Abstract
Rapid detection of methicillin-resistant Staphylococcus aureus (MRSA) nasal colonization is crucial for the prevention and control of MRSA infections in health care settings. The LightCycler MRSA Advanced Test (Roche Diagnostics) is a commercially available real-time PCR assay for direct detection of MRSA nasal colonization by targeting of the staphylococcal cassette chromosome mec (SCCmec)-orfX junction. The diagnostic performance of the assay was compared with that of ChromID MRSA agar (bioMérieux) culture and an in-house duplex real-time PCR assay. Among 1,246 nasal swab specimens collected from 2 general hospitals in Hong Kong, 174 (14%) were considered true positive for MRSA. Chromogenic culture and the in-house real-time PCR assay identified 147 (84.5%) and 133 (76.4%) true-positive cases with specificities of 100% and 98.6%, respectively. Based on the target melting temperature (Tm) values (57.0 to 62.0°C) defined by the manufacturer, the LightCycler MRSA Advanced Test identified only 85 (48.9%) true-positive specimens. Interestingly, an additional 60 (34.5%) true-positive specimens were detected despite atypical Tm values of 55°C, providing overall sensitivity and specificity values of 83.3% and 99%, respectively. Among isolates with Tm values of 55°C, most were typed as clonal complex 45 (CC45). By sequence analysis of the SCCmec-orfX junction, characteristic single-nucleotide polymorphisms (SNPs) were identified only in isolates with Tm values of 55°C and not in those with typical Tm values. It is conceivable that those SNPs were located inside the target region of the proprietary hybridization probes, which resulted in a Tm shift in the melting curve analysis. Our study highlights the importance of a global evaluation of commercial kits so that the interpretation algorithm covers different lineages of MRSA clones prevalent in various geographical regions.
INTRODUCTION
Methicillin-resistant Staphylococcus aureus (MRSA) has been established as a health care-associated pathogen in many countries worldwide since the 1980s (1, 2). Reports of MRSA carriage or acquisition in the community have become a major focus of medical and public concern during the past decade (3, 4). Nasal colonization is known to be a risk factor for the development of MRSA-associated disease (5). Rapid diagnosis of MRSA nasal colonization facilitates early implementation of control measures to prevent ongoing infection and transmission (6). The LightCycler MRSA Advanced Test (Roche Diagnostics, Basel, Switzerland) is one of the commercially available real-time PCR assays that are designed for direct detection of MRSA nasal colonization by targeting of the genetic region between staphylococcal cassette chromosome mec (SCCmec) and the S. aureus chromosomal orfX gene, based on hybridization probe technology. According to the product information, the range of melting temperature (Tm) values observed in evaluation studies with various epidemic MRSA clones was 57.0 to 62.0°C. However, because of the enormous sequence diversity of SCCmec, MRSA clones with rarely encountered SCCmec types might yield abnormal Tm values or even false-negative signals. A previous study reported a single-nucleotide polymorphism (SNP) in the SCCmec-orfX junction found in MRSA clonal complex 398 (CC398), resulting in a shift of the Tm value to 55°C in the LightCycler MRSA Advanced Test (7). Although the result could still be manually interpreted as “positive for MRSA DNA (presumed MRSA nasal colonization)” according to the manufacturer's guidelines, the calculation algorithms embedded in the automated assay interpretation software (Micro Analysis Software) of the LightCycler MRSA Advanced Test reported the result as “MRSA result: not detected,” with a specific comment of “peak(s) outside target TM range” (7).
The LightCycler MRSA Advanced Test has been introduced in 2 general hospitals in Hong Kong since 2010 for the intended purpose of direct detection of MRSA in nasal swabs. This study aimed to compare the diagnostic performance of the LightCycler MRSA Advanced Test with that of ChromID MRSA agar culture and an in-house MRSA duplex real-time PCR assay for the detection of MRSA clones that are commonly circulating in Hong Kong.
MATERIALS AND METHODS
Specimen collection.
Between December 2010 and December 2011, a total of 1,246 nonduplicated nasal swabs were collected from 2 general hospitals in Hong Kong. The subjects included patients in hospital intensive care units (ICUs), non-ICU settings, or dialysis units and patients who were admitted from nursing homes.
Molecular methods.
The swab heads were added to sample preparation buffer for swab extraction and mechanical lysis using a MagNA Lyser instrument (Roche Diagnostics), according to the manufacturer's guidelines. The LightCycler MRSA Advanced Test also was performed according to the manufacturer's instructions.
