Abstract
Newcastle disease, one of the most important health problems that affects the poultry industry around the world, is caused by virulent strains of Newcastle disease virus. Newcastle disease virus is considered to be endemic in several countries in the Americas, including Mexico. In order to control Newcastle disease outbreaks and spread, intensive vaccination programs, which include vaccines formulated with strains isolated at least 60 years ago, have been established. These vaccines are dissimilar in genotype to the virulent Newcastle disease viruses that had been circulating in Mexico until 2008. Here, 28 isolates obtained between 2008 and 2011 from different regions of Mexico from free-living wild birds, captive wild birds, and poultry were phylogenetically and biologically characterized in order to study the recent epidemiology of Newcastle disease viruses in Mexico. Here we demonstrate that, until recently, virulent viruses from genotype V continued to circulate and evolve in the country. All of the Newcastle disease viruses of low virulence, mostly isolated from nonvaccinated free-living wild birds and captive wild birds, were highly similar to LaSota (genotype II) and PHY-LMV42 (genotype I) vaccine strains. These findings, together with the discovery of two virulent viruses at the Mexican zoo, suggest that Newcastle disease viruses may be escaping from poultry into the environment.
INTRODUCTION
Newcastle disease (ND) is one of the most important health problems that affects the poultry industry around the world (1). It is caused by virulent strains of Newcastle disease virus (NDV), also known as avian paramyxovirus type 1 (APMV-1), a nonsegmented, negative, single-stranded RNA virus that is part of the genus Avulavirus (2), family Paramyxoviridae. The NDV genome contains six genes which encode at least seven proteins: nucleoprotein (NP), phosphoprotein (P), matrix protein (M), fusion protein (F), hemagglutinin-neuraminidase (HN), RNA-dependent RNA polymerase or large protein (L), and the V protein produced through editing of the phosphoprotein mRNA (1). NDV has been classified into two major classes, I and II; class II contains most of the virulent viruses circulating worldwide, subclassified into at least 16 genotypes (I to XVI) (3, 4). According to the clinical manifestations and the tropism of the virus, Newcastle disease virus has been classified into the following five pathotypes: subclinical enteric, lentogenic, mesogenic, viscerotropic velogenic, and neurotropic velogenic. Lentogenic NDV have low virulence, mesogenic NDV have middle virulence, and velogenic strains are highly virulent. Both mesogenic and velogenic NDV are considered virulent by the World Organization for Animal Health (OIE), and any isolate from birds infected with these strains is reportable (1, 5).
In order to control ND outbreaks and spread, intensive vaccination programs have been established in different countries around the world. The most widely used vaccine strains during the last 60 years have been LaSota and B1, which are class II, genotype II viruses (1, 6). Some other vaccines used commercially, such as the Ulster and PHY-LMV42 strains, form part of genotype I. Vaccination prevents disease in chickens but does not prevent viral infection and replication; therefore, virulent NDV (vNDV) continues to circulate on vaccinated animals (7).
NDV is considered to be endemic in several countries of the Americas, including Mexico (5, 8). In 1946, velogenic Newcastle disease virus was first reported in Mexico and it was detected in 1-day-old chicks imported from the United States of America (8–10). Most of the recent NDV isolates from Mexico belong to class II, genotype V (8, 10, 11), and have a divergence of approximately 16% in the amino acid sequence compared with those of the genotype II vaccines. The dissimilarity among virulent genotype V viruses and the vaccine strains that facilitate viral shedding, besides the persistence of NDV in backyard poultry and free-living wild birds, may explain why vNDV caused sporadic outbreaks in the Mexican poultry industry until recently (7, 12).
In the present study, NDV isolated from captive wild birds, free-living wild birds, and poultry in Mexico were analyzed in order to better understand the current epidemiology of NDV in the country and the relation of these viruses to older isolates.
MATERIALS AND METHODS
Isolates.
The viruses described here were isolated from different species of free-living wild birds, wild birds kept in captivity as part of a zoo exhibition, and commercial poultry in different regions of Mexico from 2008 to 2011. Table 1 contains detailed information about the GenBank accession number, year of isolation, affected avian species, and region of isolation for each isolate. In the text, the isolates are referred to by a shortened name described by the GenBank accession number, the species, and the year of isolation.
Table 1.
