Abstract
Aggregatibacter actinomycetemcomitans-induced localized aggressive periodontitis (LAP) in African-American adolescents has been documented but is poorly understood. Two thousand fifty-eight adolescents aged 11 to 17 years were screened for their periodontal status and the presence of A. actinomycetemcomitans in their oral cavity. Seventy-one A. actinomycetemcomitans-negative and 63 A. actinomycetemcomitans-positive periodontally healthy subjects were enrolled, sampled, examined, and radiographed yearly for 3 years. Gingival and periodontal pocket depth and attachment levels were recorded. Disease presentation was characterized by bone loss (BL). Subgingival sites were sampled every 6 months to assess (i) the role of A. actinomycetemcomitans in BL and (ii) the association of A. actinomycetemcomitans and other microbes in their relationships to BL. Sixteen of 63 subjects with A. actinomycetemcomitans developed BL (the other 47 subjects with A. actinomycetemcomitans had no BL). No A. actinomycetemcomitans-negative subjects developed BL. Human oral microbe identification microarray (HOMIM) was used for subgingival microbial assessment. On a subject level, pooled data from A. actinomycetemcomitans-positive subjects who remained healthy had higher prevalences of Streptococcus and Actinomyces species, while A. actinomycetemcomitans-positive subjects with BL had higher prevalences of Parvimonas micra, Filifactor alocis, A. actinomycetemcomitans, and Peptostreptococcus sp. human oral taxon 113 (HOT-113). At vulnerable sites, A. actinomycetemcomitans, Streptococcus parasanguinis, and F. alocis levels were elevated prior to BL. In cases where the three-organism consortium (versus A. actinomycetemcomitans alone) was detected, the specificity for detecting sites of future BL increased from 62% to 99%, with a sensitivity of 89%. We conclude that detecting the presence of A. actinomycetemcomitans, S. parasanguinis, and F. alocis together indicates sites of future BL in LAP. A synergistic interaction of this consortium in LAP causation is possible and is the subject of ongoing research.
INTRODUCTION
The relationship between Aggregatibacter actinomycetemcomitans and localized aggressive periodontitis (LAP) is well established (1–5). Initially, these linkages were ascertained by identifying A. actinomycetemcomitans in patients who presented to the clinic with a diagnosis of LAP (6, 7). These young adults harbored A. actinomycetemcomitans in their diseased gingival and periodontal pocket sites, while A. actinomycetemcomitans was not found in other clinic patients (8). Subsequently, it was shown that A. actinomycetemcomitans produced a virulence factor, a leukotoxin, which dampens the host response, thus providing a biologically plausible explanation for A. actinomycetemcomitans-related pathogenesis of LAP (9, 10). Other studies showed that the removal of A. actinomycetemcomitans from sites undergoing disease progression led to disease remission, adding further evidence to this association (11–14). Additional virulence attributes were discovered, and more clinical and animal data showing this relationship were collected (15, 16). More recently, case-controlled longitudinal studies have indicated that healthy subjects with A. actinomycetemcomitans develop disease, while those devoid of A. actinomycetemcomitans remain healthy (5, 14, 17, 18). The accumulating evidence has been interpreted to suggest that A. actinomycetemcomitans is causal and required to initiate LAP, which occurs with a higher frequency in children of African or Hispanic descent (18, 19).
Cross-sectional data derived from our current study indicate that >70% of those who have A. actinomycetemcomitans in their oral cavities at screening do not have LAP (5). In agreement with this finding, the longitudinal phase of this study indicates that only 25% of those who initially had A. actinomycetemcomitans developed LAP over time (5). Overall, data from both the cross-sectional and longitudinal phases of this current study raise questions as to why some individuals with A. actinomycetemcomitans develop disease while many individuals with A. actinomycetemcomitans remain healthy.
The critical data derived from the longitudinal cohort phase of this study investigated A. actinomycetemcomitans-induced LAP. The study design permitted us to distinguish those who are periodontally healthy, have A. actinomycetemcomitans, and remain healthy from those who are periodontally healthy, have A. actinomycetemcomitans, and develop disease over time. In order to recruit and retain a sufficient number of A. actinomycetemcomitans-positive subjects and matched A. actinomycetemcomitans-negative controls for this 2- to 3-year period, we screened 2,058 students for their periodontal status and the presence or absence of A. actinomycetemcomitans. Periodontally healthy A. actinomycetemcomitans carriers and A. actinomycetemcomitans-free subjects were then divided into two subgroups and enrolled into the longitudinal study. These subjects were followed every 6 months for 2 to 3 years. In the process, subgingival plaque and crevicular fluid samples from all first molar sites were collected, stored, and analyzed retrospectively when bone loss was detected in any subject enrolled in the study. Over 300 microbial taxa were analyzed using 16S rRNA human oral microbe identification microarray (HOMIM) technology (20, 21).
The microbiological portion of the study has two intertwined goals: (i) to determine the role of A. actinomycetemcomitans in the development of LAP, and (ii) to understand the link between A. actinomycetemcomitans and other members of the subgingival flora that might be involved in the development of LAP. This longitudinal model clearly demonstrates that microbiological data can discriminate between sites that remain healthy and sites that develop disease within the same subject, and that individual sites show well-defined microbiological differences before and after breakdown.
This is the first study to show that a consortium of three microbes that includes A. actinomycetemcomitans, Streptococcus parasanguinis, and Filifactor alocis is present in the advancement of disease at subgingival sites that develop disease and suggests that A. actinomycetemcomitans is necessary but insufficient by itself to be linked to the development of LAP. Further, our data indicate that A. actinomycetemcomitans can form a critical partnership with S. parasanguinis and F. alocis prior to disease development.
MATERIALS AND METHODS
Study approval was obtained from the institutional review board of the University of Medicine and Dentistry of New Jersey. Both assent and consent were received from each student's parent(s) or guardian(s) prior to participation. The initial screening visit was performed in a mobile dental van and consisted of a full oral and dental examination, review of medical and dental histories, and collection of saliva and buccal epithelial cells (BECs) from each subject as described previously (5, 22). Subjects were excluded if they required prophylactic antibiotics, had any bleeding disorder, were otherwise medically vulnerable, or if they had extensive dental caries as defined by decayed, missing, or filled surfaces (23).
Clinical parameters.
After collecting buccal cells and saliva, a full periodontal examination was performed, and plaque and crevice fluid were collected from each of the four first molars (5, 22). The periodontal examination consisted of soft tissue measurements of pocket depth and attachment-level recordings. Pockets were measured at six sites per tooth and for all teeth present. Any site with a pocket of ≥4 mm was reexamined for the clinical attachment level (CAL). As a result of the epidemiological nature of the study, all pockets were measured, but only pockets of ≥5 mm were recorded. Potential disease was defined as a pocket of ≥6 mm with an associated CAL loss of ≥2 mm. Diagnosis of disease was determined by radiographic evidence of bone loss as opposed to soft tissue measurement. This decision was based on data indicating that soft tissue measurements in this mixed-dentition age group might produce shifting landmarks, while radiographs could provide a more stable basis for diagnosis (24, 25). Standardized horizontal bitewing radiographs of the molar region were taken within 1 month of the screening visit. The detection of vertical crestal bone loss of the lamina dura and the bone directly beneath the dura fit the diagnosis of bone loss. A calibration exercise was conducted for pocket probing, CAL measurement, and radiographic measurement of bone loss as reported (5).
Sample collection. (i) Saliva for analysis of host factors.
Up to 5 ml of saliva was collected by having each student expectorate into a 50-ml-wide mouthed polypropylene tube held over ice. Saliva samples were divided into aliquots and then stored at 20°C for future analysis of host factors.
(ii) Buccal collections for identification of A. actinomycetemcomitans status at screening.
Buccal epithelial cell (BEC) scrapings were obtained by means of a wooden tongue depressor, and the sloughed BECs obtained were placed into 2 ml of phosphate-buffered saline (PBS) for classification of each student's A. actinomycetemcomitans status. After vigorous vortex agitation, a 100-μl aliquot was removed from BECs for plating on A. actinomycetemcomitans growth medium (AAGM) agar (Sigma, St. Louis, MO) consisting of Trypticase soy broth supplemented with 0.4% sodium bicarbonate and bacitracin (5 μg/ml) and vancomycin (75 μg/ml) in agar used for the detection and enumeration of A. actinomycetemcomitans (5). The remaining BECs were stored at −80°C. Cultural identification of A. actinomycetemcomitans was confirmed by PCR (See “Microbiological methods”).
(iii) Plaque and gingival crevicular fluid collection.
After gentle supragingival debridement, subgingival plaque and crevicular fluid were collected from all (healthy or pocketed) first molar sites by paper point. Plaque and crevicular fluid were given a coded number, labeled by individual and site, and stored frozen at −80°C for retrospective analysis. All plaque samples were collected and stored, but HOMIM analysis was only performed with subjects who were entered into the longitudinal study. Crevicular fluid was handled in a similar manner. However, after molar sampling for HOMIM analysis, teeth other than first molars with pockets of >5 mm were sampled for plating on selective agar for A. actinomycetemcomitans (5) to determine the relationship of A. actinomycetemcomitans to these pocket sites.
(iv) Recall visit.