The in-house MRSA duplex real-time PCR assay was designed to target a segment of the mecA gene and the S. aureus-specific genomic fragment sau. A total of 5 μl of the remaining DNA extract was added to the PCR mixture containing 1× LightCycler TaqMan Master reaction mixture (Roche Diagnostics, Germany), 0.5 μM primers for sau and mecA (sau-F, 5′-GACCTAATTGCTGCAACTGATCG-3′; sau-R, 5′-ACTCGTGGTGGTCATATGGAAGC-3′; mecA-F, 5′-GCCAATTCCACATTGTTTCG-3′; mecA-R, 5′-ACGGTAACATTGATCGCAACG-3′), and 0.2 μM TaqMan probes for sau and mecA (sau-P, 5′-6-carboxyfluorescein-AAATCTTCAGCATTATGAATAAATTCGTAACG-black hole quencher 1 [BHQ1]-3′; mecA-P, 5′-Yakima yellow [YAK]-TTACCACGTTCTGATTTTAAATTTTCAATATGTATGC-BHQ1-3′). Real-time PCRs were carried out in a LightCycler 2.0 system (Roche Diagnostics, Germany) under the following conditions: initial denaturation at 95°C for 10 min and 40 cycles of 95°C for 10 s, 60°C for 30 s, and 72°C for 1 s. The threshold crossing point (Cp) value for both sau and mecA targets was defined as 38 cycles by a receiver operating characteristic (ROC) curve with maximal area under the curve (AUC) and Jouden index. Samples with cycle numbers of ≤38 were considered to have positive amplification. Samples with Cp values of >38 cycles were required to be retested by real-time PCR assay. If the retest showed a Cp value of ≤38 cycles, then the sample was considered to have positive amplification. Alternatively, samples with cycle numbers of >38 in retests were regarded as negative. To minimize the risk of false-positive results caused by nasal cocolonization with methicillin-resistant coagulase-negative staphylococci (MRCNS) and methicillin-susceptible Staphylococcus aureus (MSSA), real-time PCR assays were considered positive for MRSA only when both sau and mecA targets showed positive amplification, with a difference in Cp values (ΔCp) of ≤3 cycles.
Culture methods.
The swab heads were streaked onto ChromID MRSA agar (bioMérieux, France) directly before they were cut and added to the sample preparation buffers, which were used for DNA extraction for both LightCycler MRSA Advanced Test and in-house duplex real-time PCR analyses. The plates were incubated for up to 48 ± 4 h at 37°C. Mauve colonies obtained after 24 to 48 h of incubation were presumed to be MRSA, followed by confirmation using Gram staining, latex agglutination (Slidex Staph Plus; bioMérieux, France), and tube coagulase testing.
Definition of true-positive MRSA results.
Samples were considered true positive for MRSA when they showed positive results for culture on ChromID MRSA agar. Culture-negative samples that showed positive results in both molecular assays (i.e., LightCycler MRSA Advanced Test and in-house duplex real-time PCR assay) also were considered true positive for MRSA.
Molecular typing of MRSA.
Representatives of MRSA strains with different Tm values in the LightCycler MRSA Advanced Test were randomly selected for further characterization by molecular typing methods, including S. aureus protein A gene (spa) typing and multilocus sequence typing (MLST), using standard protocols (8, 9). Sequencing analysis of the SCCmec-orfX junction was performed to screen for SNPs, according to the method described by Hagen et al. (10).
Statistical analysis.
The results of direct ChromID MRSA agar culture, the LightCycler MRSA Advanced Test, and the in-house duplex real-time PCR assay were compared with each other. Statistical significance was determined by the chi-square test. P values of ≤0.05 were considered statistically significant. The diagnostic performance parameters (sensitivity, specificity, positive predictive value [PPV], and negative predictive value) of the three assays also were determined, and the results were expressed as percentages, with 95% confidence intervals, using an adjusted Wald method. For the in-house real-time PCR assay, Cp values for false-positive samples were compared with those for true-positive samples using the Mann-Whitney U test. Differences were considered statistically significant when P values were ≤0.05.
RESULTS
Among 1,246 nasal swabs, 174 (14%) were considered true positive for MRSA based on the overall results from cultures and real-time PCR assays (Table 1). Direct culture on ChromID MRSA agar, which was considered the conventional method for MRSA detection with 100% specificity, identified 147 (84.5%) true-positive cases in this study (Table 2).
Table 1.