Isolates per year, species, and region of the country
| Isolate name | GenBank accession no. | Isolation date | Common name | Scientific name | Place of isolation | Living environment |
|---|---|---|---|---|---|---|
| Oropendola/Mex(CS)/661-ZM01/2008 | KC808487 | July 2008 | Oropendola | Zacua sp. | Chiapas | Wildlife |
| Flycatcher/Mex(CS)/662-ZM02/2008 | KC808488 | July 2008 | Mexican crested flycatcher | Myiarchus sp. | Chiapas | Wildlife |
| Short-tailed Hawk/Mex(CS)/663-ZM03/2008 | KC808489 | August 2008 | Short-tailed hawk | Buteo brachyurus | Chiapas | Zoo |
| Yellow-naped Parrot/Mex(CS)/664-ZM04/2008 | KC808490 | August 2008 | Yellow-naped parrot | Amazona auropalliata | Chiapas | Zoo |
| Robin/Mex(CS)/665-ZM05/2008 | KC808491 | August 2008 | Robin | Turdus grayi | Chiapas | Wildlife |
| Woodpecker/Mex(CS)/666-ZM06/2008 | KC808492 | August 2008 | Woodpecker | Melanerpes sp. | Chiapas | Wildlife |
| Gamefowl/Mex(Mex)/616/2008 | KC808508 | 2008 | Gamefowl | Not specified | State of Mexico | Unknown |
| Gamefowl/Mex(D.F)/619/2008 | KC808509 | 2008 | Gamefowl | Not specified | Mexico City | Unknown |
| Red-lored Parrot/Mex(CS)/667-ZM07/2009 | KC808493 | March 2009 | Red-lored Amazon parrot | Amazona autumnalis | Chiapas | Pet |
| Chachalaca/Mex(CS)/668-ZM08/2009 | KC808494 | April 2009 | Chachalaca | Ortalis vetula | Chiapas | Wildlife |
| Chachalaca/Mex(CS)/669-ZM09/2009 | KC808495 | April 2009 | Chachalaca | Ortalis vetula | Chiapas | Wildlife |
| Chachalaca/Mex(CS)/670-ZM10/2009 | KC808496 | April 2009 | Chachalaca | Ortalis vetula | Chiapas | Wildlife |
| Great-Egret/Mex(CS)/671-ZM11/2009 | KC808497 | April 2009 | Great egret | Ardea alba | Chiapas | Wildlife |
| Scarlet Macaw/Mex(CS)/672-ZM12/2009 | KC808510 | April 2009 | Scarlet macaw | Ara macao | Chiapas | Zoo |
| Highland-Guan/Mex(CS)/674-ZM14/2009 | KC808498 | April 2009 | Highland guan | Penelopina nigra | Chiapas | Zoo |
| Tree-Duck/Mex(CS)/675-ZM15/2009 | KC808511 | May 2009 | Tree-duck | Dendrocygna autumnalis | Chiapas | Zoo |
| Quail/Mex(Mex)/615/2009 | KC808512 | 2009 | Quail | Coturnix coturnix | State of Mexico | Poultry |
| Chicken/Mexico/612/2009 | KC808499 | 2009 | Chicken | Gallus gallus | State of Mexico | Poultry |
| Chicken/Mexico/613/2009 | KC808500 | 2009 | Chicken | Gallus gallus | State of Mexico | Poultry |
| Chicken/Mex(PU)/634/2010 | JQ697743 | 2010 | Chicken | Gallus gallus | Puebla | Backyard |
| Broiler/Mex(AG)/635/2010 | JQ697744 | 2010 | Broiler chicken | Gallus gallus | Aguascalientes | Poultry |
| Rooster/Mex(HG)/676/2010 | KC808501 | September 2010 | Chicken | Gallus gallus | Hidalgo | Poultry |
| Chicken/Mex(QT)/678/2010 | KC808502 | November 2010 | Broiler chicken | Gallus gallus | Queretaro | Poultry |
| Chicken/Mex(GT)/679/2010 | KC808503 | December 2010 | Layer hen | Gallus gallus | Guanajuato | Poultry |
| Chicken/Mex(HG)/682/2010 | KC808504 | December 2010 | Broiler chicken | Gallus gallus | Hidalgo | Poultry |
| Chicken/Mex(PU)/684/2010 | KC808505 | 2010 | Layer hen | Gallus gallus | Puebla | Poultry |
| Chicken/Mex(AG)/685/2011 | KC808506 | 2011 | Broiler breeder | Gallus gallus | Aguascalientes | Poultry |
| Chicken/Mex(AG)/686/2011 | KC808507 | 2011 | Broiler breeder | Gallus gallus | Aguascalientes | Poultry |
Virus isolation.
Virus isolation was performed from oropharyngeal and cloacal swabs or from tissue samples by inoculating 9- to 11-day-old specific-pathogen-free (SPF) embryonating chicken eggs (ECEs) into the allantoic cavity as has been described before (5, 13). After the incubation was completed or after the embryos died, the allantoic fluids were collected from chilled eggs and tested for hemagglutination activity with chicken red blood cells as previously described (5, 13). The hemagglutinin (HA)-positive samples were tested for hemagglutination inhibition (HI) with specific NDV antibodies (5, 13).
