensity. Notably, the whole-genome sequence of Beijing0811 was most closely related to another Chinese CVB3 strain, Fuyang19, isolated from a severe case of HFMD in the city of Fuyang in Anhui Province in 2008 (98% nucleotide identity). Fuyang19 can be considered the closest variant of Bejing0811.
Fig 1.
Maximum-likelihood phylogenetic trees based on the complete VP1 nucleotide sequences of HEV-B members (A), the partial VP1 sequences of 89 CVB3 China strains (B), and the complete nucleotide sequences of the P3 regions of Chinese CVB3 strains (C) generated by MEGA5.0. The bootstrap values are shown as percentages of 1,000 replicates for the major lineages within the trees.
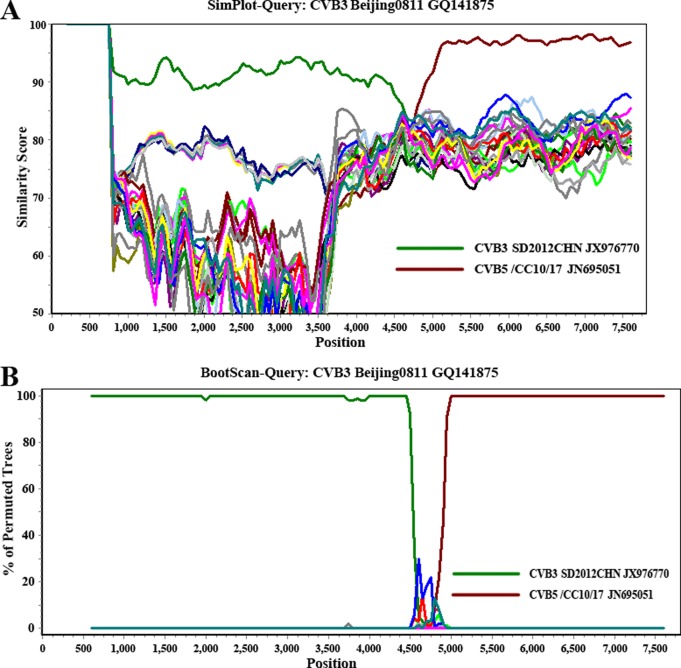
Pairwise similarity and phylogenetic analysis based on the P3 regions (Fig. 1C) showed that Beijing0811 was most closely related to Chinese CVB5 strains (95% to 97% nucleotide identity) and SD2012CHN was most closely related to Chinese CVB3 strains (95% nucleotide identity). This finding indicates that RNA recombination occurred between CVB3 and CVB5. Using SimPlot software (9), similarity plot (Fig. 2A), and bootscan analysis (Fig. 2B) demonstrated that Beijing0811 had the highest degree of similarity to SD2012CHN and other Chinese CVB3 strains in the P1 region but was most similar to Chinese CVB5 strain CVB5/CC10/17 and its related strains in the P3 region. Recombination was confirmed by detecting putative breakpoints within the genomes using GARD (10) and substantiated by positive KH test results, which demonstrated significant incongruent topologies before and after each breakpoint. Nine breakpoints were found for the alignment of CVB3 Beijing0811, CVB3 SD2012CHN, CVB5/CC10/17, and CVB5/SD/09 (at positions 650, 702, 744, 1311, 3353, 3371, 4170, 4601, 4784), but only the topology flanking position at nt 4601 was significantly discordant via the KH test (P < 0.05), supporting that it is a recombination breakpoint. The ΔAICc (improvement in Bozdogan's consistent Akaike information criterion of the breakpoint-partitioned model over a no-recombination single-phylogeny model) for this alignment was 8.60179. The GARD analysis results were consistent with the similarity plot and bootscan analysis results.
Fig 2.
SimPlot (A) and bootscan (B) analyses based on the full-length genomes of the two CVB3 strains and other related HEV-B prototype strains. CVB3 strain Beijing0811 was used as the query sequence in these analyses. A sliding window of 400 nucleotides moving in 50-nucleotide steps was used.
From the phylogenetic tree based on the P3 regions (Fig. 1C), ECoV6 strain FJLY2010327 exhibited a close relationship with the clade of Beijing0811 and related Chinese CVB5 strains, and ECoV9 strain DM exhibited a close relationship with the clade of SD2012CHN and related Chinese CVB3 strains. Recombination events were found between ECoV6 and Bejing0811 and related Chinese CVB5 strains by using similarity plot and bootscan analysis (Fig. 3). Another recombination event was also found between SD2012CHN and related Chinese CVB3 strains. This finding indicates that ECoV6 and ECoV9 act as recombinant counterparts for Chinese CVB3 and CVB5 strains among molecular evolution and recombination events in circulating enteroviruses.
Fig 3.