ChromID MRSA agar culture, LightCycler MRSA Advanced Test, and in-house duplex real-time PCR assay results
| True MRSA result (n)a | Direct ChromID MRSA agar culture resultb,c | Real-time PCR results for: |
No. of samples (total n = 1,246) | ||
|---|---|---|---|---|---|
| LightCycler MRSA Advanced Test atb,d: |
In-house duplex (sau + mecA) assayc,d | ||||
| 57–62°C | 55°C | ||||
| Positive (174) | Positive | Positive | Negative | Positive | 56 |
| Positive | Positive | Negative | Negative | 13 | |
| Positive | Negative | Positive | Positive | 42 | |
| Positive | Negative | Positive | Negative | 7 | |
| Positive | Negative | Negative | Positive | 5 | |
| Positive | Negative | Negative | Negative | 20 | |
| Positive | Invalide | Invalide | Positive | 3 | |
| Positive | Invalide | Invalide | Negative | 1 | |
| Negative | Negative | Positive | Positive | 11 | |
| Negative | Positive | Negative | Positive | 16 | |
| Negative (1,072) | Negative | Negative | Negative | Positive | 15 |
| Negative | Negative | Positive | Negative | 4 | |
| Negative | Positive | Negative | Negative | 7 | |
| Negative | Negative | Negative | Negativef | 1,046 | |
Samples with positive results for ChromID MRSA agar culture and samples with positive results in both the LightCycler MRSA Advanced Test and the in-house duplex real-time PCR assay were considered true positive for MRSA.
P < 0.001 for culture versus LightCycler MRSA Advanced Test results.
P < 0.001 for culture versus in-house duplex assay results.
P < 0.001 for LightCycler MRSA Advanced Test versus in-house duplex assay results.
The results were defined as invalid because of a failure of internal control amplification due to the presence of inhibitors.
A total of 18 samples with positive amplification for both sau and mecA genes were considered negative in the in-house duplex PCR assay because their ΔCp values were >3 cycles.
Table 2.
ChromID MRSA agar culture, LightCycler MRSA Advanced Test, and in-house duplex real-time PCR assay performance in detection of MRSA
| Assay | No. of samples with the indicated resulta |
Performance (% [95% CI])h |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Positive | Negative | True positiveb | False positive | False negative | True negativec | Sensitivity | Specificity | Positive predictive value | Negative predictive value | |
| Direct ChromID MRSA agar culture | 147 | 1,099 | 147 | 0 | 27 | 1,072 | 84.5 (78.3–89.2) | 100 (99.7–100.0) | 100 (97.8–100.0) | 97.5 (96.4–98.3) |
| LightCycler MRSA Advanced Test | 156d | 1,090 | 145e | 11f | 29 | 1,061 | 83.3 (77.0–88.5) | 99.0 (98.2–99.5) | 92.9 (87.7–96.1) | 97.3 (96.2–98.2) |
| In-house duplex real-time PCR assay | 148 | 1,098g | 133 | 15 | 41 | 1,057 | 76.4 (69.6–82.2) | 98.6 (97.7–99.2) | 89.9 (83.9–93.9) | 96.3 (95.0–97.3) |
A total of 1,246 samples were used to evaluate the performance of culture and real-time PCR assays for the detection of MRSA nasal colonization.
A total of 174 samples were considered true positive for MRSA based on the overall results of cultures and real-time PCR assays (also see Table 1).
A total of 1,072 samples were considered true negative for MRSA based on the overall results of cultures and real-time PCR assays (also see Table 1).
Among 156 positive samples from the LightCycler test, 92 showed Tm values of 59°C, whereas 64 showed Tm values of 55°C.
Among 145 true-positive samples indicated by the LightCycler test, 85 showed Tm values of 59°C, whereas 60 showed Tm values of 55°C.
The 11 false-positive samples from the LightCycler test included 7 samples with Tm values of 59°C and 4 samples with Tm values of 55°C.
A total of 18 samples with positive amplification for both sau and mecA genes were considered negative in the in-house duplex PCR assay because their ΔCp values were >3 cycles.
Results were determined as percentages and 95% confidence intervals (CIs) by an adjusted Wald method.
The in-house duplex real-time PCR assay showed positive amplification for both sau and mecA targets in 166 specimens. However, 18 of those were immediately considered negative in the in-house duplex real-time PCR assay as their ΔCp values were >3 cycles (range, 3.8 to 14.5 cycles). ChromID MRSA agar culture and the LightCycler MRSA Advanced Test confirmed that none of the 18 samples was positive for MRSA, although cultures showed the coexistence of MSSA and MRCNS in 7 of those samples. Among the 148 specimens that were positive in the in-house duplex real-time PCR assay, 133 were true positive for MRSA and 15 were considered false-positive cases, as they were negative in both ChromID MRSA agar culture and the LightCycler MRSA Advanced Test. The cultures of 6/15 false-positive samples showed positive results for MRCNS, with one showing coexistence with MSSA. All remaining false-positive samples had negative culture results. False-positive samples were found to have greater Cp values than true-positive samples (P = 0.008, Mann-Whitney U test). The diagnostic sensitivity and specificity of the in-house duplex real-time PCR assay were 76.4% (133/174 samples) and 98.6% (1,057/1,072 samples), respectively (Table 2).
