Abstract
Accurate point-of-care (POC) diagnostic tests for Chlamydia trachomatis infection are urgently needed for the rapid treatment of patients. In a blind comparative study, we evaluated microwave-accelerated metal-enhanced fluorescence (MAMEF) assays for ultrafast and sensitive detection of C. trachomatis DNA from vaginal swabs. The results of two distinct MAMEF assays were compared to those of nucleic acid amplification tests (NAATs). The first assay targeted the C. trachomatis 16S rRNA gene, and the second assay targeted the C. trachomatis cryptic plasmid. Using pure C. trachomatis, the MAMEF assays detected as few as 10 inclusion-forming units/ml of C. trachomatis in less than 9 min, including DNA extraction and detection. A total of 257 dry vaginal swabs from 245 female adolescents aged 14 to 22 years were analyzed. Swabs were eluted with water, the solutions were lysed to release and to fragment genomic DNA, and MAMEF-based DNA detection was performed. The prevalence of C. trachomatis by NAATs was 17.5%. Of the 45 samples that were C. trachomatis positive and the 212 samples that were C. trachomatis negative by NAATs, 33/45 and 197/212 were correctly identified by the MAMEF assays if both assays were required to be positive (sensitivity, 73.3%; specificity, 92.9%). Using the plasmid-based assay alone, 37/45 C. trachomatis-positive and 197/212 C. trachomatis-negative samples were detected (sensitivity, 82.2%; specificity, 92.9%). Using the 16S rRNA assay alone, 34/45 C. trachomatis-positive and 197/212 C. trachomatis-negative samples were detected (sensitivity, 75.5%; specificity, 92.9%). The overall rates of agreement with NAAT results for the individual 16S rRNA and cryptic plasmid assays were 89.5% and 91.0%, respectively. Given the sensitivity, specificity, and rapid detection of the plasmid-based assay, the plasmid-based MAMEF assay appears to be suited for clinical POC testing.
INTRODUCTION
Chlamydia trachomatis infection is the most common bacterial sexually transmitted infection (STI) in the world and the STI most frequently reported to the Centers for Disease Control and Prevention (CDC) (1). In 2011, a total of 1,412,791 chlamydial infections were reported to the CDC from the United States and the District of Columbia, representing an 8.0% increase from 2010 (1). Most chlamydial infections involved female patients 15 to 19 years (3,416.5 cases per 100,000 population) or 20 to 24 years (3,722.5 cases per 100,000 population) of age (1). Since chlamydial infections are most often asymptomatic, the CDC and other professional organizations recommend yearly screening for chlamydia among all sexually active women ≤25 years of age (2, 3). However, the Healthcare Effectiveness Data and Information Set (HEDIS) measure assessing screening coverage among female patients receiving medical care through private insurance or Medicaid indicated that only 43.1% of sexually active female patients 16 to 24 years of age in commercial plans and 57.5% of female patients covered by Medicaid received screening tests in 2010 (4). Barriers to screening include lack of awareness by clinicians and limited clinical resources.
There are several excellent commercial systems available for performing nucleic acid amplification tests (NAATs) for detection of chlamydia (5–8). However, new assays and new platforms that are able to be used at the time of the patient visit are urgently needed. Although NAATs are now recommended by the CDC as the tests of choice, these laboratory-based tests may require several days for final results, and many patients do not return for their results (9, 10). Additionally, delayed returns to clinics for treatment often contribute to pelvic inflammatory disease morbidity (11). The development of accurate rapid point-of-care (POC) tests is urgently needed in order to increase the ease of test performance and to provide treatment for patients before they leave the site of care. We report here the development of a new C. trachomatis test, a microwave-accelerated metal-enhanced fluorescence (MAMEF) test, and its performance with clinical specimens in a blind study. The MAMEF technology was developed by Geddes and colleagues (12–27). It combines the significant benefits of low-power microwave acceleration to accelerate biological reactions to completeness within seconds with those of metal-enhanced fluorescence (MEF), whereby the close proximity of silver nanoparticles (plasmon-supporting particles) amplifies the fluorescence or luminescence of labels in the near field, i.e., less than 1 wavelength of light away (28–35). The resulting technology, MAMEF, allows for combined ultrafast and ultrasensitive detection of DNAs (12–20), RNAs (36), and proteins (24–27).
MATERIALS AND METHODS
Assay description.
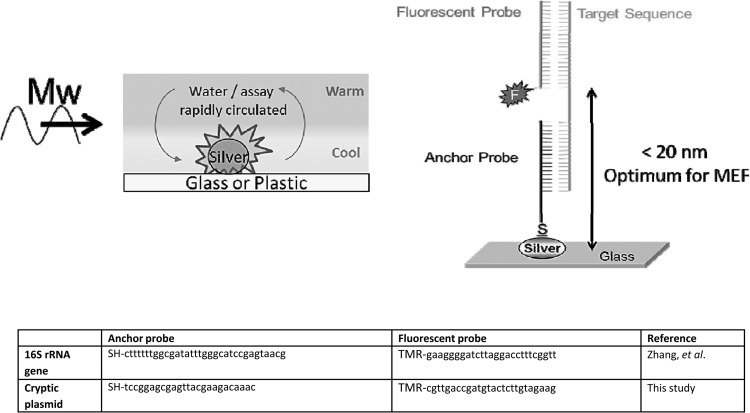
When a liquid sample is deposited on metallic nanoparticles and exposed to microwaves, the small volume of fluid above the metal is quickly heated. However, the metallic nanoparticles remain cool because they do not absorb microwaves due to their subwavelength size (typically 100 to 200 nm in diameter), rapidly creating a temperature gradient between the cool nanoparticles and the warm fluid. This temperature gradient facilitates the mass transport of target DNA molecules toward the MEF assay surface, allowing for faster biorecognition kinetics observed near metallic nanoparticles (17, 18). The expedited transport of target DNA molecules to the assay surface not only accelerates the complementary binding of target sequences to fluorescently labeled tags but also increases assay sensitivity through the amplification of fluorescence signals (MEF) (17, 18). As shown in Fig. 1, target DNA sequences bind to a fluorophore-labeled probe and to an anchor probe that is covalently bound to the assay surface. When the target DNA sequence is present, the three-piece assay construct (target DNA, fluorophore-labeled probe, and anchor probe) can form, thereby allowing the fluorophore label to come into close proximity to metallic nanoparticles and metal-enhanced fluorescence-based optical enhancement to occur (12–16). MEF assays are carried out in silver-coated microtiter plates (Fig. 2) containing the anchor probe and a fluorescent probe that includes a 6-carboxytetramethylrhodamine (TAMRA) dye. The TAMRA dye was selected to match specific wavelength requirements (32, 37) as well as to enhance transmission through clinical samples (which may contain blood) for optical sensing, i.e., it emits in the therapeutic optical window (38, 39).
