Abstract
Misidentifications of Burkholderia pseudomallei as Burkholderia cepacia by Vitek 2 have occurred. Multidimensional scaling ordination of biochemical profiles of 217 Malaysian and Australian B. pseudomallei isolates found clustering of misidentified B. pseudomallei isolates from Malaysian Borneo. Specificity of B. pseudomallei identification in Vitek 2 and potentially other automated identification systems is regionally dependent.
TEXT
Burkholderia pseudomallei is a saprophytic soil bacterium that causes melioidosis, a disease endemic in northern Australia and Southeast Asia affecting humans and animals (1). The clinical presentations of melioidosis range from skin infections without sepsis to disseminated infection with sepsis and high mortality. Pneumonia is present in around half of cases, and chronic infections, relapsed disease, and activation from latency are all recognized (1, 2).
Confirmation of diagnosis of melioidosis requires a positive culture of B. pseudomallei from clinical samples such as blood, sputum, urine, pus, joint aspirate, or swabs from throat or rectum (1). B. pseudomallei has been identified by combining the commercial API 20NE biochemical kit (bioMérieux) with a simple screening system involving Gram stain, oxidase reaction, typical growth characteristics, and resistance to gentamicin (3). Susceptibility to amoxicillin-clavulanate (AMC) has also been used to differentiate B. pseudomallei from Burkholderia cepacia, which is resistant to AMC (4). Unfamiliarity with B. pseudomallei and problems with inaccurate species identification using some automated commercial biochemical identification systems have resulted in laboratories misidentifying the bacterium as a Pseudomonas or other Burkholderia species (5–9). Confirmation of B. pseudomallei identity by real-time PCR of DNA extracted from cultured bacterial colonies is increasingly the standard for many laboratories (10). Various genetic targets have been published for PCR identification of B. pseudomallei from bacterial cultures and also for direct detection from clinical samples, with a recent review showing the type III secretion system (TTS1)-orf2 assay to be superior in detecting B. pseudomallei directly from clinical specimens (11). Apart from molecular methods, B. pseudomallei from cultures can also be confirmed by antigen detection assays, such as latex agglutination (12). More recently, matrix-assisted laser desorption ionization–time of flight mass spectrometry (MALDI-TOF MS) has been adapted to identify cultured bacteria based on protein fingerprint profiles (13).
A particular problem has been the misidentification of B. pseudomallei as Burkholderia cepacia by the Vitek 2 automated biochemical system (bioMérieux) (5–8). B. cepacia belongs to a group of 17 phenotypically and genotypically similar species which form the B. cepacia complex, with B. cepacia specifically noted as an opportunistic pathogen infecting and causing progressive pulmonary deterioration in patients with cystic fibrosis (14, 15). Other organisms that have been reportedly misidentified by the Vitek 2 system include Candida albicans being misidentified as Gram-negative bacilli (16) and Candida parapsilosis being misidentified as Candida famata (17).
We have compared the Vitek 2 system biochemical profiles of 68 confirmed B. pseudomallei clinical strains from hospitals in Sabah and Sarawak, Malaysian Borneo, with 149 B. pseudomallei and 18 B. cepacia isolates from the Royal Darwin Hospital (RDH) in Northern Territory, Australia. One isolate per patient was analyzed. All isolates were collected between September 2010 and June 2012, except for 17 isolates collected in 1994 from Sabah.
All isolates were subcultured on horse blood agar (HBA) before testing was performed on the Vitek 2 according to the manufacturer's instructions (bioMérieux). The Vitek 2 system utilizes a panel of biochemical and enzymatic tests which results in a biochemical profile that is compared against the manufacturer's bacterial taxa database. All B. pseudomallei isolates were confirmed by both real-time PCR targeting the well-validated B. pseudomallei TTS1 (10) and by a latex agglutination test (12). Of the isolates from Sarawak, 15/43 (35%) had been initially identified as B. cepacia by the Vitek 2 system but were subsequently confirmed as B. pseudomallei by both the TTS1 real-time PCR and the latex agglutination test (Table 1). These 15 patients were from hospitals from different regions in Sarawak, none had cystic fibrosis, and melioidosis was suspected clinically, with a diversity of clinical presentations, including subcutaneous infection, community-acquired pneumonia, and sepsis. Only 2/25 B. pseudomallei isolates from Sabah and 3/149 B. pseudomallei isolates from Darwin were misidentified as B. cepacia (Table 1).