A total of 92 specimens were positive in the LightCycler MRSA Advanced Test, with Tm values of 59°C (within the range of 57 to 62.0°C suggested by the embedded Micro Analysis Software). Only 85 of those specimens were true-positive MRSA cases, which resulted in a low diagnostic sensitivity of 48.9% (85/174 samples). Interestingly, another 64 samples showed positive amplification of the SCCmec-orfX junction, with atypical Tm values of 55°C in the melting curve analysis. The same results were obtained when the test was repeated with the corresponding cultured isolates. Among those samples, 60 were considered true positive for MRSA, as indicated either by culture followed by biochemical tests or by positive results from our in-house duplex real-time PCR assay. Regardless of the postamplification Tm values, overall the LightCycler MRSA Advanced Test identified 145 true-positive MRSA specimens by amplifying the SCCmec-orfX junction. Among 11 false-positive cases, 3 demonstrated MRCNS and 8 showed negative results. The overall sensitivity and specificity of the LightCycler MRSA Advanced Test were 83.3% (145/174 samples) and 99.0% (1,061/1,072 samples), respectively (Table 2).
A total of 35 and 24 specimens that showed Tm values of 55°C and 59°C, respectively, in the LightCycler MRSA Advanced Test were randomly selected for further characterization (Table 3). Of the isolates with Tm values of 55°C, a majority (32/35 isolates [91%]) were typed as clonal complex 45 (CC45) sequence type 45 (ST45), with spa patterns of t1081 (n = 23), t1857 (n = 2), t7317 (n = 2), t7854 (n = 2), t1768 (n = 1), and t8404 (n = 1). One CC45 isolate had an unknown spa type due to repeated PCR failure (Table 3). Among the 32 CC45 strains, 23 (72%) were isolated from patients admitted from nursing homes, whereas the rest were evenly distributed among ICU (n = 3), non-ICU (n = 3), and dialysis unit (n = 3) patients. The remaining 3 strains with Tm values of 55°C belonged to CC398 (ST398, n = 2) or CC672 (ST672, n = 1). All of those strains were isolated from patients in non-ICU wards. The strains characterized by Tm values of 59°C were typed to 4 clonal complexes, i.e., CC5 (ST5, n = 3), CC8 (ST8, n = 2; ST239, n = 2), CC9 (ST9, n = 1), and CC22 (ST22, n = 16) (Table 3). CC22 strains were isolated mainly from ICU (n = 6) and non-ICU (n = 6) wards, whereas the other identified lineages were evenly distributed among the collection sites.
Table 3.
Molecular characteristics of investigated MRSA strains with Tm values of 55°C and 59°C in the LightCycler MRSA Advanced Test
| No. of tested strains | Tm (°C) | spa type | MLST type | Clonal complex | SNPs in the SCCmec-orfX junction |
|---|---|---|---|---|---|
| 23 | 55 | t1081 | ST45 | CC45 | C→T at position 417, T→C at position 441 |
| 1 | 55 | t1768 | ST45 | CC45 | C→T at position 417, T→C at position 441 |
| 2 | 55 | t1857 | ST45 | CC45 | C→T at position 417, T→C at position 441 |
| 2 | 55 | t7317 | ST45 | CC45 | C→T at position 417, T→C at position 441 |
| 2 | 55 | t7854 | ST45 | CC45 | C→T at position 417, T→C at position 441 |
| 1 | 55 | t8404 | ST45 | CC45 | C→T at position 417, T→C at position 441 |
| 1 | 55 | PCR failure | ST45 | CC45 | C→T at position 417, T→C at position 441 |
| 2 | 55 | t4677 | ST398 | CC398 | A→G at position 312, G→A at position 318, G→A at position 366, T→C at position 441 |
| 1 | 55 | t1309 | ST672 | CC672 | G→A at position 435 |
| 3 | 59 | t002 | ST5 | CC5 | No SNP found |
| 2 | 59 | t008 | ST8 | CC8 | A→T at position 402 |