Students who had pockets of ≤5 mm with no CAL loss were deemed healthy and were considered for enrollment in the longitudinal study. Subjects who had no bone loss after an X-ray evaluation were recalled within 6 months of the baseline X-ray for a repeat of the initial visit, consisting of oral and dental examinations, collection of samples, periodontal examination, and X rays. A. actinomycetemcomitans-positive subjects were entered into the study first and were matched for age, gender, and race with A. actinomycetemcomitans-negative students. Both groups were treated in an identical manner. Examiners were blinded as to the A. actinomycetemcomitans status of the subjects. Buccal scrapings, saliva, and molar samples were obtained and treated as described previously. The initial, recall, and additional recall visits all were used for sample collection and clinical evaluation. To prevent any consequences of aggressive disease, any subject seen at recall with a pocket of ≥6 mm was recalled within a 3-month period to determine pocket depth, CAL changes, and radiographic evidence of bone loss (5).
(v) Exit strategy.
Any student with X-ray evidence of vertical bone loss and loss of lamina dura or of bone adjacent to the dura was considered to have disease, exited from the study, and offered treatment at no expense (5).
Microbiological methods. (i) Determination of A. actinomycetemcomitans status of subjects.
A 100-μl aliquot of the resuspended BEC sample was streaked on AAGM agar as described above, and plates were incubated for 3 to 4 days at 37°C with 10% CO2 (26). Colonies with the star-positive morphology of A. actinomycetemcomitans that were catalase positive were subcultured and confirmed to be A. actinomycetemcomitans by PCR using specific primers for A. actinomycetemcomitans leukotoxin promoter DNA (27). Briefly, DNA was obtained using the DNeasy tissue kit (Qiagen, Inc., Valencia, CA) for Gram-negative bacteria (5). Classification of subjects was based on the presence or absence of A. actinomycetemcomitans. In addition, subgingival samples collected from sites other than first molars that were ≥5 mm on probing (as described above) were also plated for positive identification of A. actinomycetemcomitans with PCR confirmation. PCR was also was used to determine if individuals were A. actinomycetemcomitans negative, as described below. If no growth occurred after plating BECs on selective AAGM agar, DNA was extracted from BEC or plaque samples using the Gram-negative protocol described above. In this procedure, we again used PCR with specific primers for A. actinomycetemcomitans leukotoxin promoter DNA for A. actinomycetemcomitans identification. This PCR procedure was done a minimum of two times. If the subject was both culture and PCR negative for A. actinomycetemcomitans DNA from both BEC and pocket sites, we classified the subject as A. actinomycetemcomitans negative (5). All samples were given a coded number to maintain blinding during the laboratory phases of the study.
(ii) Determination of microbial communities.
Subgingival samples taken by paper point were stored frozen at −80°C until needed. As mentioned, samples for HOMIM analysis were derived from subjects who participated in the longitudinal study. First, paper point samples were eluted in 180 μl of ATL buffer (Qiagen DNeasy tissue kit, Gram-negative bacterial protocol) at 37°C for 60 min before proceeding with the rest of the extraction. DNA was isolated in 400 μl of kit elution buffer (5). Ten microliters of the solution was then run on a 0.8% agarose gel electrophoresis to check for concentration (using a standardized lambda DNA-HindIII ladder) for both DNA quantity and quality. The extracted DNA solution was lyophilized, placed into a tube with a coded number, and sent to the Forsyth Institute for HOMIM analysis (28).
(iii) Human oral microbe identification microarray analyses.
Microbial profiles were generated from image files of scanned HOMIM microarrays (http://biofinformatics.forsyth.org/homim/). In brief, concentration levels of approximately 300 oral taxa were determined by microarray hybridization using a fluorescent readout reverse-capture method (29). Fluorescently labeled sample microbial DNA was captured by 16S rRNA-based probes attached to glass slides. The fluorescent intensity for each probe was scanned, normalized, and scaled as previously reported (29). Signals of <2× background were considered to be negative and assigned a HOMIM level score of 0. Positive hybridization signals were categorized into 5 levels, with 1 indicating a signal that was just detectable, to 5 indicating maximum signal intensity.
Data Analysis. Clinical data.
Cross-sectional analysis was performed on all 2,058 subjects who were screened. Demographic assessment was descriptive in nature and presented as number of subjects in the A. actinomycetemcomitans-positive versus the A. actinomycetemcomitans-negative group; subjects were divided into groups by age at time of screening visit, gender, and ethnic origin. Subjects were considered periodontally healthy if they had pockets that were ≤5 mm. Only sites of ≥5 mm were recorded. Subjects with pockets of ≥6 mm with CAL loss of ≥2 mm at one site or more were considered to have periodontal disease. Subjects with ≥2 sites with pockets of ≥6 mm and CAL loss of ≥2 mm were considered to have the Löe-Brown definition of periodontal disease (30). Pocket depths of ≥6 mm were calculated by ethnic group, as was CAL loss of ≥2 mm. Differences between the African-American subjects and Hispanic subjects with respect to the age of individuals with disease were determined. The mean ages and standard deviations for subjects with disease were calculated, and Student's t test was used to determine statistical differences in ages when groups (African-American versus Hispanic) were compared. Bone loss was recorded as present or absent and evaluated by ethnic group (African-American versus Hispanic), and the age of detection was compared in the two groups. In addition, a chi-square analysis was performed to determine the differences in the number of subjects with one or two sites of ≥6 mm and ≥2 mm of CAL loss, and comparing the Hispanic and African-American students with respect to all clinical parameters.
Analysis of longitudinal data was performed on the 134 periodontally healthy subjects who were followed for >2 years. Subjects were divided into an A. actinomycetemcomitans-negative group consisting of 71 subjects and an A. actinomycetemcomitans-positive group consisting of 63 subjects. The demographic data presented are descriptive in nature and are divided by group and presented as described for the cross-sectional portion of the study. Presence and degrees of pockets, CAL, and bone loss were recorded and reported by group and analyzed as described for the cross-sectional study (see above).
A. actinomycetemcomitans microbiological data analysis for longitudinal study.
Periodontal disease (as measured by pockets, CAL, and bone loss) was related to A. actinomycetemcomitans status at the time of screening, and statistical differences were determined. The numbers of African-American subjects with either one pocketed site, two or more pocketed sites, or bone loss were compared to those of the Hispanic subjects. Significant differences between these two groups in each category were determined by chi-square analysis and Fisher's exact test with a P value of < 0.05 as the level of significance. A. actinomycetemcomitans serotype analysis was performed in a similar manner, comparing the two groups of (African-American and Hispanic) subjects.
HOMIM data for longitudinal study.
The initial analysis was performed on 17 of the subjects, which included seven subjects in the periodontally diseased category and five subjects in each of the groups of those who remained healthy. A. actinomycetemcomitans-positive and A. actinomycetemcomitans-negative healthy subjects were matched to subjects with disease by ethnicity and age. For subjects in each group (A. actinomycetemcomitans-negative healthy combined with A. actinomycetemcomitans-positive healthy and subjects with periodontal disease), each pocket sample taken from each first molar site (typically 4 per subject) was evaluated for the presence or absence of each of the 300 taxa, as well as the level of each taxon by the HOMIM. All site HOMIM values were determined individually and then summed and averaged. The prevalence or percentage of individuals of the 17 studied who harbored each taxon was determined and then the average HOMIM levels were calculated for each of the most prevalent species. The species were ranked in terms of prevalence and HOMIM level of organisms. This first analysis allowed us to determine the most-prevalent organisms and their levels when both health and disease were combined.
In a second analysis, samples from the healthy group (both A. actinomycetemcomitans positive and A. actinomycetemcomitans negative) and the diseased group (those subjects who went on to develop at least one bone loss site) were analyzed separately; after analysis, all data were pooled for each group (healthy versus diseased) and groups were compared. In the prevalence calculations, organisms were ordered to show which organisms demonstrated the largest increases or decreases in prevalence when comparing the healthy group to the diseased group. HOMIM levels were also calculated for each group, and the average HOMIM score ± the standard deviation was calculated by summing all sites within a specific group. These calculations enabled us to demonstrate differences in HOMIM scores between the healthy group and the diseased group. The data were considered preliminary and descriptive because the data were pooled and site-related data were not presented independently. In the HOMIM analyses described above (initial and secondary), data were derived from samples obtained at one time point (6 months prior to bone loss) and were therefore not longitudinal in nature, although the data were derived from the longitudinal study.
Site-specific data.
For site-specific data, we focused on 16 subjects who developed disease and compared healthy sites that remained healthy to those 18 sites in the 16 subjects who developed bone loss. For each site, both the presence and HOMIM level of each HOMIM species was determined. For comparing groups of subjects, changes in terms of prevalence (chi-square) and HOMIM level (analysis of variance [ANOVA] and Student's t test) were differentiated for all organisms assessed. Of primary interest were organisms that demonstrated significantly different prevalences and HOMIM probe levels when the groups were compared. Here, comparison was made between (i) healthy sites in LAP subjects that never broke down, and (ii) healthy sites in LAP subjects that broke down prior to breakdown at that site. Comparisons were also made between HOMIM probe prevalence and levels in healthy sites from healthy subjects at visits that occurred in parallel with LAP subjects at times before and after breakdown.
Sites that developed bone loss.