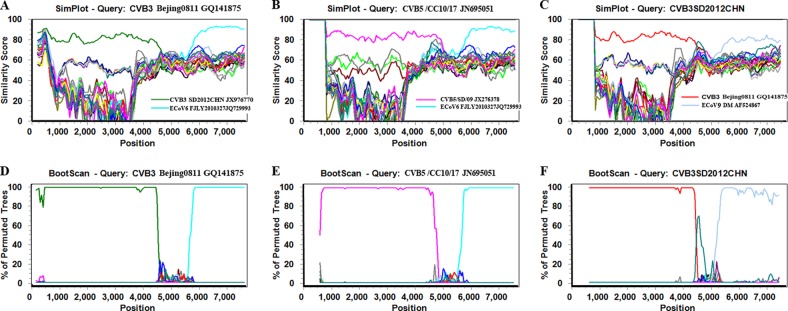
SimPlot and bootscan analyses based on the full-length genomes of CVB3 strain Beijing0811, CVB5 strain CVB5/CC10/17, CVB3 strain SD2012CHN, and related HEV-B prototype strains. CVB3 strain Beijing0811 (A, D), CVB5 strain CVB5/CC10/17 (B, E), and CVB3 strain SD2012CHN (C, F) were used as query sequences in these analyses. A sliding window of 400 nucleotides moving in 50-nucleotide steps was used.
In this study, the analysis of CVB3 strains Beijing0811 and SN2012CHN revealed two separate evolutionary tendencies of CVB3 in mainland China. The existence of SD2012CHN and previously reported Chinese CVB3 strains represents a traditional evolutionary lineage of CVB3 in China (11, 12). These Chinese CVB3 strains are distinct from CVB3 strains outside mainland China, and ECoV9 may act as a recombinant counterpart for these Chinese CVB3 strains. The existence of Bejing0811 and Fuyang19 indicate that recombination events occurred in noncapsid regions between CVB3, CVB5, and ECoV6 strains circulating in mainland China over the past several years, and a distinct, novel evolutionary lineage of CVB3 formed as a result of these recombination events. These findings indicate that more studies are required to investigate the pathogenic potentials and pathogenic roles of emerging recombinant CVB3 strains.
ACKNOWLEDGMENT
This work was supported by the National S&T Major Project, “China Mega-Project for Infectious Disease” (grant no. 2011ZX10004-001), from the People's Republic of China.
Footnotes
Published ahead of print 26 June 2013
REFERENCES
- 1.Tebruegge M, Curtis N. 2009. Enterovirus infections in neonates. Semin. Fetal Neonatal Med. 14:222–227 [DOI] [PubMed] [Google Scholar]
- 2.Oberste MS, Maher K, Michele SM, Belliot G, Uddin M, Pallansch MA. 2005. Enteroviruses 76, 89, 90 and 91 represent a novel group within the species human enterovirus A. J. Gen. Virol. 86:445–451 [DOI] [PubMed] [Google Scholar]
- 3.Cihakova D, Rose NR. 2008. Pathogenesis of myocarditis and dilated cardiomyopathy. Adv. Immunol. 99:95–114 [DOI] [PubMed] [Google Scholar]
- 4.Tracy S, Chapman NM, Drescher KM, Kono K, Tapprich W. 2006. Evolution of virulence in picornaviruses. Curr. Top. Microbiol. Immunol. 299:193–209 [DOI] [PubMed] [Google Scholar]
- 5.Yuan J, Cheung PK, Zhang HM, Chau D, Yang D. 2005. Inhibition of coxsackievirus B3 replication by small interfering RNAs requires perfect sequence match in the central region of the viral positive strand. J. Virol. 79:2151–2159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nix WA, Oberste MS, Pallansch MA. 2006. Sensitive, seminested PCR amplification of VP1 sequences for direct identification of all enterovirus serotypes from original clinical specimens. J. Clin. Microbiol. 44:2698–2704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Du J, Wu Z, Xue Y, Zhang T, Yang F, Jin Q. 2013. Complete genome sequence of a human coxsackievirus b3 from a child with myocarditis in Beijing, China. Genome Announc. 1(1):e00163–12. 10.1128/genomeA.00163-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. 2011. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 28:2731–2739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hu YF, Yang F, Du J, Dong J, Zhang T, Wu ZQ, Xue Y, Jin Q. 2011. Complete genome analysis of coxsackievirus A2, A4, A5, and A10 strains isolated from hand, foot, and mouth disease patients in China revealing frequent recombination of human enterovirus A. J. Clin. Microbiol. 49:2426–2434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kosakovsky Pond SL, Posada D, Gravenor MB, Woelk CH, Frost SD. 2006. Automated phylogenetic detection of recombination using a genetic algorithm. Mol. Biol. Evol. 23:1891–1901 [DOI] [PubMed] [Google Scholar]
- 11.Tao Z, Song Y, Li Y, Liu Y, Jiang P, Lin X, Liu G, Song L, Wang H, Xu A. 2012. Coxsackievirus B3, Shandong Province, China, 1990-2010. Emerg. Infect. Dis. 18:1865–1867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.He W, Lu H, Zhao K, Song D, Gai X, Gao F. 2012. Complete genome sequence of a coxsackievirus B3 isolated from a Sichuan snub-nosed monkey. J. Virol. 86:13134–13135 [DOI] [PMC free article] [PubMed] [Google Scholar]