Characterization.
The intracerebral pathogenicity index (ICPI) was determined as previously described (5, 13). Briefly, 1-day-old SPF chicks were inoculated with 50 μl of a 1:10 dilution of infected allantoic fluid by an intracerebral route. The chicks were monitored every 24 h for 8 days, scoring the birds as 0 if normal, 1 if sick, or 2 if dead (5, 13). The intravenous pathogenicity index (IVPI) was also determined in 6-week-old SPF chickens which were inoculated with 100 μl of a 1:10 dilution of infectious allantoic fluid; birds were examined every 24 h for 10 days and scored as 0 if normal, 1 if sick, 2 if paralyzed, or 3 if dead (14). Mean death time (MDT) in eggs was determined by inoculating 9- to 11-day-old SPF ECEs as previously described (5, 13).
RNA extraction and sequencing.
Total RNA was extracted by mixing 250 μl of allantoic fluid with 750 μl of TRIzol LS reagent (Invitrogen, Carlsbad, CA) by following the manufacturer's instructions or from FTA cards as previously described (15). The fusion (F) gene was amplified by reverse transcription-PCR (RT-PCR) using the SuperScript III one-step RT-PCR system with Platinum Taq DNA polymerase (Invitrogen, Carlsbad, CA). Amplicons were sequenced with fluorescence dideoxynucleotide terminators in an ABI 3700 automated sequencer (Applied Biosystems, Inc., Fosters City, CA). Assembly and editing of sequencing data were performed using the DNAStar LaserGene software package, version 10.0.
Phylogenetic analysis.
The F gene sequences from the received samples and from samples published in GenBank were used to construct the phylogenetic trees; a total of 93 sequences were aligned using the 374-bp region, and 70 sequences were used to construct the full fusion gene tree. Additionally, 11 sequences of the full fusion were used to estimate the evolutionary divergence. The analyses were performed on January 2013 and May 2013 by using the MEGA software, version 5.0 (16).
Nucleotide sequence accession numbers.
All the sequences from the isolates characterized in this study are available in GenBank under the accession numbers KC808487 to KC808512 (Table 1).
RESULTS
In February 2008, Newcastle disease was diagnosed in a captive barn owl (Tyto alba) from a zoo located in Tuxtla Gutierrez, Chiapas, Mexico (lat 16.723957, long −93.093362), which presented no clinical signs before death and from which a vNDV with an ICPI of 1.36 was isolated, but no more samples from this isolate were received for further characterization. This diagnosis obliged the veterinarians from the zoo and the animal health authorities to start an epidemiological surveillance in that region and to temporarily close the zoo to the public.
As a result of the epidemiological surveillance in Chiapas, the NDV isolates KC808489-Hawk/2008 (Buteo brachyurus), KC808490-Yellow-naped Parrot/2008 (Amazona auropalliata), KC808493-Red-lored Parrot/2009 (Amazona autumnalis), KC808510-Scarlet Macaw/2009 (Ara macao), KC808498-Highland-Guan/2009 (Penelopina nigra), and KC808511-Tree-Duck/2009 (Dendrocygna autumnalis) were obtained from captive wild birds in the zoo, and the majority of those isolates were lentogenic, except the isolates KC808510-Scarlet Macaw/2009 and KC808511-Tree-Duck/2009, which were identified as vNDV. The isolates KC808487-Oropendola/2008 (Psarocolius montezuma), KC808488-Flycatcher/2008 (Myiarchus sp.), KC808491-Robin/2008 (Turdus grayi), KC808492-Woodpecker/2008 (Melanerpes sp.), KC808494-Chachalaca/2009, KC808495-Chachalaca/2009, and KC808496-Chachalaca/2009 (Ortalis vetula), and KC808497-Great-Egret/2009 (Ardea alba), all lentogenic NDV, were obtained from free-living wild birds found within the territory of the zoo or in the surrounding areas (Tables 1 and 2).
Table 2.