For the group of subjects with sites that showed bone loss as mentioned above, we determined the most prevalent organisms that were significantly elevated prior to breakdown compared to levels at the three other first molar sites that remained healthy in the same subject who developed LAP (at healthy sites that remained healthy at times prior to and at the time of breakdown in the LAP subjects). In these calculations, the most prevalent microbes related to bone loss (BL) were noted, and specificity and sensitivity testing were done for each microbe in the list of the most prevalent microbes. Ultimately, the 3 most prevalent organisms and their relationships to specificity and sensitivity were calculated. Sensitivity and specificity calculations were made using general estimating equations with robust variance estimation in order to account for the clustering of sites within each patient.
RESULTS
Cross-sectional data.
Two thousand fifty-eight students were screened. Of those screened, 910 were Hispanic and 924 were African-American, together representing 89% of the screened population (Table 1). The demographics of the students, including mean age and A. actinomycetemcomitans carriage, are shown in Table 1. One thousand ten subjects were between the ages of 11 and 14 years and another 705 were between the ages of 15 and 16 years. One thousand, two hundred twenty-four were female and 824 were male. Eleven percent or 227 subjects were A. actinomycetemcomitans positive at screening. As for serotypes of the A. actinomycetemcomitans strains 170 strains were of the b or c serotypes and 46 strains were of serotype a (Table 2). While the serotypes were equally distributed between b and c in African-American subjects, the Hispanic population had a predominance of serotype c (P < 0.05). Only 9 A. actinomycetemcomitans strains of the b serotype had the JP2-like promoter region (high leukotoxin-producing type), while the rest had the 652 promoter type (data not shown). The disease characteristics of the screened population are shown in Table 3. About 3% of those screened had at least one pocket of ≥6 mm with CAL loss of ≥2 mm. Sixty-seven percent of those with one diseased site had A. actinomycetemcomitans present at screening. Using the Löe-Brown definition of disease, which requires ≥2 sites with ≥6 mm or pocketing and ≥2 mm in CAL loss, 1.3% (27 subjects) had disease (30). Of those 27 with ≥2 diseased sites, 18 subjects were of African-American descent, while 6 were of Hispanic descent. Eighty-one percent of these individuals had A. actinomycetemcomitans. Using bone loss to define disease, 12 subjects (0.6%) had bone loss, with seven being African-American and five being Hispanic. A. actinomycetemcomitans was detected in 11 of these 12 (91.7%) subjects.
Table 1.
Screened population demographics
| Population group | No. (%) of subjectsb |
Age (mean ±SD) (yr)a |
No. (%) of A. actinomycetemcomitans-positive subjectsc |
|||
|---|---|---|---|---|---|---|
| Female | Male | Female | Male | Female | Male | |
| Ethnic group | ||||||
| African-American | 584 (47) | 340 (41) | 12.9 ± 1.7 | 12.6 ± 1.6 | 65 (11.1) | 44 (12.9) |
| American Indian or Alaskan Native | 0 | 4 | 11.8 ± 0.8 | 0 | 0 | |
| Asian or Pacific Islander | 28 | 29 | 14.2 ± 1.8 | 13.5 ± 2.1 | 4 | 2 |
| Caucasian | 28 | 18 | 13.8 ± 1.9 | 13.4 ± 1.4 | 1 | 0 |
| Hispanic | 519 (42) | 391 (47) | 12.5 ± 1.6 | 12.6 ± 1.6 | 54 (10.4) | 47 (12.0) |
| Not reported | 46 | 23 | 12.6 ± 1.5 | 12.8 ± 1.8 | 7 | 3 |
| Other | 29 | 19 | 13.2 ± 2.0 | 12.6 ± 1.8 | 0 | 0 |
| Male/female, total | 1,234 | 824 | 12.7 ± 1.7 | 12.7 ± 1.7 | 131 (10.6) | 96 (11.7) |
SD, standard deviation.
Combined total of 2,058 subjects.
Combined total of 227 A. actinomycetemcomitans-positive subjects.
Table 2.
A. actinomycetemcomitans serotypes detected at screening
| Ethnicity (n) | No. of subjects with A. actinomycetemcomitans serotype: |
|||||
|---|---|---|---|---|---|---|
| a | b | C | d | f | Nontypeable | |
| African-American (109) | 27 | 40 | 37 | 0 | 1 | 4 |
| Hispanic (101) | 15 | 32 | 53a | 1 | 0 | 0 |
| Asian/Caucasian (7) | 4 | 1 | 1 | 1 | 0 | 0 |
| Other (10) | 1 | 3 | 4 | 0 | 0 | 2 |
| Total (227) | 47 | 76 | 95 | 2 | 1 | 6 |
Significantly higher frequency of the c serotype within the Hispanic group when c is compared to either a or b serotypes by chi-square (P < 0.01). No other comparisons between or within groups were significant.
Table 3.
Screened population disease characterization
| Population group | ≥1 sitea |
≥2 sitesa,c |
Bone lossb |
|||
|---|---|---|---|---|---|---|
| No. of subjects (no. A. actinomycetemcomitans positive) | Age (mean ± SD) (yr) | No. of subjects (no. A. actinomycetemcomitans positive) | Age (mean ± SD) (yr) | No. of subjects (no. A. actinomycetemcomitans positive) | Age (mean ± SD) (yr) | |
| Ethnic group | ||||||
| Hispanic | ||||||
| Female | 14 (8) | 14.9 ± 2.1 | 3 (2) | 14.0 ± 0.0 | 2 (2) | 15.6 ± 0.7 |
| Male | 17 (9) | 15.9 ± 1.8 | 3 (2) | 14.3 ± 0.6 | 3 (2) | 14.5 ± 1.4 |
| Total | 31 (17) | 15.4 ± 1.9 | 6 (4) | 14.2 + 0.3 | 5 (4) | 14.9 ± 1.2 |
| African-American | ||||||
| Female | 18 (14) | 16.2 ± 1.8 | 11 (9) | 16.5 ± 1.1 | 4 (4) | 15.5 ± 1.5 |
| Male | 14 (11) | 15.8 ± 1.6 | 7 (6) | 15.4 ± 1.1 | 3 (3) | 14.7 ± 1.3 |
| Total | 32 (25) | 16.0 ± 1.7 | 18 (15) | 16.1 ± 1.2 | 7 (7) | 14.9 ± 1.5 |
| Asian | ||||||
| Female | 0 | 0.0 | 0 | 0.0 | 0 | |
| Male | 1 (1) | 18.0 | 1 (1) | 18.0 | 0 | |
| Total | 1 (1) | 18.0 | 1 (1) | 18.0 | 0 | |
| Not reported | ||||||
| Female | 0 | 1 (1) | 16.0 | 0 | ||
| Male | 0 | 1 (1) | 15.0 | 0 | ||
| Total | 0 | 2 (2) | 15.5 ± 0.7 | 0 | ||
| All ethnic groups combined | ||||||
| Female | 32 (22) | 15.6 ± 2.0 | 15 (12) | 15.8 ± 1.4 | 6 (6) | 15.2 ± 1.2 |
| Male | 32 (21) | 15.9 ± 1.8 | 12 (10) | 15.3 ± 1.3 | 6 (5) | 14.6 ± 1.5 |
| Total | 64 | 15.8 ± 1.9 | 27 | 15.6 ± 1.3 | 12 | 14.9 ± 1.3 |
| No. A. actinomycetemcomitans positive (%) | 43 (67.2) | 22 (81.2) | 11 (91.7) | |||
Site refers to a first molar pocket of ≥6 mm with attachment loss of ≥2 mm detected by probing.
Bone loss refers to radiographic evidence of vertical bone loss in a first molar interproximal area.
There was a significantly greater number of African-American subjects having two or more sites with attachment loss compared to Hispanic subjects (chi-square, P < 0.05).
Longitudinal study of 134 initially periodontally healthy students who completed the study.
The demographics and disease characteristics of longitudinal study subjects are shown in Table 4. Of the 134 subjects enrolled, 63 were A. actinomycetemcomitans positive and 71 were A. actinomycetemcomitans negative. Of the 23 students who developed one site with pocketing of ≥6 mm and CAL loss of ≥2 mm, 20 were A. actinomycetemcomitans positive (86.9%) and 14 were of African-American descent (Table 4). Eight students developed two or more pockets of ≥6 mm with CAL loss of ≥2 mm. All of these students were A. actinomycetemcomitans positive. All 16 students who developed bone loss were A. actinomycetemcomitans positive, which amounted to 25.4% of the total of 63 students who started the study. Eleven of the 16 who showed bone loss were African-American. Those 11 came from a total of 31 African-American students who were A. actinomycetemcomitans positive. Of those eleven, five students had A. actinomycetemcomitans serotype b, five had serotype c, and one had serotype f. Of those with the b serotype, only one had the JP2 promoter type. All three Hispanic subjects with bone loss had serotype c. Of the two in the nonreported group who had A. actinomycetemcomitans, one had serotype a and one had serotype b (data not shown). Thirty-nine African-American students were A. actinomycetemcomitans negative, none of whom developed bone loss.
Table 4.