Table 1.
Number of isolates tested with the Vitek 2 system
| Sample origin | Total no. of B. pseudomallei isolatesa | Total no. of B. cepacia isolates tested | No. of B. pseudomallei isolates correctly identified as B. pseudomalleib,e | No. of isolates with low discriminationc,e | No. of B. pseudomallei isolates misidentified as B. cepaciad,e | No. of B. cepacia isolates correctly identifiede |
|---|---|---|---|---|---|---|
| Sabah, Malaysian Borneo | 25 | Not done | 22 (88) | 1 (4) | 2 (8) | Not done |
| Sarawak, Malaysian Borneo | 43 | Not done | 23 (53) | 5 (12) | 15 (35) | Not done |
| Darwin, Australia | 149 | 18 | 146 (98) | 0 | 3 (2) | 18 (100) |
Positive by TTS1 and agglutination tested.
With a 90 to 99% probability of being B. pseudomallei.
Low discrimination between B. cepacia and B. pseudomallei.
With a 90 to 99% probability of being B. cepacia.
Numbers in parentheses refer to the percentages of total isolates of the same state/country origin.
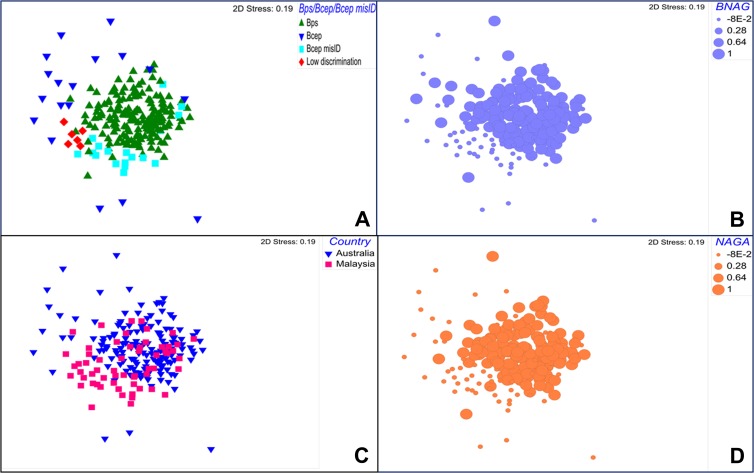
Using Primer version 6 (Primer-E Ltd., Plymouth Marine Laboratory, United Kingdom), we performed a nonmetric multidimensional scaling (nMDS) ordination on the Euclidean distance resemblance matrix of the Vitek 2 biochemical profiles of these 235 isolates. The nMDS (stress value of 0.19) showed a distinct clustering of the 15 B. pseudomallei isolates from Sarawak that were misidentified as B. cepacia (Fig. 1A). The nMDS ordination also revealed a tight clustering of the correctly identified B. pseudomallei isolates regardless of country of origin, while the B. cepacia isolates were more diverse (Fig. 1A and C). A permutation-based, nonparametric analysis of similarities (ANOSIM) confirmed this finding, with strong evidence that the biochemical profiles of the misidentified B. pseudomallei isolates were distinct from correctly identified B. pseudomallei (R statistic of 0.345, P < 0.001).
Fig 1.
Nonmetric multidimensional scaling (nMDS) ordination on the Euclidean distance resemblance matrix of the Vitek 2 biochemical profile of 235 B. pseudomallei and B. cepacia isolates from Australia and Malaysian Borneo. (A) Samples were identified as either B. pseudomallei, B. cepacia, B. pseudomallei misidentified as B. cepacia, or isolates with low discrimination; (B) the bubble size reflects the presence (large) or absence (small) of BNAG substrate in an isolate; (C) analysis based on isolates from both countries, Australia and Malaysia; (D) the bubble size reflects the presence (large) or absence (small) of NAGA substrate in an isolate. Abbreviations: Bps, B. pseudomallei; Bcep, B. cepacia; Bcep misID, B. pseudomallei misidentified as B. cepacia; BNAG, β-N-acetyl-glucosaminidase; NAGA, β-N-acetyl-galactosaminidase.