| 1 | 59 | t899 | ST9 | CC9 | No SNP found |
| 16 | 59 | t032 | ST22 | CC22 | G→A at position 354 |
| 2 | 59 | t037 | ST239 | CC8 | No SNP found |
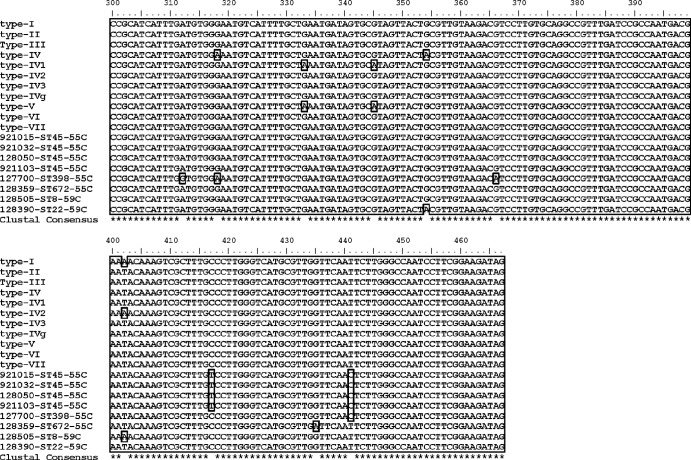
By systematic sequencing of the SCCmec-orfX junction, all CC45 MRSA strains were found to have identical sequences in multiple alignments (Fig. 1). A detailed investigation of the sequences revealed some polymorphisms that are uncommon among S. aureus GenBank sequences; one characteristic SNP, C→T at position 417, was found exclusively in all investigated CC45 MRSA strains. Another SNP, T→C at position 441, was identified in all selected MRSA strains with Tm values of 55°C except for CC672, which harbored another unique SNP, G→A at position 435. The previously described SNP (G→A at position 366) associated with livestock-associated MRSA CC398 (7) was identified in both of our CC398 MRSA strains. Among MRSA strains with Tm values of 59°C, CC8 (ST8) harbored a SNP at position 402, whereas CC22 showed a nucleic acid substitution at position 354. All remaining selected isolates did not show any SNPs throughout the investigated SCCmec-orfX junction.
Fig 1.
Multiple sequence alignment of the selected SCCmec-orfX junction of various MRSA clones. type-I, type I SCCmec (GenBank accession no. AB033763); type-II, type II SCCmec (GenBank accession no. D86934); type-III, type III SCCmec (GenBank accession no. AB047089); type-IV, type IV SCCmec (GenBank accession no. AB425823); type-IV1, type IVa SCCmec (GenBank accession no. AB063172); type-IV2, type IVb SCCmec (GenBank accession no. AB063173); type-IV3, type IVc SCCmec (GenBank accession no. AB096217); type-IVg, type IVg SCCmec (GenBank accession no. DQ106887); type-V, type V SCCmec (GenBank accession no. AB121219); type-VI, type VI SCCmec (GenBank accession no. AF411935); type-VII, type VII SCCmec (GenBank accession no. AB373032); 921015-ST45-55C, 921032-ST45-55C, 128050-ST45-55C, and 921103-ST45-55C, CC45 lineage MRSA clones with Tm values of 55°C; 127700-ST398-55C, CC398 lineage MRSA clones with Tm values of 55°C; 128359-ST672-55C, CC672 lineage MRSA clones with Tm values of 55°C; 128505-ST8-59C, CC8 lineage MRSA clones with Tm values of 59°C; 128390-ST22-59C, CC22 lineage MRSA clones with Tm values of 59°C. Numbers indicate nucleotide positions, asterisks indicate nucleotide positions showing consensus among all the selected SCCmec-orfX sequences, and the boxes indicate the nucleotide substitutions.
DISCUSSION
MRSA nasal colonization is a known risk factor for staphylococcal infections, and carriers exhibit an increased risk of infection with the strain they carry (11, 12). Direct culture from nasal swabs is considered the conventional standard method for detection of MRSA nasal colonization. Despite very high specificity, the sensitivity of this method is always dependent on the quantities of viable MRSA on the swab samples, which in turn depends on whether the swab heads reached and sampled the particular nasal regions with high densities of viable MRSA. In our study, only 84% of true-positive samples were detected by ChromID MRSA agar culture after 48 h of incubation. Similar results were reported by Peterson et al. (13); in that study, the sensitivities of directly plated cultures performed in 5 different evaluation centers ranged from 78.6% to 95.1%, indicating that considerable proportions of false-negative results could be obtained from directly plated cultures without the supplement of molecular assays.