Longitudinal study demographics and disease characterization
| Population group |
A. actinomycetemcomitans positive |
A. actinomycetemcomitans negative |
≥1 sitea |
≥2 sitesa |
Bone lossb |
|||||
|---|---|---|---|---|---|---|---|---|---|---|
| No. of subjects | Age (mean ± SD) (yr) | No. of subjects | Age (mean ± SD) (yr) | No. of subjects (no. of A. actinomycetemcomitans positive) | Age (mean ± SD) (yr) | No. of subjects (no. of A. actinomycetemcomitans positive) | Age (mean ± SD) (yr) | No. of subjects (no. of A. actinomycetemcomitans positive) | Age (mean ± SD) (yr) | |
| Ethnic group | ||||||||||
| Hispanic | ||||||||||
| Female | 14 | 12.9 ± 1.1 | 16 | 12.4 ± 1.4 | 7 (6) | 13.4 ± 1.0 | 4 (4) | 13.8 ± 0.96 | 3 (3) | 14.0 ± 1.0 |
| Male | 14 | 12.8 ± 0.9 | 16 | 12.9 ± 1.4 | 2 (2) | 14.0 ± 1.4 | 0 | 0 | 0 | |
| Total | 28 | 12.9 ± 1.5 | 32 | 12.7 ± 1.4 | 9 (8) | 13.5 ± 1.1 | 4 (4) | 13.8 ± 0.96 | 3 (3) | 14.0 ± 1.0 |
| African-American | ||||||||||
| Female | 15 | 12.7 ± 2.3 | 18 | 12.3 ± 1.4 | 8 (7) | 12.5 ± 1.2 | 2 (2) | 14.5 ± 2.1 | 5 (5) | 14.6 ± 1.5 |
| Male | 16 | 12.5 ± 0.9 | 21 | 12.6 ± 1.7 | 6 (5) | 13.5 ±.71 | 2 (2) | 13.5 ± 0.70 | 6 (6) | 14.3 ± 1.8 |
| Total | 31 | 12.6 + 1.7 | 39 | 12.5 ± 1.4 | 14 (12) | 12.9 ±.96 | 4 (4) | 14.0 ± 1.4 | 11 (11) | 14.4 ± 1.6 |
| Not reported | ||||||||||
| Female | 2 | 12.0 ± 1.0 | 0 | 0 | 0 | 2 (2) | 12.5 ± 0.50 | |||
| Male | 2 | 13.0 ± 1.0 | 0 | 0 | 0 | 0 | ||||
| Total | 4 | 12.5 ± 1.2 | 0 | 0 | 0 | 2 (2) | 12.5 ± 0.50 | |||
| All ethnic groups combined | ||||||||||
| Female | 31 | 12.7 ± 1.7 | 34 | 12.3 ± 1.6 | 15 (13) | 12.9 ± 1.1 | 6 (6) | 14.0 ± 1.3 | 10 (10) | 14.0 ± 1.2 |
| Male | 32 | 12.7±.90 | 37 | 12.7 ± 1.6 | 8 (7) | 13.6±.87 | 2 (2) | 14.5 ± 2.5 | 6 (6) | 14.3 ± 1.8 |
| Total | 63 | 12.7 ± 1.3 | 71 | 12.5 ± 1.6 | 23 | 13.1 ± 1.0 | 8 | 14.1 ± 1.2 | 16 | 14.1 ± 1.3 |
| No. A. actinomycetemcomitans positive (%) | 20 (86.9) | 8 (100) | 16 (100) | |||||||
Site refers to a first molar pocket of ≥6 mm with attachment loss of ≥2 mm detected by probing.
Bone loss refers to radiographic evidence of vertical bone loss in a first molar interproximal area. A significantly higher number of African-American subjects had bone loss compared to Hispanic subjects (chi-square, P < 0.05).
Microbial assessment for healthy versus LAP patients, pooled data using HOMIM.
Seven subjects who developed LAP were compared to 10 subjects who remained healthy (5 control healthy A. actinomycetemcomitans-negative subjects and 5 control healthy A. actinomycetemcomitans-positive subjects). These samples were obtained from sites analyzed in students who were enrolled in the longitudinal study and taken at the time prior to detection of bone loss in the LAP group and were compared with matched samples from 10 subjects who were healthy at that time point. The rationale for the first analysis was to determine whether any trends could be observed by analyzing the overall microflora obtained from all subjects in all groups. Data from these healthy and diseased sites were summed and averaged to develop a pooled score for each microorganism in all groups. Table S1 in the supplemental material shows the prevalence ranking and average HOMIM score of the most prevalent species regardless of the subject's periodontal status. As has commonly been seen, the major bacterial species fell into the genera Streptococcus, Veillonella, Gemella, Granulicatella, Fusobacterium, and Campylobacter.
In the second analysis as seen in Fig. 1, we measured the prevalence of taxa that increased in healthy subjects compared to those who developed periodontal disease using summed data from each site in each of the two groups (healthy versus diseased). The data underlying Fig. 1 are presented in Table S2 in the supplemental material. Actinomyces massiliensis, Actinomyces gerencseriae, several Streptococcus spp., Granulicatella elegans, Gemella haemolysans, Gemella morbillorum, Rothia dentocariosa, and Rothia mucilaginosa were among the Gram-positive species that increased in healthy subjects. Prevotella loescheii, other Prevotella spp., Megasphaera sp. human oral taxon 123 (HOT-123), and Kingella oralis were among the Gram-negative species that increased in prevalence in healthy compared to LAP subjects. Table S2 in the supplemental material shows the mean and standard deviation of the HOMIM score for each microbe assessed.
Fig 1.
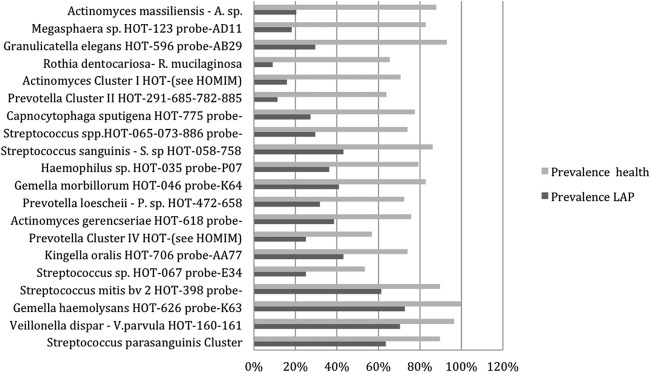
Prevalence of bacterial species that increased when samples of healthy and LAP subjects were compared. The bar graph shows the prevalence of taxa, which increased most significantly when comparing all subjects who remained healthy to those with LAP. The pairs are arranged with greatest change between health and LAP groups at the top. For each probe, the taxon(a) name(s) and human oral taxon(a) number(s) (HOT) is given.
Shown in Fig. 2 are taxa that increased significantly in prevalence in diseased subjects when comparing all healthy subjects to those who developed LAP. A. actinomycetemcomitans, F. alocis, Treponema socranskii, Tannerella forsythia, Porphyromonas gingivalis, Eubacterium nodatum, Eubacterium infirmum, Eubacterium brachy, Selenomonas sputigena, and Prevotella intermedia are among the prominent named species with increased prevalence in disease (prior to BL) compared to microbes obtained from healthy sites in healthy subjects. The unnamed taxa Peptostreptococcaceae[XIII][G-1] sp. HOT-113, Lachnospiraceae[G-5] sp. HOT-080, and Desulfobulbus sp. HOT-041 also increased significantly (Fig. 2; see also Table S3 in the supplemental material). It is important to reemphasize that the data in Fig. 1 and 2 presented above were based on pooled sample analyses with data selected from one time point (6 months prior to disease in LAP subjects and matched time points for the healthy subjects who remained healthy).
Fig 2.
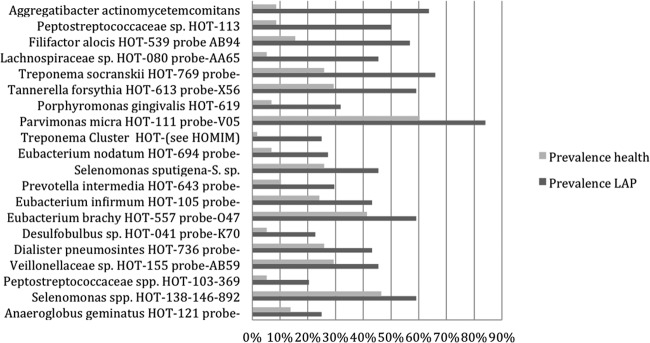
Prevalence of bacterial species that increased in disease when samples of healthy and diseased subjects were compared. The bar graph is as for Fig. 1 except that this figure identifies species in diseased subjects that increased most significantly compared to all subjects who remained healthy.
Microbial assessment of LAP site-specific data taken from subjects and sites before and after bone loss.
In this analysis, we had 16 subjects in the longitudinal study that went from health to bone loss at a specific tooth site. Of the 16 subjects, there were 18 sites where bone loss occurred during the course of the study (2 sites in 2 subjects broke down); there were a number of first molar sites from these subjects that remained healthy, and these were also studied. In the 16 subjects who developed LAP, microbial assessment was performed at the diseased site that developed BL and at that same site in the visit prior to BL. In addition, there were typically three healthy first molar sites distant from the BL site in these LAP subjects that were assessed for microbial composition. In these cases, the healthy sites were assessed at the time disease was detected and at these same sites at the time prior to BL. We recovered samples from 84 of the 96 molar sites from the 16 subjects studied. Twelve of the 96 healthy site samples were unavailable and were lost to analysis over time. Our main comparison in these site-specific studies was between the breakdown site 6 months prior to BL and sites that remained healthy before and at the time BL was detected. Subsequently, these data were compared to data obtained from these same breakdown sites (18 in total) when X-ray evidence of BL was observed.