Fig 1.
(Top left) Cartoon depicting microwave-accelerated heating above MEF substrates. (Top right) MAMEF assay construct for detection of chlamydial DNA. (Bottom) Anchor and fluorescent probe sequences. SH, sulfhydryl group for attachment of DNA to the silver surface; TMR, TAMRA NHS ester dye; Mw, microwave heating; MEF, metal-enhanced fluorescence; F, fluorophore.
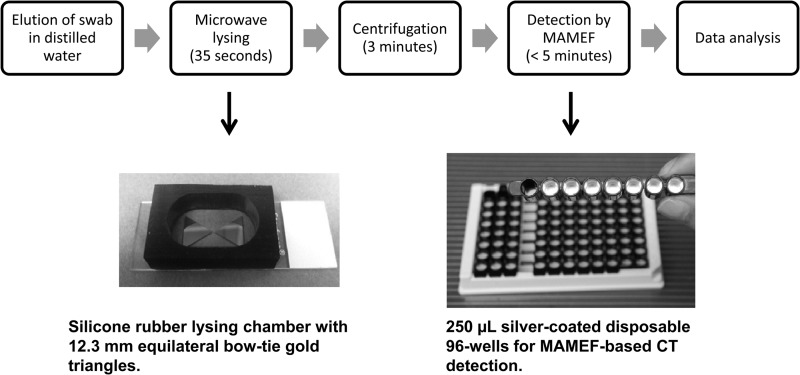
Fig 2.
Flow chart showing the sequence of events and timeline for a MAMEF-based C. trachomatis test.
The sample preparation step associated with extraction of nucleic acids is a significant bottleneck in current PCR-based approaches, in addition to the time required for PCR-based detection (40–42). In contrast to the traditional lysis techniques, which can take several hours (43–45), our previously developed lysis approach can rapidly (typically in 5 to 10 s) lyse virtually any bacteria, enabling the genetic material (e.g., C. trachomatis DNA) to be rapidly collected for analysis with the MAMEF platform. This approach employs gold bowtie geometries (Fig. 2), which highly focus microwaves at 2.45 GHz onto eluted clinical samples. The sample lysis chambers have been theoretically designed and modeled using numerical simulations (finite different time domain) (12–14, 16). Thus, we can lyse samples using a $30 commercial low-power microwave oven with only a few slight modifications made inside for sample mounting. The rapid heating of the water (both around and within the C. trachomatis organism) rapidly disrupts the membranes, allowing the sample lysate to be extracted and subsequently used for the MAMEF assay, where the extent of heating is controlled simply by the gap size between the triangles, the salt concentration, and the microwave exposure time (12–14, 16). Our lysis technology has been likened to how popcorn pops in a microwave oven; water, which absorbs at 2.45 GHz, heats rapidly, expands as a gas, and pops the corn. Our lysis approach also has the particular advantage of thermally fragmenting genomes into smaller sequences (<100 bp) (12–16), the extent of which is determined by the lysis conditions (including temperature, geometry, and salt concentration) and which is ideal for high-capacity DNA surface sensing (12–16). Further, this approach has the advantage that the high temperatures necessary for lysis (∼95°C) also perturb enzymes that may destroy low-copy-number targets (26, 27).
Microbial species.
The analytical sensitivity of the assay was tested through the use of 10-fold serial dilutions of Chlamydia trachomatis grown in McCoy cells, yielding final concentrations of 0 to 106 inclusion-forming units (IFU)/ml. Each dilution was tested in quadruplicate. The limit of detection of the assays was calculated based on IFU/ml values. DNA from a masked panel of 18 microorganisms likely to be present in human genital samples or closely genetically related to C. trachomatis also was tested, to determine the analytical specificity of the assays.
Clinical samples.
Vaginal swabs collected at the Cincinnati Children's Hospital Medical Center Teen Health Center (Cincinnati, OH) in December 2010 through March 2012 were included in the study. Duplicate swabs were tested locally with the ProbeTec assay (Becton Dickinson, Sparks, MD). The specimens for MAMEF assays were collected and stored frozen at −80°C as dry swabs, as part of a study looking at the accuracy of new POC Chlamydia test devices among adolescents and young women. The frozen vaginal swabs initially were shipped to the Johns Hopkins School of Medicine (Baltimore, MD) and then were given to the Institute of Fluorescence as masked samples for testing with the Chlamydia MAMEF assays.
DNA extraction and fragmentation.
Vaginal swabs were transferred frozen to 15-ml conical tubes, and 2 ml of autoclaved deionized water was added to each swab. Following a 20-min incubation, the swabs were vortex-mixed for 10 s and excess liquid was removed by pressing the swab against the side of the tube. A 200-μl aliquot of each sample was transferred to Gen-Probe medium and stored frozen for future testing in the event of discordant results between the ProbeTec assay and the MAMEF assay. DNA from the rest of the sample (approximately 1.5 ml) was extracted using the previously described gold-bowtie/focused-microwave lysis approach, with minor modifications (12, 16). Briefly, two equilateral gold triangles (12.5 mm long and 100 nm thick) were deposited on glass slides using a vapor disposition system, and a self-adhesive silicon isolator (31 mm by 9 mm) was placed over the bowtie region, creating a lysis chamber (Fig. 2). The swab eluate was then placed in the lysis chamber and exposed to 35 s of microwave irradiation, at a power corresponding to 270 W, over the entire microwave cavity. The lysed sample was then collected and centrifuged at 4,580 × g for 3 min before undergoing MAMEF testing. Gels confirmed that all sample DNA had been fractured into <100-bp fragments, which is ideal for high-capacity MAMEF-based sensing (12–16).