An analysis of similarity percentages (SIMPER) calculating the average contribution of each biochemical test to the overall observed dissimilarity between clusters revealed that, in particular, two enzymatic tests, the β-N-acetyl-glucosaminidase (BNAG) and β-N-acetyl-galactosaminidase (NAGA), which hydrolyze polysaccharides, were distinct between correctly and misidentified B. pseudomallei isolates. A total of 88% of correctly identified B. pseudomallei isolates contained BNAG substrates resulting in a positive test as opposed to 13% of misidentified isolates. This is also evident in Fig. 1B and D. The exopolysaccharide (EPS) poly-β-(1-6)-N-acetylglucosamine (PNAG) is a substrate of the enzyme BNAG and is produced by Burkholderia spp. (18). PNAG has been reported to be an important component in biofilm formation in Burkholderia species, potentially contributing to multidrug resistance (18). N-Acetylgalactosamine, a derivative of NAGA, has also been documented as one of the basic components for EPS of B. pseudomallei (19). The implications for virulence and immune response of these different biochemical profiles remains uncertain, but it has been suggested that the amount of capsular polysaccharide in B. pseudomallei compared to that in other Burkholderia species may well contribute to its relative virulence (20).
As an environmental bacterium adapted to a diverse range of tropical and subtropical habitats globally, B. pseudomallei is known to harbor a vast intraspecies genomic diversity as a result of high recombination frequency (21). It is therefore not surprising that the biochemical database of the Vitek 2 system performs variably based on geographical location. That there was 98% accuracy for the recent Australian strains tested in this study shows substantial improvement since prior studies (5, 6). The Sarawak data are supported by the recent report from China of the same misidentification in a case of melioidosis imported from Malaysia (8).
In conclusion, clinicians and laboratory scientists need to be aware of continuing potential misidentification of B. pseudomallei as B. cepacia by the Vitek 2 automated biochemical identification system, especially in patients with suspected melioidosis acquired in exotic locations, such as Malaysian Borneo. Similar difficulties are likely to be encountered with other automated identification systems, such as MALDI-TOF MS, as they are increasingly developed and utilized for patients infected in diverse geographical locations. PCR using validated targets (11) and ultimately whole-genome sequencing can confirm correct identification of species. Alternatively, for laboratories with limited resources, a combination of latex agglutination and AMC susceptibility testing assists in distinguishing B. pseudomallei from B. cepacia (4).
ACKNOWLEDGMENTS
We thank Vanessa Theobald, Charles Goh, Irene Tan, Chien Su Lin, Suria Wahap, Desiree Wong, Reginal Bantin, Chua Wen Yi, and Alexander Mijen for their laboratory assistance. We also acknowledge the support of Sarawak Health Department and the hospitals involved in this study.
Footnotes
Published ahead of print 19 June 2013
REFERENCES
- 1.Wiersinga WJ, Currie BJ, Peacock SJ. 2012. Melioidosis. N. Engl. J. Med. 367:1035–1044 [DOI] [PubMed] [Google Scholar]
- 2.Cheng AC, Currie BJ. 2005. Melioidosis: epidemiology, pathophysiology, and management. Clin. Microbiol. Rev. 18:383–416 (Erratum, 20:533, 2007.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dance DA, Wuthiekanun V, Naigowit P, White NJ. 1989. Identification of Pseudomonas pseudomallei in clinical practice: use of simple screening tests and API 20NE. J. Clin. Pathol. 42:645–648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hodgson K, Engler C, Govan B, Ketheesan N, Norton R. 2009. Comparison of routine bench and molecular diagnostic methods in identification of Burkholderia pseudomallei. J. Clin. Microbiol. 47:1578–1580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lowe P, Engler C, Norton R. 2002. Comparison of automated and nonautomated systems for identification of Burkholderia pseudomallei. J. Clin. Microbiol. 