Abstract
Cryptococcus gattii consists of four cryptic species, VGI, VGII, VGIII, and VGIV. Herein, a duplex PCR assay using two primer pairs targeting the vacuolar membrane gene and the intergenic spacer region was developed. It successfully distinguished the cryptic species according to the distinct size of the amplicons.
TEXT
The basidiomycetous yeast Cryptococcus gattii is a primary pathogen that has a predilection for infecting immunocompetent individuals. A wide geographic distribution, other than restriction in tropical and subtropical areas, has been recognized for this species in many studies (1–7). Based on genetic differences, four molecular types, VGI to VGIV, within C. gattii have been determined (8). The four molecular types are also considered cryptic species due to sequence divergence and no nuclear recombination events observed between them (1, 9). The cryptic species differ in their biologies, epidemiologies, virulence characteristics, antifungal susceptibilities, and geographic distributions (1–16). Among the four cryptic species, VGII is more concerning because of the outbreak of the C. gattii VGII genotype in the Pacific Northwest and Australia, as well as its higher virulence in experimental models, mating efficiency, and decreased susceptibility to antifungal agents (1, 10–14, 16). In addition, VGI accounts for the most cryptococcal cases caused by C. gattii around the world and has the widest geographic distribution (1, 2, 7); VGIII and VGIV are more geographically restricted and usually associated with HIV-infected cases (5, 15).
Several molecular tools have been developed to distinguish the four cryptic species (1, 7, 8, 16–21). However, these techniques include time-consuming and laborious PCR-based methods or molecular methods requiring expensive instruments. PCR analyses of differences in intron size or intron loss have been established to differentiate closely related species of Candida and the Cryptococcus neoformans-C. gattii species complex and have been confirmed to be simple, inexpensive, and reliable methods (22, 23). Recently, genomic studies revealed that intron loss was also a phylogenetic marker for distinct cryptic species within C. gattii (24).
In this study, adjacent introns within protein-coding genes were detected and compared between type strains WM276 (VGI) and R265 (VGII) by the LAGAN tool as previously described (22). The putative vacuolar membrane gene was chosen due to its different types of intron loss among the type strains of WM276 (VGI), R265 (VGII), and WM779 (VGIV), as well as WM161 (VGIII), which is identical to R265 (VGII) (Fig. 1). No intron difference among all four cryptic species was found in our study, which corresponded with a recent report (24). Additionally, a VGII-specific primer pair which could specifically yield a 156-bp amplicon for VGII strains by PCR analysis (25) was utilized herein to distinguish VGII from VGIII.
Fig 1.
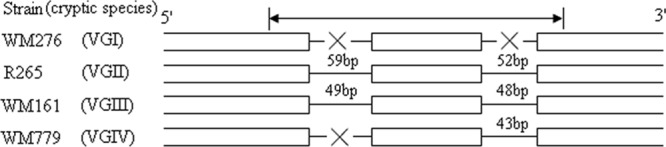
Schematic representation of the amplified regions of the putative vacuolar membrane gene. Open boxes represent exons (exon2, exon3, and exon4, in turn), while lines between exons represent introns. “×” represents intron loss; intron length is indicated by the numeral above the line; the region between the arrows represents the amplified fragment.
A C. gattii-specific primer pair targeting the vacuolar membrane gene, VACF (5′ AGCCCACGGCAAAATAGTG 3′) and VACR (5′ CACGGTCCAAAACTTGATTGTT 3′), was designed. A duplex PCR system using two primer pairs, VACF and VACR and IGSF (5′ CCGAGGCAGGACACACATAC 3′) and IGSR (5′ GGCGGAATACAAATACTACTTACCT 3′), was established. PCR was performed in a final volume of 50 μl containing 50 ng DNA, 1× PCR buffer with 1.5 mM MgCl2, 0.2 mM (each) dATP, dCTP, dGTP, and dTTP, 0.2 μM of each primer, and 1.5 U of Taq polymerase. PCR was conducted in a Bio-Rad thermal cycler at 94°C for 5 min for initial denaturation, followed by 30 cycles of 94°C for 30 s, 56°C for 30 s, and 72°C for 20 s, with a final extension step at 72°C for 6 min. PCR products were separated on a 2% (wt/vol) agarose gel at 90 V for 2 h. All PCRs were conducted in duplicate. Strains tested included C. gattii (n = 80), C. neoformans (n = 16), and other pathogenic yeast species (n = 42). The latter were involved to detect the specificity of the primer pairs. Genomic DNA was prepared from each strain as described previously (22). Detailed information regarding the strains tested here is presented in Table S1 in the supplemental material.