We evaluated all species at all sites and focused on the levels of the most prevalent probes in sites that remained healthy (both pre- and post-BL in that subject) and in sites that broke down (pre-BL). In Table 5, statistical analysis was determined by chi-square and Fisher's exact test (P < 0.05). We also focused on those organisms where probes demonstrated both statistically significant higher prevalences and higher HOMIM scale levels, and whose mean HOMIM level was ≥1 (Table 5, data in bold type; statistical analysis determined by ANOVA and Tukey-Kramer post hoc testing [P ≤ 0.05]). In Table 5, those species not in bold type represent species that demonstrated significant differences either in prevalence, HOMIM probe levels, or both, but where the mean HOMIM level change was <1.
Table 5.
Bacterial prevalence and levels by HOMIM probe analysis in LAP subjects: sites with bone loss versus sites that remained healthya
| Bacterial taxon/a and probe for hybridization | Breakdown presence by: |
Mean HOMIM leveld | |
|---|---|---|---|
| Significantly higher prevalenceb | Significantly higher HOMIM levelc | ||
| Aggregatibacter actinomycetemcomitans HOT-531 probe-AA84 | + | + | High |
| Filifactor alocis HOT-539 probe-AA69 | + | + | High |
| Porphyromonas gingivalis HOT-619 probe-AA93 | + | + | High |
| Streptococcus parasanguinis cluster HOT-057, -411, -721 probe-V77 | + | + | High |
| Veillonella dispar-Veillonella parvula-Veillonella rogosae HOT-158, -160, -161 probe-D96 | + | + | High |
| Campylobacter concisus-Campylobacter rectus HOT-575, -748 probe-T86 | + | High | |
| Campylobacter curvus-C. rectus-Campylobacter showae HOT-580, -748, -763 probe-T87 | + | High | |
| C. curvus-C. rectus-C. showae HOT-580-748-763 probe-X37 | + | High | |
| Eubacterium [XI][G-6] minutum HOT-673 probe-AC65 | + | High | |
| Porphyromonas endodontalis-Porphyromonas sp. HOT-273, -285, -395 probe-W78 | + | High | |
| Tannerella forsythia HOT-613 probe-X56 | + | High | |
| Desulfobulbus sp. HOT-041 probe-K70 | + | + | Low |
| Leptotrichiaceae [G-1] sp. HOT-210, -220 probe-Y92 | + | + | Low |
| Peptostreptococcus [XIII][G-1] sp. HOT-113 probe-P72 | + | + | Low |
| Streptococcus anginosus HOT-543 probe-AB84 | + | + | Low |
| Veillonella atypica HOT-524 probe-W04 | + | + | Low |
| Bacteroidales [G-2] sp. HOT-274 probe-K76 | + | Low | |
| Bifidobacterium animalis HOT-895 probe-AC26 | + | Low | |
| Brevundimonas diminuta HOT-590 probe-AC90 | + | Low | |
| Corynebacterium matruchotii HOT-666 probe-666 | + | Low | |
| Fretibacterium sp. HOT-362 probe-P97 | + | Low | |
| Mogibacterium timidum HOT-042 probe-AB25 | + | Low | |
| Megasphaera sp. HOT-123 probe-Y85 | + | Low | |
| Neisseria pharyngis HOT-729 probe-AD93 | + | Low | |
| Ochrobactrum anthropi HOT-544 probe-AB24 | + | Low | |
| Prevotella sp. HOT-299 probe-X15 | + | Low | |
| Propionibacterium acidifaciens HOT-191 probe-AD69 | + | Low | |
| Rothia dentocariosa-Rothia mucilaginosa HOT-587, -681 probe-E52 | + | Low | |
| Slackia exigua HOT-602 probe-AB52 | + | Low | |
| Selenomonas dianae HOT-139 probe-Q50 | + | Low | |
| Streptococcus cristatus HOT-578 probe-AD89 | + | Low | |
| Treponema denticola HOT-584 probe-O40 | + | Low | |
| Treponema genus all-HOT probe AA63 | + | Low | |
| Treponema cluster: Treponema spp. HOT-250, -251, -252, -253, -254, -255, -256, -508, -517, -518 probe-AC42 | + | Low | |
| Treponema spp. HOT-237-242 probe-F89 | + | Low | |
| Lachnoanaerobaculum saburreum- Lachnoanaerobaculum orale HOT-082, -494 probe-AB50 | − | Low | |
| Peptostreptococcus stomatis HOT-112 probe-AA55 | − | Low | |
| Solobacterium moorei HOT-678 probe-AC02 | − | Low | |
| Gemella morbillorum HOT-046 probe-AB09 | − | − | Low |
| Streptococcus spp. HOT-070, -071 probe-N20 | − | − | Low |
| Streptococcus downei HOT-594 probe-AD67 | − | Low | |
| Prevotella genus most-HOT probe-AA44 | − | Low | |
| Granulicatella elegans HOT-596 probe-AB29 | − | High | |
| Fusobacterium cluster HOT-200, -201, -202, -203, -205, -370, -420, -689 probe-AD99 | − | − | High |
| V. dispar HOT-160 probe-AD63 | − | − | High |
Comparison of sites that show bone loss versus sites that do not in LAP subjects.
A boldface “+” indicates that the corresponding taxon or taxa were among the most prominent taxa at 18 sites that showed the most significant increase in prevalence 6 months prior to bone loss, as opposed to the time bone loss was detected; a lightface “+” indicates that the corresponding taxon or taxa were prevalent but less significant. A boldface “−” indicates that the corresponding taxon or taxa were among the most prominent taxa that showed increased prevalence at the time of bone loss, as opposed to 6 months prior to bone loss; a lightface “−” indicates that the corresponding taxon or taxa were found to be less prevalent at the sites at the time of bone loss than 6 months prior to bone loss. All results were analyzed by chi-square and Fisher's exact tests (P < 0.05).
A boldface “+” indicates that the corresponding taxon or taxa were among the most prominent taxa at 18 sites that showed the most significant increase in HOMIM level 6 months prior to bone loss, as opposed to the time bone loss was detected; a lightface “+” indicates that there was still an increase in HOMIM level but that is was of lesser significance. A boldface “−” indicates that the corresponding taxon or taxa were among the most prominent taxa that showed the most significantly increased HOMIM level at the time of bone loss, as opposed to 6 months prior to detection of bone loss; a lightface “−” indicates that there was found to be less of a HOMIM level increase when levels at the sites at the time of bone loss were compared to levels 6 months prior to bone loss. All results were analyzed by analysis of variance and Tukey-Kramer post hoc testing (P < 0.05).
“High” indicates that HOMIM levels for all sites analyzed for a given organism when averaged were calculated to be ≥1.0. “Low” indicates that HOMIM levels for all sites analyzed for a given organism when averaged were calculated to be <1.0.
In Table 5, there were five microbial species that showed highly elevated probe levels in sites prior to breakdown, including A. actinomycetemcomitans, F. alocis, P. gingivalis, Veillonella spp., and S. parasanguinis. In addition, several species were elevated to a lesser extent in sites prior to bone loss, including Desulfobulbus sp. HOT-113, Peptostreptococcaceae[XIII][G-1] sp. HOT-113, Streptococcus anginosus, and Veillonella atypica. The Fusobacterium cluster (Fusobacterium nucleatum all subspecies, Fusobacterium naviforme, Fusobacterium periodonticum, and clustered Fusobacterium spp.) and Veillonella dispar were statistically elevated in sites that were healthy to start and remained healthy in subjects where other sites showed LAP (Table 5, indicated in bold type as breakdown−).
We also assessed the relationship between the presence of A. actinomycetemcomitans on BECs at screening and the presence of A. actinomycetemcomitans at subgingival sites in subjects who started healthy and developed disease (n = 16) and in subjects who started healthy and remained healthy (n = 16). All sites that developed LAP (n = 18) came from subjects who had A. actinomycetemcomitans on BECs at screening. Not all sites with subgingival A. actinomycetemcomitans in these same subjects developed LAP. Thus, while 100% of sites that developed BL had A. actinomycetemcomitans on BECs at screening, suggesting a subject at risk, these same subjects had another 38% of their subgingival sites that remained healthy show A. actinomycetemcomitans (Table 6; approximately 62% of the remaining healthy sites assessed did not show A. actinomycetemcomitans). In addition, in individuals who had A. actinomycetemcomitans at screening and who started and remained healthy, only 10% of those subjects showed A. actinomycetemcomitans in their subgingival molar sites (data not shown).
Table 6.