Design of DNA probes.
Two different MAMEF assays were used during this study. The first assay targets the C. trachomatis 16S rRNA gene, and the details of this assay have been reported previously (16). A second assay targeting the C. trachomatis cryptic plasmid was developed to increase the sensitivity of the MAMEF platform and for use as a confirmatory assay. Similar to the 16S rRNA assay, the C. trachomatis cryptic plasmid assay involves two probes; the anchor probe is composed of 26 nucleotides with a terminal thiol group that readily binds to the silver surface, and the fluorescent probe is composed of 27 nucleotides and is labeled with a TAMRA N-hydroxysuccinimide (NHS) ester dye at the first nucleotide, which corresponds to the position closest to the metal surface. When C. trachomatis DNA is present, the 3-piece DNA assay construct is complete (Fig. 1). In both assays, the DNA probes are complementary to the C. trachomatis target sequence. The anchor and fluorophore-labeled probes were designed to bind to the negative strand.
MAMEF-based C. trachomatis DNA assay.
MAMEF-based DNA detection is mediated by the complementary binding of two probes to the target DNA sequence. The anchor probe is chemically linked to the silver nanoparticles on the wells of the microtiter plate via a thiol group. The fluorophore-labeled probe is added to the anchor probe-containing wells with the sample prior to microwave irradiation. In the presence of the target DNA sequence, the 3-piece assay construct is complete, resulting in an enhanced fluorescence signal due to the close proximity of the fluorescent label to the silver nanoparticles (Fig. 1). MAMEF-based DNA detection involves four steps: (i) elution of the sample from the swab, (ii) microwave-based cell lysis and DNA fragmentation, (iii) separation of DNA and cellular debris by centrifugation, and (iv) MAMEF-based DNA detection. Following the previously described sample elution and centrifugation steps, sample testing was carried out in silver-coated microtiter plates (37) (Fig. 2). DNA detection was carried out by combining 50 μl of 50 nM fluorescent probe with 200 μl of sample in the anchor probe-containing wells and heating the sample in a microwave cavity for 3 min. All samples were tested in duplicate, using both the 16S rRNA and cryptic plasmid C. trachomatis MAMEF assays. A negative-control sample, consisting of pooled C. trachomatis-negative specimens, and a C. trachomatis-positive sample were tested in parallel with the unknown samples. Prior to fluorescence detection, the silver-coated wells were subjected to a primary washing step with deionized water to remove excess unbound fluorescent probe and sample. A secondary washing step was performed for all samples with elevated fluorescence signals, as outlined below.
Post-MAMEF analysis.
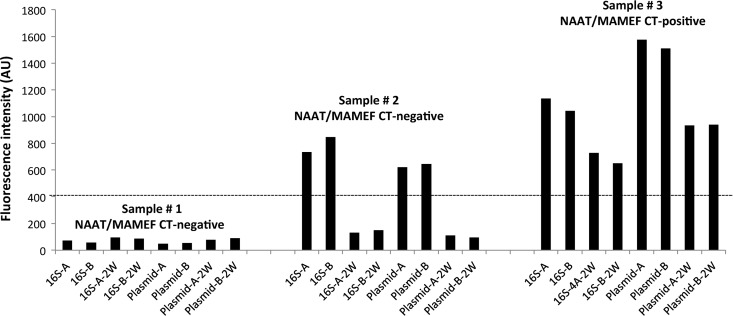
All samples were tested masked. For determination of positivity, fluorescence data from the unknown sample were compared to fluorescence data from the Chlamydia-negative control sample, as shown in Fig. 3. Samples with fluorescence signals equal to or below the value for the standardized negative-control sample were reported as C. trachomatis negative and were not subjected to the secondary washing step. All samples with fluorescence signals above the value for the standardized negative-control sample were subjected to a secondary washing step to remove residual unbound fluorescent probe. Initial characterization of the assay revealed that the secondary washing step can help to remove excess unbound probe, the presence of which can result in false-positive results. Furthermore, the secondary washing step does not disrupt the 3-piece DNA construct when target DNA is present (Fig. 3). Samples were considered to be C. trachomatis positive if the level of fluorescence was above the threshold for negativity following the secondary washing step.
Fig 3.
Classification of MAMEF assay results according to fluorescence intensity. CT, C. trachomatis; AU, arbitrary units; 16S, 16S rRNA MAMEF assay; plasmid, cryptic plasmid MAMEF assay; A and B, duplicate tests; 2W, second wash. Sample 1 showed low fluorescence intensity below the threshold of positivity (dashed line), characteristic of a C. trachomatis-negative sample. For sample 2, elevated signal intensity was detected initially but intensity decreased below the threshold of positivity after a second washing step. Sample 3 showed high signal intensity before and after the second washing step, indicative of strong binding of the target C. trachomatis DNA to the DNA capture probes.
MEF detection.