Electrophoretic and sequence analysis revealed an expected 166-bp amplicon for C. gattii VGI, a combination of 277-bp and 156-bp amplicons for C. gattii VGII, a 263-bp amplicon for C. gattii VGIII, and a 209-bp amplicon for C. gattii VGIV (Fig. 2). As a result, all C. gattii strains tested were exactly assigned to the cryptic species level, and none of the other pathogenic yeast species, including the sibling species C. neoformans, resulted in an amplification product by using this approach. Moreover, the rarely reported C. neoformans × C. gattii hybrids may be further characterized by the singleplex PCR as previously described (22).
Fig 2.
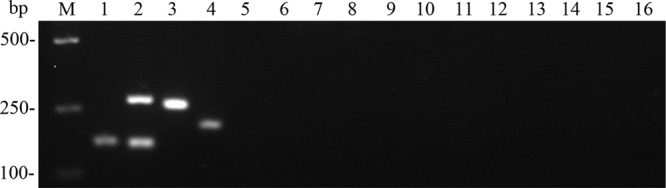
Partial results in agarose gel electrophoresis analysis. Lanes (species and cryptic species are shown in parentheses): M, DL2000 ladder; 1, WM179 (C. gattii VGI); 2, WM178 (C. gattii VGII); 3, WM161 (C. gattii VGIII); 4, WM779 (C. gattii VGIV); 5, WM148 (C. neoformans VNI); 6, WM626 (C. neoformans VNII); 7, WM628 (C. neoformans VNIII); 8, WM629 (C. neoformans VNIV); 9, Y2090 (Cryptococcus albidus); 10, Y2536 (Cryptococcus laurentii); 11, ATCC 10231 (Candida albicans); 12, ATCC 750 (Candida tropicalis); 13, CBS604 (Candida parapsilosis); 14, ATCC 90030 (Candida glabrata); 15, Y17953 (Trichosporon asahii); 16, negative control.
Differences in intron loss in the protein-coding genes were found not only among species or varieties within the C. neoformans-C. gattii species complex but also among the monophylogenetic clusters within C. gattii (24, 26). The latter was utilized herein for molecular typing of C. gattii. Thus, we report a duplex PCR assay which performed well in distinguishing among the four cryptic species belonging to C. gattii.
Nucleotide sequence accession numbers.
The sequences of the vacuolar membrane gene fragment of strains WM161 and WM779 were deposited in GenBank under accession numbers KF010296 and KF010297, respectively.
Supplementary Material
ACKNOWLEDGMENTS
We thank Joseph Heitman, John R. Perfect, Anastasia P. Litvintseva, Wiley A. Schell, and Anna Floyd (Duke University Medical Center, Durham, NC), Wieland Meyer and Dee Carter (University of Sydney, Sydney, Australia), C. De Vroey (Prince Leopold Institut of Tropical Medicine, Belgium), Maria Anna Viviani (Istituto di Igiene e Medicina Preventiva, Milan, Italy), Kyung J. Kwon-Chung (National Institutes of Health, Bethesda, MD), James W. Kronstad (University of British Columbia, Vancouver, BC, Canada), Teun Boekhout and Ferry Hagen (Centraalbureau voor Schimmelcultures–Fungal Biodiversity Centre, Utrecht, the Netherlands), Jianping Xu (McMaster University, Hamilton, Canada), James Swezey (ARS Culture Collection, United States), and Orazio Romeo (University of Messina, Italy) and Claudete R. Paula (University of São Paulo, São Paulo, Brazil) for the strains.
This work was supported by grants from the National Natural Science Foundation of China (no. 31000549), Shanghai Science and Technology Commission Fund (no. 10dz2220100), and Shanghai Key Laboratory of Molecular Medical Mycology Fund (no. 20110001).