Predictive value of six bacterial taxa at sites 6 months prior to bone loss compared to sites that remained healthy
| Bacterial taxa and HOMIM probe no. | Probe presence | No. of sites breakdown+a | No. of sites breakdown−b | Performance characteristic (%) |
|||
|---|---|---|---|---|---|---|---|
| Sensitivity | Specificity | Positive predictive value | Negative predictive value | ||||
| Aggregatibacter actinomycetemcomitans | + | 18 | 32 | ||||
| HOT-531 probe-AA84 | − | 0 | 52 | 100 | 62 | 36 | 100 |
| Porphyromonas gingivalis | + | 9 | 17 | ||||
| HOT-619 probe-AA93 | − | 9 | 67 | 50 | 80 | 35 | 88 |
| Fusobacterium cluster | + | 4 | 39 | ||||
| HOT-200, -201, -202, -203, -205, -370, -420, -689 probe-AD99 | − | 14 | 45 | 22 | 54 | 9 | 76 |
| Filifactor alocis | + | 18 | 33 | ||||
| HOT-539 probe-AA69 | − | 0 | 51 | 100 | 61 | 35 | 100 |
| Streptococcus parasanguinis cluster | + | 14 | 13 | ||||
| HOT-057, -411, -721 probe-V77 | − | 4 | 71 | 78 | 85 | 52 | 95 |
| Veillonella dispar-Veillonella parvula-Veillonella rogosae | + | 16 | 18 | ||||
| HOT-158-160-161 probe-D96 | − | 2 | 66 | 89 | 79 | 47 | 97 |
Eighteen sites in 16 subjects that showed bone loss used to calculate sensitivity, specificity, and predictive values.
Eighty-four healthy sites that remained healthy and thus did not show bone loss in these same 16 subjects used to calculate sensitivity, specificity, and predictive values.
We next calculated the sensitivities and specificities of the most significant bacterial taxa (those that were most prevalent with the greatest increase in levels) comparing breakdown to nonbreakdown sites 6 months prior to breakdown (Table 6). Both A. actinomycetemcomitans and F. alocis achieved maximum sensitivity with low specificity. We ruled out the Fusobacterium cluster and P. gingivalis because of the low sensitivity values (22% and 50%, respectively). We ruled out the Veillonella complex because the probe recognized three differing species ( V. dispar, Veillonella parvula, and Veillonella rogosa) and our goal was to find organisms at the species level. A. actinomycetemcomitans has often been suspected as the key organism responsible for LAP (5, 17, 19). However, when we calculated sensitivity and specificity with respect to the relationship of A. actinomycetemcomitans and the development of LAP, our results indicated that although the level of sensitivity was 100%, the level of specificity was much lower (i.e., 62%; see Table 6).
Our next effort was to examine the consortium of the three most elevated and prevalent bacterial taxa with respect to conversion to disease (Table 7). In this case, we selected the three most likely candidate probes (A. actinomycetemcomitans, S. parasanguinis, and F. alocis) based on the highest prevalence and elevated HOMIM levels (Table 7). When doing these calculations, we found that the sensitivity of these three probes was reduced from 100% (for A. actinomycetemcomitans and F. alocis alone) to 89% (for all three microbes); however, the specificity increased from 62% (for A. actinomycetemcomitans and F. alocis alone) to 99% when these 3 microbial species were combined in the calculations compared to using A. actinomycetemcomitans alone (compare Tables 6 and 7). In a separate series of calculations, we replaced the S. parasanguinis probe with the broad Veillonella probe we had previously ruled out and still found that the best results occurred when we included A. actinomycetemcomitans, S. parasanguinis, and F. alocis.
Table 7.
Predictive value of simultaneous detection of A. actinomycetemcomitans, F. alocis, and S. parasanguinis for LAP breakdown at 18 sites in 16 subjects
| No. of bacterial species detected | Breakdown + | Breakdown − | Performance characteristic (%) |
|||
|---|---|---|---|---|---|---|
| Sensitivity | Specificity (% CI)b | Positive predictive value | Negative predictive value | |||
| All 3a | 16 | 1 | 89 | 99 (96–100) | 94 | 98 |
| ≤2 | 2 | 83 | ||||
| Total | 18 | 84 | ||||
A. actinomycetemcomitans, F. alocis, and S. parasanguinis.
CI, 95% confidence interval.
DISCUSSION
The main goal of this study was to determine the role of A. actinomycetemcomitans and its associated microflora with respect to the development of LAP. Along those lines, our data confirm the importance of A. actinomycetemcomitans in disease susceptibility but question A. actinomycetemcomitans as being the exclusive cause of LAP (31–34). While our data show that A. actinomycetemcomitans is present at all sites that develop bone loss, 30 to 40% of sites that start healthy and fail to develop bone loss in the same subject also have A. actinomycetemcomitans present. More specifically, our findings indicate that those individuals with sites that harbor A. actinomycetemcomitans in consortium with S. parasanguinis and F. alocis are likely to develop bone loss, while those subjects with sites that harbor A. actinomycetemcomitans alone are much less likely to develop bone loss. As such, this is the first report to present evidence implicating a partnership between A. actinomycetemcomitans, S. parasanguinis, and F. alocis at sites that develop bone loss.
While we have ignored the effect of the host response in the presentation of these data, we firmly believe that a variety of host factors can influence microbial patterns related to periodontal disease. For example, a lactotransferrin lysine-to-arginine polymorphism at amino acid position 47 has been shown to affect the microbial composition of dental plaque by influencing the survival of acid-producing bacteria related to caries (35). We will report in the future on host factors that were collected for analysis in this longitudinal study.
Our cross-sectional data on the prevalence of LAP in the African-American adolescent agree well with the National Health and Nutrition Examination Survey (NHANES) data set. We find that 1.9% of the African-American adolescents from Newark have the Löe-Brown soft tissue definition of disease (36) compared to 2.01% of African-American children in the NHANES data (5, 36). In our studies of LAP, African-American and Hispanic children were recruited to increase the chances of finding subjects who will develop LAP. Our data indicate that the chance of detecting a subject at risk for LAP increases by about 10-fold by combining A. actinomycetemcomitans detection at screening with subject ethnicity compared to using ethnicity alone (from 2% to about 25%). Of the 227 A. actinomycetemcomitans-positive subjects at screening, 43 (18.9%) were found to have LAP defined as soft tissue pocketing. Our longitudinal data showed that about 25% of A. actinomycetemcomitans-positive African-American and Hispanic students who entered as periodontally healthy subjects developed LAP as defined by bone loss. These longitudinal data closely resemble the data derived from the cross-sectional analysis (see above). Thus, ethnicity and A. actinomycetemcomitans carriage appear to be strong prognostic indicators of susceptibility to LAP.
The general microbial differences we found when we compared the microbiota in healthy subjects with those who developed LAP using HOMIM analysis are supported by many previous studies of periodontal disease (21, 28). The prevalences of many Gram-positive genera, including Streptococcus, Granulicatella, Gemella, Rothia, and Actinomyces, were lower in LAP subjects than in healthy subjects. A large number of proven and suspected periodontal pathogens were shown to increase in diseased subjects, including A. actinomycetemcomitans, F. alocis, T. forsythia, P. gingivalis, P. micra, E. nodatum, E. infirmum, and E. brachy. It should be noted that the majority of these potential pathogens are from Firmicutes, so the older concept that periodontitis represents a shift from Gram-positive to Gram-negative microbiota is oversimplistic. There appear to be beneficial and detrimental microorganisms in essentially all taxonomic groups from the phylum to genus levels; thus, species-level identifications are essential to differentiate host-compatible from disease-inducing microbiomes. Our results also confirm data indicating that LAP is not the result of infection with a single pathogen but rather is due to a shift of the microbiome from one that has a prevalence of host-compatible microbes to one with a higher prevalence of more-pathogenic microbes. This type of microbe shift has sometimes been called dysbiosis (37, 38).
The results from our examination of bacterial species that increase at sites prior to bone loss, as opposed to results summarized above that related to subjects, suggest that A. actinomycetemcomitans, F. alocis, and S. parasanguinis are key members of the LAP disease complex. Their copresence has high sensitivity and specificity for predicting bone loss. As indicated by their high negative predictive values (95% to 100%), a lack of any of these organisms is strongly associated with a site not developing bone loss. At this time, the nature of the pathogenic interaction of these bacteria with each other and the host are only partially understood and require further study.
Evidence is emerging that F. alocis is a member of pathogenic complexes in periodontitis (39). F. alocis is known to produce sialidases that can alter the glycosylation of immunoglobulins and thus theoretically reduce pathogen neutralization by means of immunoglobulin-complement-polymorphonuclear (PMN) interactions. F. alocis produces numerous proteases that can cause tissue destruction (40). It also produces a substance known as poly(A), which is capable of actively recruiting PMNs from the vasculature underlying the junctional epithelium into the pocket domain (40). Pocket inflammation appears to favor the growth of F. alocis, which has been shown to exist in the deeper and more-anaerobic portions of the pocket environment (mid- to apical third of pocket) adjacent to the pocket epithelium (39). It is plausible that A. actinomycetemcomitans and F. alocis cooperate in attacking host defenses. It is known that A. actinomycetemcomitans leukotoxin transcription is increased under the anaerobic growth conditions found in deeper pockets (41). Leukotoxin can cause PMNs to lyse and release lactoferrin, lysozyme, and H2O2, which can produce a subgingival environment that can repress the growth of the competing commensal microflora (42). F. alocis is resistant to these host defense factors (40).