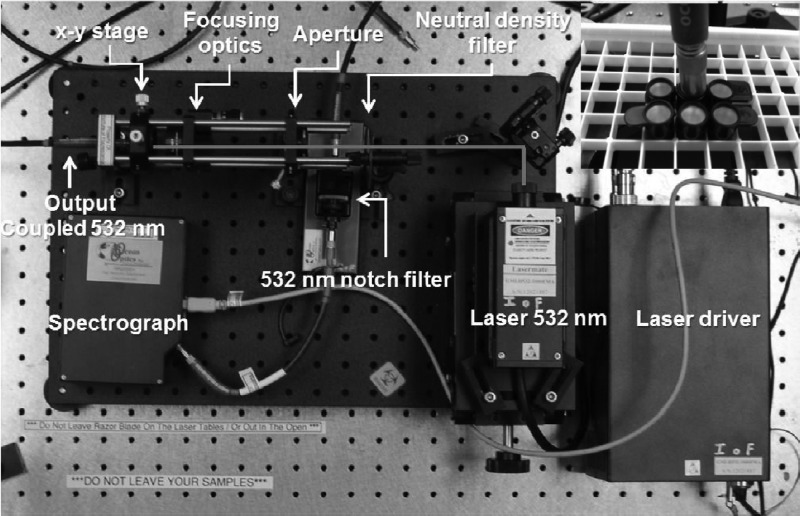
Our MEF reader consists of a 532-nm continuous-wave laser (LaserMate), for which the excitation power is adjusted using an absorbing neutral-density filter wheel (Edmund Optics, Barrington, NJ), with focusing optics (Thorlabs) in a 600-μm bitruncated fiber (Ocean Optics, Dunedin, FL). A 532-nm notch filter blocks the excitation light through the emission channel of the bitruncated fiber, which falls incident onto an Ocean Optics HD2000 spectrometer (Fig. 4).
Fig 4.
Inner configuration of the optical reader for the C. trachomatis assays. The bitruncated fiber both excites and collects metal-enhanced fluorescence emission from the silver-coated wells.
Patient demographics.
A total of 260 vaginal swabs were collected from 248 subjects, and MAMEF assay results were available for 257 samples. Twelve subjects had swabs from two different visits. The age distribution of the 245 subjects was as follows: 17.1%, 14 to 16 years of age; 58.4%, 17 to 19 years of age; 24.5%, 20 to 22 years of age. The majority of subjects were African-American (88.6%), followed by Caucasian (8.9%), Hispanic (1.6%), and other (0.8%). The majority of subjects with NAAT-confirmed chlamydial infections were African-American (95.6%).
RESULTS
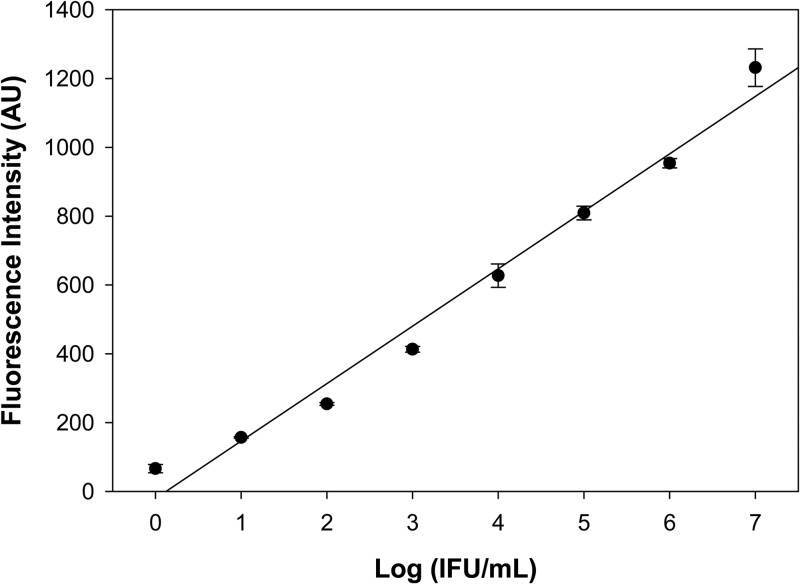
The limit of detection (3 × the standard mean value) of the cryptic plasmid MAMEF assay was determined by testing serial dilutions of C. trachomatis, in the form of cultured C. trachomatis tested in tissue culture, by visualization of inclusion-forming units (IFU). The assay showed high analytical sensitivity, on the order of 10 IFU/ml (Fig. 5). The 16S rRNA-based assay correctly identified all of the C. trachomatis strains tested, but it also showed cross-reactivity with two different strains of Chlamydia pneumoniae. The cryptic plasmid-based assay also correctly identified its target, but it did not show cross-reactivity against any other microbial species (Table 1).
Fig 5.
Serial dilution plot for the cryptic plasmid-based MAMEF assay. The log(IFU/ml) values for C. trachomatis were plotted against fluorescence intensity (R2 = 0.9801).
Table 1.