Recently developed PCR-based assays, either developed in-house or available commercially, provide rapid diagnosis (within 2 h) of MRSA colonization in patients. Current PCR approaches are generally classified into two categories, (i) single detection of the SCCmec-orfX integration site or (ii) duplex PCR for coamplification of the mecA gene and a S. aureus-specific region such as nuc (12, 14), femA (15), or sau (16).
Most of the commercially available rapid tests, such as the LightCycler MRSA Advanced Test, the GeneXpert MRSA test (Cepheid, Sunnyvale, CA), and the GeneOhm MRSA assay (BD, Franklin Lakes, NJ), are based on detection of the SCCmec-orfX junction by using one primer specific for different types of SCCmec elements but not targeting the mecA gene specifically and another primer specific for S. aureus chromosomal sequences flanking the SCCmec integration site. Although these tests usually provide high specificities (>95%), positive predictive values generally range from 72% to 78% (17–20). False-positive cases in Europe have been reported, as 4.6% of the investigated MSSA strains were positive for SCCmec elements but lacked the mecA gene in a previous study (18). In the present study, 11 (7%) specimens that showed positive results with the LightCycler MRSA Advanced Test were defined as false-positive cases. However, none of those was positive for MSSA as indicated by culture and latex agglutination. A similar false-positive rate was reported by Peterson et al., who demonstrated that the LightCycler MRSA Advanced Test and the BD GeneOhm MRSA assay showed false-positive rates of 6% and 25%, respectively (13).
It would be expected that MSSA with SCCmec elements could be resolved by amplification of the mecA gene together with a S. aureus-specific region. However, the specificity and positive predictive value (PPV) of this method were usually compromised by MRCNS and MSSA nasal cocolonization. A previous study reported that MSSA and MRCNS cocolonization was detected for 3.4% of nasal swab specimens and an unacceptable PPV of about 40% was obtained with simultaneous detection of femA and mecA (21). The performance of this method was significantly improved in a subsequent study, which showed that ΔCp values for femA versus mecA were ≤3 cycles in more than 80% of true-positive MRSA cases (15, 22). A recent study demonstrated that the Cp differences between SCCmec-orfX and mecA were >3 cycles for mixtures of genomic DNA from MRCNS and MSSA (23). The observation was confirmed by our pilot study, which showed that the presence of MRSA could be distinguished from MSSA/MRCNS cocolonization on the basis of ΔCp values of >3 cycles for sau versus mecA unless there were <10-fold differences in concentrations between MSSA and MRCNS (data not shown). In this study, only one sample with MSSA/MRCNS cocolonization showed a ΔCp value of ≤3, indicating that the differences between MSSA and MRCNS populations in the nasal cavity usually were large enough to be detected on the basis of ΔCp values of >3, resulting in a PPV of about 92%. Nevertheless, given the high sequence similarities and the ability to exchange genetic material among staphylococcal species inside the nasal cavity, a false-positive rate of 10% was noted for our in-house assay. False-positive samples were found to exhibit greater Cp values than true-positive samples (P = 0.008, Mann-Whitney U test), indicating that the “wrong results” were mainly attributable to samples with higher Cp values, which might result from nonspecific amplification of homologous genetic regions in MRCNS.
Our study reported an unexpectedly large proportion of MRSA-positive specimens with atypical Tm values of 55°C in the LightCycler MRSA Advanced Test. Most of these strains belonged to the CC45 lineage, which accounts for most MRSA colonization in Hong Kong, especially among residents of nursing homes (20). In addition to nasal colonization, the CC45 lineage is significantly associated with the increasing multisusceptible MRSA isolated from blood cultures in our region (24). In this study, we noted a characteristic SNP in the SCCmec-orfX junction that was found in all investigated CC45 MRSA isolates. It was thought that this particular mutated position was located inside the binding regions of the proprietary hybridization probes for the LightCycler MRSA Advanced Test, resulting in a Tm shift from 59°C to 55°C. In addition to CC45, MRSA CC398 and CC672, which harbor SNPs at different nucleotide positions of the SCCmec-orfX junction, were associated with Tm values of 55°C. The three MRSA clones collectively accounted for 34.5% of identified MRSA cases in this study.