S. parasanguinis, as a viridans group streptococcus, has been associated with endocarditis and oral biofilm formation but has not been considered a periodontal pathogen or member of a periodontopathic complex (43). It has been shown previously that A. actinomycetemcomitans can participate in a trophic food chain with viridans group streptococci (44). In the experiments reported, Streptococcus gordonii (a viridans group streptococcus) outcompeted A. actinomycetemcomitans for glucose but produced lactate, which A. actinomycetemcomitans then utilized as its main carbon source (45). This allowed A. actinomycetemcomitans to reduce its dependence on glucose used by S. parasanguinis (46). In addition to providing lactate as the primary carbon source for A. actinomycetemcomitans, the viridans group streptococci produce hydrogen peroxide (H2O2), which suppresses many oral bacteria, particularly anaerobes. These findings suggest a biological rationale for the association of A. actinomycetemcomitans with S. parasanguinis in the periodontal pocket in the earlier stages of disease (44). Further, when A. actinomycetemcomitans is confronted with H2O2, it upregulates katA (a catalase) and apiA (an adhesin that provides resistance to complement) through an oxyR-dependent mechanism (46). Moreover, F. alocis possesses a superoxide reductase (GenBank accession no. EFE28874) and has been shown to grow in the presence of H2O2 (40). Taken together, these data suggest that A. actinomycetemcomitans, F. alocis, and S. parasanguinis would survive in the pocket. However, while it has been proposed that S. parasanguinis would be found in the upper third of the pocket, both A. actinomycetemcomitans and F. alocis have been shown to be present in the deeper portions of the pocket resting closer to the pocket epithelium (39). These temporal and anatomical associations need additional in vivo confirmation.
A shortcoming of our study is the fact that we used a fixed time interval of 6 months to examine the microbial relationship with disease status. Earlier sampling might help clarify speculations regarding the association of A. actinomycetemcomitans and S. parasanguinis in the earlier stages of disease pathogenesis (44). More-frequent sampling (at 3-month intervals) would also help clarify findings indicating that A. actinomycetemcomitans and F. alocis appear in deeper pocket sites possibly in the later stages of disease pathogenesis (39). Thus, in future experiments, subjects will be scheduled for recall visits at 3-month intervals in an attempt to catch the disease at earlier and more-frequent time points; hopefully, this will clarify the time course and localization of the A. actinomycetemcomitans/S. parasanguinis/F. alocis interbacterial and bacterium-host interactions.
A second shortcoming of the study is the failure of 15 A. actinomycetemcomitans-positive and 10 A. actinomycetemcomitans-negative students to maintain their recall schedules, such that they were dropped from the study. Fewer than 5% of these students attended more than one visit and thus they were not included in the calculations and had little impact on the study results. Nevertheless, within the context of the limitations described above, the data that emerged provided powerful differences between healthy and diseased sites and subjects.
A significant strength of the study is its longitudinal design, which allowed us to follow individuals and sites within individuals from health to disease. The success of this study might be attributed to the fact that LAP is a disease that occurs in subjects that are distinct by age and ethnicity, at specific sites, within a limited time frame (2 to 3 years), in the absence of significant confounders (e.g., smoking), and in association with a particular type of bacteria, A. actinomycetemcomitans. As a result of these attributes, the comparison of first molar plaque collected from A. actinomycetemcomitans-positive versus A. actinomycetemcomitans-negative African-American or Hispanic adolescents, over a 3-year period, provided us with our strongest data.
Data from this study shed little new information regarding the relationship between A. actinomycetemcomitans serotypes and disease, although we do confirm previous reports indicating that the c serotype predominates in subjects of Hispanic descent while serotypes are equally distributed in subjects of African-American descent (5). Because we had a limited number of subjects with the JP2 promoter type (n = 9, eight of whom were of African-American descent), we cannot address whether JP2 strains are more pathogenic or whether these strains require the presence or absence of a consortium of other microbes (4, 17, 47). Further, we cannot address the hypothesis that LAP comes in 2 forms, one caused by an exogenous strain of A. actinomycetemcomitans that produces a high level of leukotoxin (JP2 promoter-induced disease) and a second form associated with A. actinomycetemcomitans strains that produce lower levels of leukotoxin, since all but one of our BL subjects (15 of 16 subjects) had the purported low-level (652 promoter type) A. actinomycetemcomitans leukotoxin producers (10).
In conclusion, data from our study reinforce the importance of A. actinomycetemcomitans in the LAP disease process but also indicate a potential synergistic partnership of A. actinomycetemcomitans with F. alocis and S. parasanguinis in localized aggressive periodontitis in non-JP2-related disease, as this consortium is strongly associated with bone loss. Moreover, we still need to determine if additional microbes, identified by open-end methods like next-generation sequencing, can also be added to this newly established consortium or bacterial complex. While it seems clear that A. actinomycetemcomitans does not act alone, we hypothesize that A. actinomycetemcomitans is a keystone pathogen and thus sets up the subgingival environment by producing toxins (A. actinomycetemcomitans leukotoxin and cytolethal distending toxin), resulting in immune paralysis and permitting the overgrowth of specific organisms at specific time points that would otherwise be controlled and regulated by the host (15). The roles of each of these organisms, either independently and/or in concert, in the pathogenesis of this rapidly progressing disease need further examination. However, based on data derived from this study, identification of the consortium of A. actinomycetemcomitans, S. parasanguinis, and F. alocis at a specific subgingival site represents an improved method for predicting that a site is at risk for future disease.
Supplementary Material
ACKNOWLEDGMENTS
We thank the Office of Health Services of the Newark Public Schools. In addition we thank all the teachers, nurses, public school elementary students, parents, and guardians who have played a vital role in the success of this study. We thank J. Gunsolley for his help with the statistical portion of the study. We also thank the Colgate Palmolive company for providing the mobile dental van.
Research reported in this publication was supported by the National Institute of Dental and Craniofacial Research of the National Institutes of Health under award no. R01DE017968 (to D.H.F.) and R37DE016937 (to F.E.D.).
Footnotes
Published ahead of print 19 June 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JCM.00729-13.
REFERENCES
- 1.Newman MG, Socransky SS, Savitt ED, Propas DA, Crawford A. 1976. Studies of the microbiology of periodontosis. J. Periodontol. 47:373–379 [DOI] [PubMed] [Google Scholar]
- 2.Slots J. 1976. The predominant cultivable organisms in juvenile periodontitis. Scand. J. Dent. Res. 84:1–10 [DOI] [PubMed] [Google Scholar]
- 3.Bogert M, Berthold P, Brightman V, Craig R, DiRienzo JM, Lai C-H, Lally E, Oler J, Rams T, Shenker B, Slots J, Taichman NS, Tisot R. 1989. Longitudinal study of LJP families—two year surveillance. J. Dental Res. 68:312 [Google Scholar]
- 4.Haubek D, Poulsen K, Westergaard J, Dahlèn G, Kilian M. 1996. Highly toxic clone of Actinobacillus actinomycetemcomitans in geographically widespread cases of juvenile periodontitis in adolescents of African origin. J. Clin. Microbiol. 34:1576–1578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fine DH, Markowitz K, Furgang D, Fairlie K, Ferrandiz J, Nasri C, McKiernan M, Gunsolley J. 2007. Aggregatibacter actinomycetemcomitans and its relationship to initiation of localized aggressive periodontitis: longitudinal cohort study of initially healthy adolescents. J. Clin. Microbiol. 45:3859–3869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Slots J, Zambon JJ, Rosling HS, Reynolds HS, Christersson LA, Genco RJ. 1982. Actinobacillus actinomycetemcomitans in human periodontal disease. Association, serology, leukotoxicity and treatment. J. Periodontal Res. 17:447–448 [DOI] [PubMed] [Google Scholar]
- 7.Zambon JJ, Christersson LA, Slots J. 1983. Actinobacillus actinomycetemcomitans in human periodontal disease. Prevalence in patient groups and distribution of biotypes and serotypes within families. J. Periodontol. 54:707–711 [DOI] [PubMed] [Google Scholar]
- 8.Zambon JJ, Christersson LA, Genco RJ. 1986. Diagnosis and treatment of localized juvenile periodontitis. J. Am. Dent. Assoc. 113:295–299 [DOI] [PubMed] [Google Scholar]