Specificity of the MAMEF assays against microbial organisms commonly present in vaginal samples
| Microbial organism | Results for: |
|
|---|---|---|
| 16S rRNA-based assay | Cryptic plasmid-based assay | |
| Chlamydia trachomatis IU | + | + |
| Chlamydia trachomatis serovar E | + | + |
| Chlamydia trachomatis serovar L | + | + |
| Chlamydia pneumoniae T4 | + | − |
| Chlamydia pneumoniae AR39 | + | − |
| Chlamydia psittaci | − | − |
| Neisseria gonorrhoeae | − | − |
| Neisseria meningitidis | − | − |
| Trichomonas vaginalis | − | − |
| Mycoplasma genitalium | − | − |
| Herpes simplex virus 1 | − | − |
| Herpes simplex virus 2 | − | − |
| Haemophilus ducreyi | − | − |
| Staphylococcus epidermidis | − | − |
| Streptococcus agalactiae | − | − |
| Acinetobacter spp. | − | − |
| Pseudomonas aeruginosa | − | − |
Among the 260 swabs tested with the ProbeTec NAAT (Becton, Dickinson, Sparks MD), 42 were C. trachomatis positive and 218 were C. trachomatis negative. Additional testing of MAMEF assay-positive samples with a second NAAT (Aptima Combo 2; Hologic Gen-Probe, San Diego, CA) identified four additional C. trachomatis-positive swabs that were negative by the first NAAT. Overall, 46 swabs were considered C. trachomatis positive and 214 were C. trachomatis negative by either of the two NAATs (ProbeTec or Gen-Probe). The overall prevalence of STIs in this sample set was 17.7% for C. trachomatis, 14% for Trichomonas vaginalis, and 5.1% for Neisseria gonorrhoeae; 8.5% of samples were positive for two STIs. MAMEF assay results were available for 257 (98.8%) of the 260 swabs. Three samples were excluded from the analysis due to loss of sample during the lysing procedure. One of the excluded samples was C. trachomatis positive and two were C. trachomatis negative, resulting in 45 C. trachomatis positive swabs and 212 negative swabs. As shown in Table 2, of the 45 samples identified as C. trachomatis positive and 212 samples identified as C. trachomatis negative in NAATs, 33/45 and 197/212 samples were correctly identified by both MAMEF assays, i.e., 16S rRNA and C. trachomatis cryptic plasmid assays. The calculated clinical sensitivities and specificities of the two MAMEF assays required to be positive in comparison to NAATs were 73.3% (33/45 samples) (95% confidence interval [CI], 60.4 to 86.2%) and 92.9% (197/212 samples) (95% Cl, 89.8 to 96.0%), respectively. Eighteen percent of samples (35/197 samples) determined to be C. trachomatis negative in NAATs and MAMEF assays were positive for at least one STI. The 16S rRNA-based MAMEF assay (Table 2) had a sensitivity of 75.5% (34/45 samples) (95% Cl, 62.9 to 88.1%) and a specificity of 92.9% (197/212 samples) (95% Cl, 89.8 to 96.0%). The cryptic plasmid-based MAMEF assay had a sensitivity of 82.2% (37/45 samples) (95% Cl, 71.0 to 93.4%) and a specificity of 92.9% (197/212 samples) (95% Cl, 89.8 to 96.0%) (Table 2). The overall agreement of MAMEF assay results with NAAT results was 89.5% (95% Cl, 85.4 to 92.8%) for the 16S rRNA-based assay and 91.0% (95% Cl, 87.3 to 94.5%) for the plasmid-based assay. The total time to detection was <9 min, which included DNA extraction by microwave lysis (35 s), centrifugation (3 min), probe hybridization, and MAMEF detection by a human operator (5 min) (Fig. 2).
Table 2.
MAMEF assay results versus NAAT results
| MAMEF assay result | No. of samples with a NAAT (ProbeTec or Gen-Probe) result of: |
Total no. of samples | |
|---|---|---|---|
| Positive | Negative | ||
| Combination of 16S rRNA- and cryptic plasmid-based MAMEF assays | |||
| Positive | 33 | 15 | 48 |
| Negative | 12 | 197 | 209 |
| Total | 45 | 212 | 257 |
| 16S rRNA-based MAMEF assay | |||
| Positive | 34 | 15 | 49 |
| Negative | 11 | 197 | 208 |
| Total | 45 | 212 | 257 |
| Cryptic plasmid-based MAMEF assay | |||
| Positive | 37 | 15 | 52 |
| Negative | 8 | 197 | 205 |
| Total | 45 | 212 | 257 |
DISCUSSION
Although the use and acceptability of NAATs for detection of C. trachomatis have increased significantly over the past decade, the utility of NAATs as point-of-care tests in clinical settings is limited and they are cost-prohibitive in low-resource settings. The World Health Organization Sexually Transmitted Diseases Diagnostics Initiative has developed the ASSURED (affordable, sensitive, specific, user-friendly, rapid and robust, equipment-free, and deliverable to those in need) criteria as benchmarks to determine whether a diagnostic test addresses certain disease control needs (46). According to a qualitative study of focus group discussions with medical care providers, C. trachomatis was identified as the priority organism for the development of a new POC test and the ideal POC test for C. trachomatis detection should be accurate and rapid (<20 min) (47). A recent review of available POC tests for detection of STIs reported disappointing sensitivity for tests for Chlamydia (3). A recent cost-effectiveness study has demonstrated that implementation of POC tests meeting the ASSURED criteria can be cost-effective, compared with traditional NAATs (48). Additionally, a POC test with moderate sensitivity (65%) can help to treat more Chlamydia-positive cases than a NAAT alone when the rate of patients returning for results and treatment is lower than 65% (49).
We previously reported on the development of a 16S rRNA-based MAMEF assay for rapid detection of C. trachomatis (16). In the present study, we have developed an additional MAMEF assay for detection of the C. trachomatis cryptic plasmid and have shown that the MAMEF assays can detect as little as 10 IFU/ml of Chlamydia trachomatis. The combined performance of the two assays (16S rRNA and cryptic plasmid) was initially evaluated to determine the diagnostic utility of a dual-assay approach. Compared to NAATs, the sensitivity was moderate (73.3%) when both MAMEF assays were required to be positive. This sensitivity was similar to that of the 16S rRNA-based assay alone (75.5%). Compared to NAATs, the sensitivity of the cryptic plasmid-based assay was higher (82.2%) than that of the 16S rRNA assay alone or of both assays if required to determine positivity. The increased sensitivity of the cryptic plasmid assay in comparison to the 16S rRNA assay was attributed to additional quantities of cryptic plasmid DNA in C. trachomatis cells, in comparison to DNA from the 16S rRNA gene. It has been estimated that C. trachomatis has an average of only 2.1 copies of the 16S rRNA gene, which is less than the 10 copies of cryptic plasmid commonly present in C. trachomatis cells.
There were several cases of discordant MAMEF assay and NAAT results. Of the 257 samples, 27 had discordant MAMEF assay and NAAT results (Table 2). Twelve samples identified as Chlamydia positive by the ProbeTec assay were MAMEF assay negative by both the 16S rRNA assay and the cryptic plasmid assay. All 12 samples were confirmed as Chlamydia positive by the Gen-Probe assay. This equates to 12 missed positive results (false-negative results). There were a total of 11 false-negative results by the 16S rRNA MAMEF assay and 8 samples that were missed by the cryptic plasmid assay (Table 3). Fifteen samples identified by the ProbeTec assay as Chlamydia negative were positive by both the 16S rRNA and cryptic plasmid MAMEF assays. All 15 samples also tested negative with the Gen-Probe assay. This equates to 15 false-positive results (Table 2). The exact reason for these false-positive results is unknown. Cross-reactivity of the probes with other STIs is unlikely, as only 20% of the NAAT-negative/MAMEF assay-positive samples (3/15 samples) were positive for another STI. Additionally, no cross-reactivity with other STIs was noted in C. trachomatis-negative samples, as 18% (35/197 samples) of the NAAT/MAMEF assay-negative samples were positive for another STI.