Footnotes
Published ahead of print 3 July 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JCM.01455-13.
REFERENCES
- 1.Fraser JA, Giles SS, Wenink EC, Geunes-Boyer SG, Wright JR, Diezmann S, Allen A, Stajich JE, Dietrich FS, Perfect JR, Heitman J. 2005. Same-sex mating and the origin of the Vancouver Island Cryptococcus gattii outbreak. Nature 437:1360–1364 [DOI] [PubMed] [Google Scholar]
- 2.Hagen F, Colom MF, Swinne D, Tintelnot K, Iatta R, Montagna MT, Torres-Rodriguez JM, Cogliati M, Velegraki A, Burggraaf A, Kamermans A, Sweere JM, Meis JF, Klaassen CH, Boekhout T. 2012. Autochthonous and dormant Cryptococcus gattii infections in Europe. Emerg. Infect. Dis. 18:1618–1624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chowdhary A, Randhawa HS, Boekhout T, Hagen F, Klaassen CH, Meis JF. 2012. Temperate climate niche for Cryptococcus gattii in Northern Europe. Emerg. Infect. Dis. 18:172–174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chowdhary A, Rhandhawa HS, Prakash A, Meis JF. 2012. Environmental prevalence of Cryptococcus neoformans and Cryptococcus gattii in India: an update. Crit. Rev. Microbiol. 38:1–16 [DOI] [PubMed] [Google Scholar]
- 5.Byrnes ER, Li W, Ren P, Lewit Y, Voelz K, Fraser JA, Dietrich FS, May RC, Chaturvedi S, Chaturvedi V, Heitman J. 2011. A diverse population of Cryptococcus gattii molecular type VGIII in southern Californian HIV/AIDS patients. PLoS Pathog. 7:e1002205. 10.1371/journal.ppat.1002205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Romeo O, Scordino F, Criseo G. 2011. Environmental isolation of Cryptococcus gattii serotype B, VGI/MATalpha strains in southern Italy. Mycopathologia 171:423–430 [DOI] [PubMed] [Google Scholar]
- 7.Feng X, Yao Z, Ren D, Liao W, Wu J. 2008. Genotype and mating type analysis of Cryptococcus neoformans and Cryptococcus gattii isolates from China that mainly originated from non-HIV-infected patients. FEMS Yeast Res. 8:930–938 [DOI] [PubMed] [Google Scholar]
- 8.Meyer W, Castaneda A, Jackson S, Huynh M, Castaneda E. 2003. Molecular typing of IberoAmerican Cryptococcus neoformans isolates. Emerg. Infect. Dis. 9:189–195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ngamskulrungroj P, Gilgado F, Faganello J, Litvintseva AP, Leal AL, Tsui KM, Mitchell TG, Vainstein MH, Meyer W. 2009. Genetic diversity of the Cryptococcus species complex suggests that Cryptococcus gattii deserves to have varieties. PLoS One 4:e5862. 10.1371/journal.pone.0005862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Byrnes ER, Li W, Lewit Y, Ma H, Voelz K, Ren P, Carter DA, Chaturvedi V, Bildfell RJ, May RC, Heitman J. 2010. Emergence and pathogenicity of highly virulent Cryptococcus gattii genotypes in the northwest United States. PLoS Pathog. 6:e1000850. 10.1371/journal.ppat.1000850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Carriconde F, Gilgado F, Arthur I, Ellis D, Malik R, van de Wiele N, Robert V, Currie BJ, Meyer W. 2011. Clonality and alpha-a recombination in the Australian Cryptococcus gattii VGII population—an emerging outbreak in Australia. PLoS One 6:e16936. 10.1371/journal.pone.0016936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kidd SE, Hagen F, Tscharke RL, Huynh M, Bartlett KH, Fyfe M, Macdougall L, Boekhout T, Kwon-Chung KJ, Meyer W. 2004. A rare genotype of Cryptococcus gattii caused the cryptococcosis outbreak on Vancouver Island (British Columbia, Canada). Proc. Natl. Acad. Sci. U. S. A. 101:17258–17263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Trilles L, Meyer W, Wanke B, Guarro J, Lazera M. 2011. Correlation of antifungal susceptibility and molecular type within the Cryptococcus neoformans/C. gattii species complex. Med. Mycol. 50:328–332 [DOI] [PubMed] [Google Scholar]