- 9.Taichman NS, Dean RT, Sanderson CJ. 1980. Biochemical and morphological characterization of the killing of human monocytes by a leukotoxin derived from Actinobacillus actinomycetemcomitans. Infect. Immun. 28:258–268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Baehni PC, Tsai CC, McArthur WP, Hammond BF, Shenker BJ, Taichman NS. 1981. Leukotoxic activity in different strains of the bacterium Actinobacillus actinomycetemcomitans isolated from juvenile periodontitis in man. Arch. Oral Biol. 26:671–676 [DOI] [PubMed] [Google Scholar]
- 11.Mandell RL. 1984. A longitudinal microbiological investigation of Actinobacillus actinomycetemcomitans and Eikenella corrodens in juvenile periodontitis. Infect. Immun. 45:778–780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mandell RL, Socransky SS. 1988. Microbiological and clinical effects of surgery plus doxycycline on juvenile periodontitis. J. Periodontol. 59:373–379 [DOI] [PubMed] [Google Scholar]
- 13.van Winkelhoff AJ, de Graaff J. 1991. Microbiology in the management of destructive periodontal disease. J. Clin. Periodontol. 18:406–410 [DOI] [PubMed] [Google Scholar]
- 14.Bueno LC, Mayer MP, DiRienzo JM. 1998. Relationship between conversion of localized juvenile periodontitis-susceptible children from health to disease and Actinobacillus actinomycetemcomitans leukotoxin promoter structure. J. Periodontol. 69:998–1007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shenker BJ, McArthur WP, Tsai CC. 1982. Immune suppression induced by Actinobacillus actinomycetemcomitans. I. Effects on human peripheral blood lymphocyte responses to mitogens and antigens. J. Immunol. 128:148–154 [PubMed] [Google Scholar]
- 16.Schreiner HC, Sinatra K, Kaplan JB, Furgang D, Kachlany SC, Planet PJ, Perez BA, Figurski DH, Fine DH. 2003. Tight-adherence genes of Actinobacillus actinomycetemcomitans are required for virulence in a rat model. Proc. Natl. Acad. Sci. U. S. A. 100:7295–7300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Haubek D, Ennibi OK, Poulsen K, Poulsen S, Benzarti N, Kilian M. 2001. Early-onset periodontitis in Morocco is associated with a highly leukotoxic clone of Actinobacillus actinomycetemcomitans. J. Dent. Res. 80:1580–1583 [DOI] [PubMed] [Google Scholar]
- 18.Shaddox L, Wiedey J, Bimstein E, Magnuson I, Clare-Salzler M, Aukhil I, Wallet SM. 2010. Hyper-responsiveness phenotype in localized aggressive periodontitis. J. Dent. Res. 89:143–148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Haubek D, DiRienzo JM, Tinoco EM, Westergaard J, López NJ, Chung CP, Poulsen K, Kilian M. 1997. Racial tropism of a highly toxic clone of Actinobacillus actinomycetemcomitans associated with juvenile periodontitis. J. Clin. Microbiol. 35:3037–3042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Paster B, Boches SK, Galvin JL, Ericson RE, Lau CN, Levanos VA, Sahasrabudhe A, Dewhirst F. 2001. Bacterial diversity in human subgingival plaque. J. Bacteriol. 183:3770–3783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kumar PS, Griffen AL, Barton JA, Paster BJ, Moeschberger ML, Leys EJ. 2003. New bacterial species associated with chronic periodontitis. J. Dent. Res. 82:338–344 [DOI] [PubMed] [Google Scholar]
- 22.Fine DH, Markowitz K, Furgang D, Fairlie K, Ferrandiz J, Nasri C, McKiernan M, Donnelly RD, Gunsolley J. 2009. Macrophage inflammatory protein-1 alpha: a salivary biomarker of bone loss in a longitudinal cohort study of children at risk for aggressive periodontal disease? J. Periodontol. 80:106–113 [DOI] [PubMed] [Google Scholar]
- 23.Markowitz K, Fairlie K, Ferrandiz J, Nasri-Heir C, Fine DH. 2012. A longitudinal study of occlusal caries in Newark New Jersey school children: relationship between initial dental findings and the development of new lesions. Arch. Oral Biol. 57:1482–1490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gjermo P, Bellini HT, Pereira Santos V, Martins JG, Ferracyoli JR. 1984. Prevalence of bone loss in a group of Brazilian teenagers assessed on bite-wing radiographs. J. Clin. Periodontol. 11:104–113 [DOI] [PubMed] [Google Scholar]
- 25.Albandar J, Rise J, Gjermo P, Johansen J. 1986. Radiographic quantification of alveolar bone level changes. J. Clin. Periodontol. 13:195–200 [DOI] [PubMed] [Google Scholar]
- 26.Fine DH, Furgang D, Schreiner HC, Goncharoff P, Charlesworth J, Ghazwan G, Fitzgerald-Bocarsly P, Figurski DH. 1999. Phenotypic variation in Actinobacillus actinomycetemcomitans during laboratory growth: implications for virulence. Microbiology 145:1335–1347 [DOI] [PubMed] [Google Scholar]
- 27.Goncharoff P, Figurski DH, Stevens RH, Fine DH. 1993. Identification of Actinobacillus actinomycetemcomitans: polymerase chain reaction amplification of lktA-specific sequences. Oral Microbiol. Immunol. 8:105–110 [DOI] [PubMed] [Google Scholar]
- 28.Paster BJ, Olsen I, Aas J, Dewhirst FE. 2006. The breadth of bacterial diversity in the human periodontal pocket and other oral sites. Periodontol. 2000 42:80–87 [DOI] [PubMed] [Google Scholar]
- 29.Colombo AP, Boches SK, Cotton SL, Goodson JM, Kent R, Haffajee AD, Socransky SS, Hasturk H, Van Dyke TE, Dewhirst F, Paster BJ. 2009. Comparisons of subgingival microbial profiles of refractory periodontitis, severe periodontitis, and periodontal health using the human oral microbe identification microarray. J. Periodontol. 80:1421–1432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Albandar JM, Brown LJ, Genco RJ, Löe H. 1997. Clinical classification of periodontitis in adolescents and young adults. J. Periodontol. 68:545–555 [DOI] [PubMed] [Google Scholar]
- 31.Zambon JJ. 1985. Actinobacillus actinomycetemcomitans in human periodontal disease. J. Clin. Periodontol. 12:1–20 [DOI] [PubMed] [Google Scholar]
- 32.Fives-Taylor PM, Meyer DH, Mintz KP, Brissette C. 1999. Virulence factors of Actinobacillus actinomycetemcomitans. Periodontol. 2000 20:136–167 [DOI] [PubMed] [Google Scholar]
- 33.Fine DH, Kaplan JB, Kachlany SC, Schreiner HC. 2006. How we got attached to Actinobacillus actinomycetemcomitans: a model for infectious diseases. Periodontol. 2000 42:114–157 [DOI] [PubMed] [Google Scholar]
- 34.Moore WE, Holdeman LV, Smibert RM, Hash DE, Burmeister JA, Ranney RR. 1982. Bacteriology of severe periodontitis in young adult humans. Infect. Immun. 38:1137–1148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fine DH, Toruner GA, Velliyagounder K, Sampathkumar V, Godboley D, Furgang D. 2013. A lactotransferrin single nucleotide polymorphism demonstrates biological activity that can reduce susceptibility to caries. Infect. Immun. 81:1596–1605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Löe H, Brown LJ. 1991. Early onset periodontitis in the United States of America. J. Periodontol. 62:608–616 [DOI] [PubMed] [Google Scholar]
- 37.Lupp ML, Robertson ML, Wickham ME, Sekirov I, Champion OL, Gaynor EC, Finlay BB. 2007. Host-mediated inflammation disrupts the intestinal microbiota and promotes the overgrowth of Enterobacteriaceae. Cell Host Microbe 2:119–129 [DOI] [PubMed] [Google Scholar]
- 38.Hajishengallis G, Darveau RP, Curtis MA. 2012. The keystone-pathogen hypothesis. Nat. Rev. Microbiol. 10:717–725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schlafer S, Riep B, Griffen AL, Petrich A, Hubner J, Berning M, Friedmann A, Göbel UB, Moter A. 2010. Filifactor alocis-involvement in periodontal biofilms. BMC Microbiol. 10:66. 10.1186/1471-2180-10-66 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Aruni AW, Roy F, Fletcher HM. 2011. Filifactor alocis has virulence attributes that can enhance its persistence under oxidative stress conditions and mediate invasion of epithelial cells by Porphyromonas gingivalis. Infect. Immun. 79:3872–3886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kolodrubetz D, Phillips L, Burgum A. 2010. Repression of aerobic leukotoxin transcription by integration host factor in Aggregatibacter actinomycetemcomitans. Res. Microbiol. 161:541–546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Taichman NS, Wilton JM. 1981. Leukotoxicity of an extract from Actinobacillus actinomycetemcomitans for human gingival polymorphonuclear leukocytes. Inflammation 5:1–12 [DOI] [PubMed] [Google Scholar]
- 43.Geng J, Chiu C-H, Tang P, Chen Y, Shieh H-R, Hu S, Chen Y-YM. 2012. Complete genome and transcriptomes of Streptococcus parasanguinis FW213: phylogenic relations and potential virulence mechanisms. PLoS One 7:e34769. 10.1371/journal.pone.0034769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ramsey MM, Rumbaugh KP, Whiteley M. 2011. Metabolite cross-feeding enhances virulence in a model polymicrobial infection. PLoS Pathog. 7:e1002012. 10.1371/journal.ppat.1002012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Brown SA, Whiteley M. 2007. A novel mechanism for carbon resource partitioning in Aggregatibacter actinomycetemcomitans. J. Bacteriol. 189:6407–6414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ramsey MM, Whiteley M. 2009. Polymicrobial interactions stimulate resistance to host innate immunity through metabolite perception. Proc. Natl. Acad. Sci. U. S. A. 106:1578–1583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kaplan JB, Schreiner HC, Furgang D, Fine DH. 2002. Population structure and genetic diversity of Actinobacillus actinomycetemcomitans strains isolated from localized juvenile periodontitis patients. J. Clin. Microbiol. 40:1181–1187 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