Table 3.
Discordant NAAT-positive and MAMEF assay-negative results
| Sample no. | Test results for: |
Comment | |||
|---|---|---|---|---|---|
| NAATa | Combined assayb | 16S rRNA assay | Cryptic plasmid assay | ||
| POCC 4222 | + | − | − | − | Retested: cryptic plasmid assay-positive |
| POCC 4228 | + | − | − | − | |
| POCC H003 | + | − | − | − | |
| POCC H005 | + | − | − | − | |
| POCC H011 | + | − | − | + | |
| POCC H018 | + | − | − | − | |
| POCC H046 | + | − | + | − | |
| POCC H122 | + | − | − | + | |
| POCC H126 | + | − | − | + | |
| POCC H135 | + | − | − | − | Retested: cryptic plasmid assay-positive |
| POCC H137 | + | − | − | − | |
| POCC H154 | + | − | − | + | |
Gen-Probe or ProbeTec assay.
Combined results for the 16S rRNA- and cryptic plasmid-based MAMEF assays.
Several rapid tests for C. trachomatis detection have been developed and evaluated. The Clearview Chlamydia immunoassay test (Inverness, Princeton, NJ) has been primarily evaluated against culture, and lack of sensitivity has been reported repeatedly (48). The Chlamydia Rapid Test has shown promising results for the sensitive detection of C. trachomatis in 25 min (50). However, a recent evaluation study in Suriname found that the assay lacks sensitivity (51). Our MAMEF assays, especially the cryptic plasmid-based assay, have shown moderate to good sensitivity and acceptable specificity in a high-prevalence population.
One of the limitations of our study is the large number of false-positive samples. Unfortunately, we were unable to do further testing on these samples due to sample availability.
In this study, we successfully demonstrated that two C. trachomatis MAMEF assays had substantial agreement (κ = 64.6% and 70.8% for the 16S rRNA and plasmid assays, respectively) with NAATs for 257 vaginal swab samples. The new cryptic plasmid assay is more sensitive than the original C. trachomatis 16S rRNA assay (16). The total time for our assays was <9 min, with assays able to be run in parallel. Additionally, the cryptic plasmid-based MAMEF assay has the ability to detect the Swedish C. trachomatis variant, as the target sequence for the MAMEF probes is located outside the 377-bp deletion region (52). We have estimated that the cost of each assay is $1.00, with an additional $1.00 per lysing procedure. Consequently, our C. trachomatis MAMEF assay is a low-cost, rapid-turnaround, specific, sensitive test for C. trachomatis detection. While our current detection device (Fig. 6) is about the size of a shoebox, work is under way to reduce the size and cost of the reader, to enable the approach to provide additional benefits in low-resource settings. Additionally, we are currently working on developing a chip-based assay for the simultaneous lysis and detection of C. trachomatis DNA with a single platform, as well as the multiplex detection of multiple STIs. We are hopeful that further improvements to our assay platform will meet most of the ASSURED criteria when the platform comes to market.
Fig 6.
Outer configuration of the optical reader for the C. trachomatis MAMEF assays. The inner workings of the reader are shown in Fig. 4.
ACKNOWLEDGMENTS
We acknowledge the NIH for support (NIBIB grant U54 EB007958) and acknowledge salary support from the Institute of Fluorescence, the Department of Chemistry and Biochemistry, and the University of Maryland Baltimore County to both C.D.G. and J.H.M. Training support for J.H.M. also was provided by NIGMS grant R25 GM55036 and NIH grant T32 GM66706.
Footnotes
Published ahead of print 26 June 2013
REFERENCES
- 1.Centers for Disease Control and Prevention 2012. Sexually transmitted disease surveillance, 2011. CDC, Atlanta, GA [Google Scholar]
- 2.Centers for Disease Control and Prevention 2010. Sexually transmitted diseases treatment guidelines, 2010. MMWR Recomm. Rep. 59(RR-12):1–110 [PubMed] [Google Scholar]
- 3.Huppert J, Hesse E, Gaydos CA. 2010. What's the point? How point-of-care STI tests can impact infected patients. Point Care 9:36–46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.National Committee for Quality Assurance 2011. The state of health care quality 2011: HEDIS measures of care, p 77–78 National Committee for Quality Assurance, Washington, DC [Google Scholar]
- 5.Gaydos CA, Quinn TC, Willis D, Weissfeld A, Hook EW, Martin DH, Ferrero DV, Schachter J. 2003. Performance of the APTIMA Combo 2 assay for the multiplex detection of Chlamydia trachomatis and Neisseria gonorrhoeae in female urine and endocervical swab specimens. J. Clin. Microbiol. 41:304–309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gaydos CA, Cartwight CP, Colaninno P, Welsch J, Holden J, Ho SY, Webb EM, Anderson C, Bertuzis R, Zhang L, Miller T, Leckie G, Abravaya K, Robinson J. 2010. Performance of the Abbott Real-time CT/NG for the detection of Chlamydia trachomatis and Neisseria gonorrhoeae. J. Clin. Microbiol. 48:3236–3243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Taylor SN, Van Der Pol B, Lillis R, Hook EW, III, Lebar W, Davis T, Fuller D, Mena L, Fine P, Gaydos CA, Martin DH. 2011. Clinical evaluation of the BD ProbeTecTM Chlamydia trachomatis (CT) Qx amplified DNA assay on the BD ViperTM system with XTRTM technology. Sex. Transm. Dis. 38:603–609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Van Der Pol B, Liesenfeld O, Williams JA, Taylor SN, Lillis RA, Body BA, Nye M, Eisenhut C, Hook EW., III 2012. Performance of the Cobas CT/NG test compared to the Aptima AC2 and Viper CTQ/GCQ assays for detection of Chlamydia trachomatis and Neisseria gonorrhoeae. J. Clin. Microbiol. 50:2244–2249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hook EW, III, Spitters C, Reichart CA, Neumann TM, Quinn TC. 1994. Use of cell culture and a rapid diagnostic assay for Chlamydia trachomatis screening. JAMA 272:867–870 [PubMed] [Google Scholar]