- 14.Ma H, Hagen F, Stekel DJ, Johnston SA, Sionov E, Falk R, Polacheck I, Boekhout T, May RC. 2009. The fatal fungal outbreak on Vancouver Island is characterized by enhanced intracellular parasitism driven by mitochondrial regulation. Proc. Natl. Acad. Sci. U. S. A. 106:12980–12985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Litvintseva AP, Thakur R, Reller LB, Mitchell TG. 2005. Prevalence of clinical isolates of Cryptococcus gattii serotype C among patients with AIDS in Sub-Saharan Africa. J. Infect. Dis. 192:888–892 [DOI] [PubMed] [Google Scholar]
- 16.Hagen F, Illnait-Zaragozi MT, Bartlett KH, Swinne D, Geertsen E, Klaassen CH, Boekhout T, Meis JF. 2010. In vitro antifungal susceptibilities and amplified fragment length polymorphism genotyping of a worldwide collection of 350 clinical, veterinary, and environmental Cryptococcus gattii isolates. Antimicrob. Agents Chemother. 54:5139–5145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Firacative C, Trilles L, Meyer W. 2012. MALDI-TOF MS enables the rapid identification of the major molecular types within the Cryptococcus neoformans/C. gattii species complex. PLoS One 7:e37566. 10.1371/journal.pone.0037566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Posteraro B, Vella A, Cogliati M, De Carolis E, Florio AR, Posteraro P, Sanguinetti M, Tortorano AM. 2012. Matrix-assisted laser desorption ionization-time of flight mass spectrometry-based method for discrimination between molecular types of Cryptococcus neoformans and Cryptococcus gattii. J. Clin. Microbiol. 50:2472–2476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Feng X, Yao Z, Liao W. 2012. Approaches for molecular identification and typing of the Cryptococcus species complex: an update. Rev. Med. Microbiol. 24:1–6 [Google Scholar]
- 20.Feng X, Yao Z, Ren D, Liao W. 2008. Simultaneous identification of molecular and mating types within the Cryptococcus species complex by PCR-RFLP analysis. J. Med. Microbiol. 57:1481–1490 [DOI] [PubMed] [Google Scholar]
- 21.Bovers M, Diaz MR, Hagen F, Spanjaard L, Duim B, Visser CE, Hoogveld HL, Scharringa J, Hoepelman IM, Fell JW, Boekhout T. 2007. Identification of genotypically diverse Cryptococcus neoformans and Cryptococcus gattii isolates by Luminex xMAP technology. J. Clin. Microbiol. 45:1874–1883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Feng X, Fu X, Ling B, Wang L, Liao W, Yao Z. 2013. Development a singleplex PCR assay for rapid identification and differentiation of Cryptococcus neoformans var. grubii, Cryptococcus neoformans var. neoformans, Cryptococcus gattii and hybrids. J. Clin. Microbiol. 51:1920–1923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Enache-Angoulvant A, Guitard J, Grenouillet F, Martin T, Durrens P, Fairhead C, Hennequin C. 2011. Rapid discrimination between Candida glabrata, Candida nivariensis, and Candida bracarensis by use of a singleplex PCR. J. Clin. Microbiol. 49:3375–3379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Croll D, McDonald BA. 2012. Intron gains and losses in the evolution of Fusarium and Cryptococcus fungi. Genome Biol. Evol. 4:1148–1161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Feng X, Yao Z, Ren D, Liao W. 2010. Rapid differentiation of VGII/AFLP6 genotype within Cryptococcus gattii by polymerase chain reaction. Diagn. Microbiol. Infect. Dis. 68:471–473 [DOI] [PubMed] [Google Scholar]
- 26.Sharpton TJ, Neafsey DE, Galagan JE, Taylor JW. 2008. Mechanisms of intron gain and loss in Cryptococcus. Genome Biol. 9:R24. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