As a Conformité Européene-approved in vitro diagnostic product, the LightCycler MRSA Advanced Test has been validated with an inclusive collection of MRSA strains. A previous evaluation study performed in the United States reported that the LightCycler MRSA Advanced Test had a sensitivity of 95.2% (13). However, the epidemiologically relevant clones frequently encountered in certain geographic regions may not be congruent with the spectrum of variant isolates circulating in other regions. In this study, we found that the characteristic SNPs that induced the Tm shift in the LightCycler MRSA Advanced Test were present in more than one-third of MRSA clones prevalent in the Hong Kong region. When the PCR results were interpreted solely based on the target range of melting temperature (Tm) values (57.0 to 62.0°C) defined by the manufacturer, the LightCycler MRSA Advanced Test exhibited diagnostic sensitivity of 48.9%. However, regardless of the postamplification Tm values, the overall sensitivity of the test could be improved to 83.3% without compromising the specificity. From a technical point of view, although positive amplifications with atypical Tm values of 55°C can be manually interpreted individually as positive, the statement of “peak(s) outside target TM range” reported by the automated assay interpretation software might confuse users who are not familiar with the test. The interpretation procedures also may be complicated in regions with high prevalence rates of different lineages of MRSA clones. We recommend that the manufacturer amend the interpretation algorithm in order to extend the detection coverage of the kit for different lineages of MRSA clones.
Footnotes
Published ahead of print 19 June 2013
REFERENCES
- 1.Ho P, Yuen K, Yam W, Sai-yin Wong S, Luk W. 1995. Changing patterns of susceptibilities of blood, urinary and respiratory pathogens in Hong Kong. J. Hosp. Infect. 31:305–317 [DOI] [PubMed] [Google Scholar]
- 2.Chambers HF. 1997. Methicillin resistance in staphylococci: molecular and biochemical basis and clinical implications. Clin. Microbiol. Rev. 10:781–791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lina G, Piemont Y, Godail-Gamot F, Bes M, Peter MO, Gauduchon V, Vandenesch F, Etienne J. 1999. Involvement of Panton-Valentine leukocidin-producing Staphylococcus aureus in primary skin infections and pneumonia. Clin. Infect. Dis. 29:1128–1132 [DOI] [PubMed] [Google Scholar]
- 4.Reischl U, Tuohy MJ, Hall GS, Procop GW, Lehn N, Linde H. 2007. Rapid detection of Panton-Valentine leukocidin-positive Staphylococcus aureus by real-time PCR targeting the lukS-PV gene. Eur. J. Clin. Microbiol. Infect. Dis. 26:131–135 [DOI] [PubMed] [Google Scholar]
- 5.Datta R, Huang SS. 2008. Risk of infection and death due to methicillin-resistant Staphylococcus aureus in long-term carriers. Clin. Infect. Dis. 47:176–181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fang H, Hedin G. 2003. Rapid screening and identification of methicillin-resistant Staphylococcus aureus from clinical samples by selective-broth and real-time PCR assay. J. Clin. Microbiol. 41:2894–2899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Reischl U, Frick J, Hoermansdorfer S, Melzl H, Bollwein M, Linde HJ, Becker K, Kock R, Tuschak C, Busch U, Sing A. 2009. Single-nucleotide polymorphism in the SCCmec-orfX junction distinguishes between livestock-associated MRSA CC398 and human epidemic MRSA strains. Euro Surveill. 14(49):pii=19436 http://www.eurosurveillance.org/ViewArticle.aspx?ArticleId=19436 [PubMed] [Google Scholar]
- 8.Ho PL, Chuang SK, Choi YF, Lee RA, Lit AC, Ng TK, Que TL, Shek KC, Tong HK, Tse CW, Tung WK, Yung RW. 2008. Community-associated methicillin-resistant and methicillin-sensitive Staphylococcus aureus: skin and soft tissue infections in Hong Kong. Diagn. Microbiol. Infect. Dis. 61:245–250 [DOI] [PubMed] [Google Scholar]
- 9.Enright MC, Day NP, Davies CE, Peacock SJ, Spratt BG. 2000. Multilocus sequence typing for characterization of methicillin-resistant and methicillin-susceptible clones of Staphylococcus aureus. J. Clin. Microbiol. 38:1008–1015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hagen RM, Seegmuller I, Navai J, Kappstein I, Lehn N, Miethke T. 2005. Development of a real-time PCR assay for rapid identification of methicillin-resistant Staphylococcus aureus from clinical samples. Int. J. Med. Microbiol. 295:77–86 [DOI] [PubMed] [Google Scholar]