- 10.Bachmann LH, Richley CM, Waites K, Schwebke JR, Hook EW., III 1999. Patterns of Chlamydia trachomatis testing and follow-up at a university hospital medical center. Sex. Transm. Dis. 26:496–499 [DOI] [PubMed] [Google Scholar]
- 11.Hook EW, III, Richey CM, Leone P, Bolan G, Spalding C, Henry K, Clarke P, Smith M, Celum CL. 1997. Delayed presentations to clinics for sexually transmitted diseases by symptomatic patients: a potential contributor to continuing STI morbidity. Sex. Transm. Dis. 24:443–448 [DOI] [PubMed] [Google Scholar]
- 12.Tennant SM, Zhang Y, Galen J, Geddes CD, Levine MM. 2011. Ultra-fast and sensitive detection of non-typhoidal Salmonella using microwave-accelerated metal-enhanced fluorescence (MAMEF). PLoS One 6:e18700. 10.1371/journal.pone.0018700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Aslan K, Previte MJR, Zhang Y, Gallagher T, Baillie L, Geddes CD. 2008. Extraction and detection of DNA from Bacillus anthracis spores and the vegetative cells within 1 minute. Anal. Chem. 80:4125–4132 [DOI] [PubMed] [Google Scholar]
- 14.Aslan K, Zhang Y, Hibbs S, Baillie L, Geddes CD. 2007. Microwave-accelerated metal-enhanced fluorescence: application to detection of genomic and exosporium anthrax DNA in <30 seconds. Analyst 132:1130–1138 [DOI] [PubMed] [Google Scholar]
- 15.Aslan K, Baillie L, Geddes CD. 2010. Ultra-fast and sensitive detection of biological threat agents using microwaves, nanoparticles and luminescence. J. Med. CBR Def. 8:1–21 [Google Scholar]
- 16.Zhang Y, Agreda P, Kelly S, Gaydos C, Geddes CD. 2011. Development of a microwave-accelerated metal-enhanced fluorescence 40 seconds, <100 cfu/ml point-of-care assay for the detection of Chlamydia trachomatis. IEEE Trans. Biomed. Eng. 58:781–784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Aslan K, Geddes CD. 2008. New tools for rapid clinical and bioagent diagnostics: microwaves and plasmonic nanostructures. Analyst 133:1469–1480 [DOI] [PubMed] [Google Scholar]
- 18.Aslan K, Geddes CD. 2008. A review of an ultra-fast and sensitive bioassay platform technology: microwave-accelerated metal-enhanced fluorescence. Plasmonics 3:89–101 [Google Scholar]
- 19.Aslan K, Previte MJR, Zhang Y, Geddes CD. 2008. Microwave-Accelerated Surface Plasmon-Coupled Directional Luminescence 2: a platform technology for ultra-fast and sensitive target DNA detection in whole blood. J. Immunol. Methods 331:103–113 [DOI] [PubMed] [Google Scholar]
- 20.Aslan K, Malyn SN, Bector G, Geddes CD. 2007. Microwave-accelerated metal-enhanced fluorescence: an ultra-fast and sensitive DNA sensing platform. Analyst 132:1122–1129 [DOI] [PubMed] [Google Scholar]
- 21.Aslan K, Malyn SN. 2007. Microwave-accelerated surface plasmon-coupled directional luminescence: application to fast and sensitive assays in buffer, human serum and whole blood. J. Immunol. Methods 323:55–64 [DOI] [PubMed] [Google Scholar]
- 22.Aslan K, Geddes CD. 2007. Microwave-accelerated ultra-fast nanoparticle aggregation assays using gold nanoparticles. Anal. Chem. 79:2131–2136 [DOI] [PubMed] [Google Scholar]
- 23.Aslan K, Previte MJR, Zhang Y, Geddes CD. 2007. Microwave-accelerated plasmonics: application to ultra-fast and ultra-sensitive clinical assays. Proc. SPIE 6450:645007. 10.1117/12.699159 [DOI] [Google Scholar]
- 24.Aslan K, Holley P, Geddes CD. 2006. Microwave-accelerated metal-enhanced fluorescence (MAMEF) with silver colloids in 96-well plates: application to ultra-fast and sensitive immunoassays, high throughput screening and drug discovery. J. Immunol. Methods 312:137–147 [DOI] [PubMed] [Google Scholar]
- 25.Aslan K, Geddes CD. 2006. Microwave-accelerated and metal-enhanced fluorescence myoglobin detection on silvered surfaces: potential application to myocardial infarction diagnosis. Plasmonics 1:53–59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Aslan K, Geddes CD. 2006. Microwave-accelerated metal-enhanced fluorescence (MAMEF): application to ultra-fast and sensitive clinical assays. J. Fluoresc. 16:3–8 [DOI] [PubMed] [Google Scholar]
- 27.Aslan K, Geddes CD. 2005. Microwave-accelerated metal-enhanced fluorescence (MAMEF): a new platform technology for ultra-fast and ultra-bright assays. Anal. Chem. 77:8057–8067 [DOI] [PubMed] [Google Scholar]
- 28.Dragan AI, Bishop ES, Casas-Finet JR, Strouse RJ, Schenerman MA, Geddes CD. 2010. Metal-enhanced picogreen fluorescence: application to fast and ultra-sensitive pg/ml DNA quantitation. J. Immunol. Methods 362:95–100 [DOI] [PubMed] [Google Scholar]
- 29.Zhang Y, Mandeng LN, Bondre N, Dragan A, Geddes CD. 2010. Metal-enhanced fluorescence from silver-SiO2 nanoburger structures. Langmuir 26:12371–12376 [DOI] [PubMed] [Google Scholar]