40:4625–4627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lowe P, Haswell H, Lewis K. 2006. Use of various common isolation media to evaluate the new VITEK 2 colorimetric GN card for identification of Burkholderia pseudomallei. J. Clin. Microbiol. 44:854–856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Koh TH, Yong Ng LS, Foon Ho JL, Sng LH, Wang GC, Tzer Pin Lin RV. 2003. Automated identification systems and Burkholderia pseudomallei. J. Clin. Microbiol. 41:1809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zong Z, Wang X, Deng Y, Zhou T. 2012. Misidentification of Burkholderia pseudomallei as Burkholderia cepacia by the VITEK 2 system. J. Med. Microbiol. 61:1483–1484 [DOI] [PubMed] [Google Scholar]
- 9.Weissert C, Dollenmaier G, Rafeiner P, Riehm J, Schultze D. 2009. Burkholderia pseudomallei misidentified by automated system. Emerg. Infect. Dis. 15:1799–1801. 10.3201/eid1511.081719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Novak RT, Glass MB, Gee JE, Gal D, Mayo MJ, Currie BJ, Wilkins PP. 2006. Development and evaluation of a real-time PCR assay targeting the type III secretion system of Burkholderia pseudomallei. J. Clin. Microbiol. 44:85–90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kaestli M, Richardson LJ, Colman RE, Tuanyok A, Price EP, Bowers JR, Mayo M, Kelley E, Seymour ML, Sarovich DS, Pearson T, Engelthaler DM, Wagner DM, Keim PS, Schupp JM, Currie BJ. 2012. Comparison of TaqMan PCR assays for detection of the melioidosis agent Burkholderia pseudomallei in clinical specimens. J. Clin. Microbiol. 50:2059–2062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Anuntagool N, Naogowit P, Petkanchanapong V, Aramsri P, Panichakul T, Sirisinha S. 2000. Monoclonal antibody-based rapid identification of Burkholderia pseudomallei in blood culture fluid from patients with community-acquired septicaemia. J. Med. Microbiol. 49:1075–1078 [DOI] [PubMed] [Google Scholar]
- 13.Dubois D, Grare M, Prere MF, Segonds C, Marty N, Oswald E. 2012. Performances of the Vitek MS matrix-assisted laser desorption ionization-time of flight mass spectrometry system for rapid identification of bacteria in routine clinical microbiology. J. Clin. Microbiol. 50:2568–2576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Coenye T, Vandamme P, LiPuma JJ, Govan JR, Mahenthiralingam E. 2003. Updated version of the Burkholderia cepacia complex experimental strain panel. J. Clin. Microbiol. 41:2797–2798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Vandamme P, Dawyndt P. 2011. Classification and identification of the Burkholderia cepacia complex: past, present and future. Syst. Appl. Microbiol. 34:87–95 [DOI] [PubMed] [Google Scholar]
- 16.Gilad J, Giladi M, Schwartz D. 2007. Candida albicans masquerading as Gram-negative bacilli in the clinical laboratory. Scand. J. Infect. Dis. 39:907–910 [DOI] [PubMed] [Google Scholar]
- 17.Burton MJ, Shah P, Swiatlo E. 2011. Misidentification of Candida parapsilosis as C. famata in a clinical case of vertebral osteomyelitis. Am. J. Med. Sci. 341:71–73 [DOI] [PubMed] [Google Scholar]
- 18.Yakandawala N, Gawande PV, LoVetri K, Cardona ST, Romeo T, Nitz M, Madhyastha S. 2011. Characterization of the poly-β-1,6-N-acetylglucosamine polysaccharide component of Burkholderia biofilms. Appl. Environ. Microbiol. 77:8303–8309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Masoud H, Ho M, Schollaardt T, Perry MB. 1997. Characterization of the capsular polysaccharide of Burkholderia (Pseudomonas) pseudomallei 304b. J. Bacteriol. 179:5663–5669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cuccui J, Milne TS, Harmer N, George AJ, Harding SV, Dean RE, Scott AE, Sarkar-Tyson M, Wren BW, Titball RW, Prior JL. 2012. Characterization of the Burkholderia pseudomallei K96243 capsular polysaccharide I coding region. Infect. Immun. 80:1209–1221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tuanyok A, Leadem BR, Auerbach RK, Beckstrom-Sternberg SM, Beckstrom-Sternberg JS, Mayo M, Wuthiekanun V, Brettin TS, Nierman WC, Peacock SJ, Currie BJ, Wagner DM, Keim P. 2008. Genomic islands from five strains of Burkholderia pseudomallei. BMC Genomics. 9:566. 10.1186/1471-2164-9-566 [DOI] [PMC free article] [PubMed] [Google Scholar]