- 11.Ho PL, Chiu SS, Chan MY, Gan Y, Chow KH, Lai EL, Lau YL. 2012. Molecular epidemiology and nasal carriage of Staphylococcus aureus and methicillin-resistant S. aureus among young children attending day care centers and kindergartens in Hong Kong. J. Infect. 64:500–506 [DOI] [PubMed] [Google Scholar]
- 12.DeLeo FR, Otto M, Kreiswirth BN, Chambers HF. 2010. Community-associated meticillin-resistant Staphylococcus aureus. Lancet 375:1557–1568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Peterson LR, Liesenfeld O, Woods CW, Allen SD, Pombo D, Patel PA, Mehta MS, Nicholson B, Fuller D, Onderdonk A. 2010. Multicenter evaluation of the LightCycler methicillin-resistant Staphylococcus aureus (MRSA) advanced test as a rapid method for detection of MRSA in nasal surveillance swabs. J. Clin. Microbiol. 48:1661–1666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Louie L, Goodfellow J, Mathieu P, Glatt A, Louie M, Simor AE. 2002. Rapid detection of methicillin-resistant staphylococci from blood culture bottles by using a multiplex PCR assay. J. Clin. Microbiol. 40:2786–2790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Francois P, Pittet D, Bento M, Pepey B, Vaudaux P, Lew D, Schrenzel J. 2003. Rapid detection of methicillin-resistant Staphylococcus aureus directly from sterile or nonsterile clinical samples by a new molecular assay. J. Clin. Microbiol. 41:254–260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Maes N, Magdalena J, Rottiers S, De Gheldre Y, Struelens MJ. 2002. Evaluation of a triplex PCR assay to discriminate Staphylococcus aureus from coagulase-negative staphylococci and determine methicillin resistance from blood cultures. J. Clin. Microbiol. 40:1514–1517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Park SH, Jang YH, Sung H, Kim MN, Kim JS, Park YJ. 2009. Performance evaluation of BD GeneOhm MRSA PCR assay for detection of nasal colonization of methicillin-resistant Staphylococcus aureus at endemic intensive care units. Korean J. Lab. Med. 29:439–447 (In Korean) [DOI] [PubMed] [Google Scholar]
- 18.Blanc DS, Basset P, Nahimana-Tessemo I, Jaton K, Greub G, Zanetti G. 2011. High proportion of wrongly identified methicillin-resistant Staphylococcus aureus carriers by use of a rapid commercial PCR assay due to presence of staphylococcal cassette chromosome element lacking the mecA gene. J. Clin. Microbiol. 49:722–724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Laurent C, Bogaerts P, Schoevaerdts D, Denis O, Deplano A, Swine C, Struelens MJ, Glupczynski Y. 2010. Evaluation of the Xpert MRSA assay for rapid detection of methicillin-resistant Staphylococcus aureus from nares swabs of geriatric hospitalized patients and failure to detect a specific SCCmec type IV variant. Eur. J. Clin. Microbiol. Infect. Dis. 29:995–1002 [DOI] [PubMed] [Google Scholar]
- 20.Lindqvist M, Isaksson B, Grub C, Jonassen TO, Hallgren A. 2012. Detection and characterisation of SCCmec remnants in multiresistant methicillin-susceptible Staphylococcus aureus causing a clonal outbreak in a Swedish county. Eur. J. Clin. Microbiol. Infect. Dis. 31:141–147 [DOI] [PubMed] [Google Scholar]
- 21.Ho PL, Lai EL, Chow KH, Chow LS, Yuen KY, Yung RW. 2008. Molecular epidemiology of methicillin-resistant Staphylococcus aureus in residential care homes for the elderly in Hong Kong. Diagn. Microbiol. Infect. Dis. 61:135–142 [DOI] [PubMed] [Google Scholar]
- 22.Jukes L, Mikhail J, Bome-Mannathoko N, Hadfield SJ, Harris LG, El-Bouri K, Davies AP, Mack D. 2010. Rapid differentiation of Staphylococcus aureus, Staphylococcus epidermidis and other coagulase-negative staphylococci and meticillin susceptibility testing directly from growth-positive blood cultures by multiplex real-time PCR. J. Med. Microbiol. 59:1456–1461 [DOI] [PubMed] [Google Scholar]
- 23.Kim JU, Cha CH, An HK, Lee HJ, Kim MN. 2013. Multiplex real-time PCR assay for detection of methicillin-resistant Staphylococcus aureus (MRSA) strains suitable in regions of high MRSA endemicity. J. Clin. Microbiol. 51:1008–1013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ho PL, Chow KH, Lo PY, Lee KF, Lai EL. 2009. Changes in the epidemiology of methicillin-resistant Staphylococcus aureus associated with spread of the ST45 lineage in Hong Kong. Diagn. Microbiol. Infect. Dis. 64:131–137 [DOI] [PubMed] [Google Scholar]