- 30.Dragan A, Bishop ES, Casas-Finet JR, Strouse RJ, Schenerman MA, Geddes CD. 2010. Metal-enhanced picogreen fluorescence: application for dsDNA quantification. Anal. Biochem. 396:8–12 [DOI] [PubMed] [Google Scholar]
- 31.Pribik R, Dragan A, Zhang Y, Gaydos C, Geddes CD. 2009. Metal-enhanced fluorescence (MEF): physical characterization of silver-island films and exploring sample geometries. Chem. Phys. Lett. 478:70–74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhang Y, Dragan A, Geddes CD. 2009. Wavelength dependence of metal-enhanced fluorescence. J. Phys. Chem. C 113:12095–12100 [Google Scholar]
- 33.Aslan K, Previte MJR, Zhang Y, Geddes CD. 2008. Metal-enhanced fluorescence from nanoparticulate zinc films. J. Phys. Chem. C 112:18368–18375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Aslan K, Malyn SN, Zhang Y, Geddes CD. 2008. Conversion of just-continuous metallic films to large particulate substrates for metal-enhanced fluorescence. J. Appl. Phys. 103:84307–843077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhang Y, Aslan K, Previte MJR, Geddes CD. 2007. Metal-enhanced fluorescence from copper substrates. Appl. Phys. Lett. 90:173116 [Google Scholar]
- 36.Aslan K, Huang J, Wilson GM, Geddes CD. 2006. Metal-enhanced fluorescence-based RNA sensing. J. Am. Chem. Soc. 128:4206–4207 [DOI] [PubMed] [Google Scholar]
- 37.Dragan AI, Mali B, Geddes CD. 2013. Wavelength-dependent metal-enhanced fluorescence using synchronous spectral analysis. Chem. Phys. Lett. 556:168–172 [Google Scholar]
- 38.Abugo OO, Herman P, Lakowicz JR. 2001. Fluorescence properties of albumin blue 633 and 670 in plasma and whole blood. J. Biomed. Opt. 6:359–365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Deng T, Li JS, Zhang LL, Jiang JH, Chen JN, Shen GL, Yu RQ. 2010. A sensitive fluorescence anisotropy method for the direct detection of cancer cells in whole blood based on aptamer-conjugated near-infrared fluorescent nanoparticles. Biosens. Bioelectron. 25:1587–1591 [DOI] [PubMed] [Google Scholar]
- 40.Foy CA, Parkers HC. 2001. Emerging homogeneous DNA-based technologies in the clinical laboratory. Clin. Chem. 47:990–1000 [PubMed] [Google Scholar]
- 41.Mulcahy G. 1999. The integration of molecular diagnostic methods into the clinical laboratory. Ann. Clin. Lab. Sci. 29:43–54 [PubMed] [Google Scholar]
- 42.Saunders G, Parkers H. 1999. Analytical molecular biology: quality and validation. RSC Publishing, Cambridge, United Kingdom [Google Scholar]
- 43.Mahalanabis M, Al-Muayad H, Kulinski MD, Altman D, Klapperich CM. 2009. Cell lysis and DNA extraction of Gram-positive and Gram-negative bacteria from whole blood in a disposable microfluidic chip. Lab Chip 9:2811–2817 [DOI] [PubMed] [Google Scholar]
- 44.Listwan P, Pedelacq JD, Lockard M, Bell C, Terwilliger TC, Waldo GS. 2010. The optimization of in vitro high-throughput chemical lysis of Escherichia coli: application to ACP domain of the polypeptide synthase ppsC from Mycobacterium tuberculosis. J. Struct. Funct. Genomics 11:41–49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bahamonde ME. 2009. Bacterial cell breakage or lysis, p 191–198 In Goldman E, Green LH. (ed), Practical handbook of microbiology, 2nd ed. CRC Press, Boca Raton, FL [Google Scholar]
- 46.Peeling RW, Holmes KK, Mabey D, Ronald A. 2006. Rapid tests for sexually transmitted infections (STIs): the way forward. Sex. Transm. Infect. 82(Suppl 5):V1–V6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hsieh YH, Gaydos CA, Hogan MT, Uy OY, Jackman J, Jett-Goheen M, Albertie A, Dangerfield DT, Neustadt CR, Weiner ZS, Rompalo AM. 2011. What qualities are most important to making a point of care test desirable for clinicians and others offering sexually transmitted infection testing? PLoS One 6:e19263. 10.1371/journal.pone.0019263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Huang W, Gaydos CA, Barnes MR, Jett-Goheen M, Blake DR. 2013. Comparative effectiveness of a rapid point-of-care test for detection of Chlamydia trachomatis among women in a clinical setting. Sex. Transm. Infect. 89:108–114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gift TL, Pate MS, Hook EW, III, Kassler WJ. 1999. The rapid test paradox: when fewer cases detected lead to more cases treated: a decision analysis of tests for Chlamydia trachomatis. Sex. Transm. Dis. 26:232–240 [DOI] [PubMed] [Google Scholar]
- 50.Mahilum-Tapay L, Laitila V, Wawrzyniak JJ, Lee HH, Alexander S, Ison C, Swain A, Barber P, Ushiro-Lumb I, Goh BT. 2007. New point of care Chlamydia rapid test—bridging the gap between diagnosis and treatment: performance evaluation study. BMJ 335:1190–1194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.van der Helm JJ, Sabajo LO, Grunberg AW, Morré SA, Speksnijder AG, de Vries HJ. 2012. Point-of-care test for detection of urogenital Chlamydia in women shows low sensitivity: a performance evaluation study in two clinics in Suriname. PLoS One 7:e32122. 10.1371/journal.pone.0032122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ripa T, Nilsson P. 2006. A variant of Chlamydia trachomatis with deletion in cryptic plasmid: implications for use of PCR diagnostic tests. Euro Surveill. 11(45):pii=3076 http://www.eurosurveillance.org/ViewArticle.aspx?ArticleId=3076 [DOI] [PubMed] [Google Scholar]