Pathotyping and characterization of free-living wild bird and captive wild bird isolates
| Isolate name | MDT (h)a | ICPIb | IVPIc | Fusion protein cleavage site | Pathotype |
|---|---|---|---|---|---|
| Oropendola/Mex(CS)/661-ZM01/2008 | NDd | 0.0 | 0.0 | G R Q G R L | Lentogenic |
| Flycatcher/Mex(CS)/662-ZM02/2008 | ND | 0.0 | 0.0 | G R Q G R L | Lentogenic |
| Short-tailed Hawk/Mex(CS)/663-ZM03/2008 | ND | 0.0 | 0.0 | G R Q G R L | Lentogenic |
| Yellow-naped Parrot/Mex(CS)/664-ZM04/2008 | ND | 0.0 | 0.0 | G R Q G R L | Lentogenic |
| Robin/Mex(CS)/665-ZM05/2008 | ND | 0.0 | 0.0 | G R Q G R L | Lentogenic |
| Woodpecker/Mex(CS)/666-ZM06/2008 | ND | 0.0 | 0.0 | G R Q G R L | Lentogenic |
| Red-lored Parrot/Mex(CS)/667-ZM07/2009 | 124 | 0.0 | 0.0 | G K Q G R L | Lentogenic |
| Chachalaca/Mex(CS)/668-ZM08/2009 | 124 | 0.0 | 0.0 | G K Q G R L | Lentogenic |
| Chachalaca/Mex(CS)/669-ZM09/2009 | 124 | 0.61 | 0.0 | G K Q G R L | Lentogenic |
| Chachalaca/Mex(CS)/670-ZM10/2009 | 124 | 0.60 | 0.0 | G K Q G R L | Lentogenic |
| Great-Egret/Mex(CS)/671-ZM11/2009 | 124 | 0.60 | 0.0 | G R Q G R L | Lentogenic |
| Scarlet Macaw/Mex(CS)/672-ZM12/2009 | 36 | 1.65 | 2.31 | R R Q K R F | Velogenic |
| Highland-Guan/Mex(CS)/674-ZM14/2009 | ND | 0.31 | ND | G K Q G R L | Lentogenic |
| Tree-Duck/Mex(CS)/675-ZM15/2009 | 56 | 1.63 | 2.11 | R R Q K R F | Velogenic |
Mean death time.
Intracerebral pathogenicity index.
Intravenous pathogenicity index.
ND, not determined.
The NDV isolates KC808508-Gamefowl/2008, KC808509-Gamefowl/2008, KC808512-Quail/2009, KC808499-Chicken/2009, KC808500-Chicken/2009, JQ697743-Chicken/2010, JQ697744-Broiler/2010, KC808501-Rooster/2010, KC808502-Chicken/2010, KC808503-Chicken/2010, KC808504-Chicken/2010, KC808505-Chicken/2010, KC808506-Chicken/2011, and KC808507-Chicken/2011 were obtained from commercial or backyard poultry as part of epidemiological surveillance programs in the states of Mexico, Puebla, Aguascalientes, Guanajuato, Hidalgo, Queretaro, and Mexico City (Table 1). Most of these isolates were identified as vNDV, but isolate KC808501-Rooster/2010 was characterized as lentogenic NDV (Table 2).
Characterization.
The ICPI, IVPI, and MDT were determined on those samples collected in Chiapas, Mexico, from free-living wild birds and captive wild birds. Most of the isolated viruses (KC808487-Oropendola/2008, KC808488-Flycatcher/2008, KC808489-Hawk/2008, KC808490-Yellow-naped Parrot/2008, KC808491-Robin/2008, KC808492-Woodpecker/2008, KC808493-Red-lored Parrot/2009, KC808494-Chachalaca/2009, KC808495-Chachalaca/2009, KC808496-Chachalaca/2009, KC808497-Great-Egret/2009, and KC808498-Highland-Guan) were identified as lentogenic NDV, with ICPIs ranging between 0.0 and 0.61 and IVPIs equal to 0.0 (Table 2) (5, 13); the MDT also classified those isolates as lentogenic. However, the isolates KC808510-Scarlet Macaw/2009 and KC808511-Tree-Duck/2009 showed ICPIs (1.65 and 1.63, respectively), MDT values (36 h and 56 h, respectively), and IVPI values (greater than 2.0) compatible with vNDV (Table 1) (5, 13).
Sequencing.
Sequencing data from the F gene showed that most of the isolates from free-living wild birds and captive wild birds (KC808487-Oropendola/2008, KC808488-Flycatcher/2008, KC808489-Hawk/2008, KC808490-Yellow-naped Parrot/2008, KC808491-Robin/2008, KC808492-Woodpecker/2008, KC808493-Red-lored Parrot/2009, KC808494-Chachalaca/2009, KC808495-Chachalaca/2009, KC808496-Chachalaca/2009, KC808497-Great-Egret/2009, and KC808498-Highland-Guan) presented fusion protein cleavage sites with an amino acid sequence compatible with lentogenic NDV strains (112 G R Q G R L117 or 112G K Q G R L117) (5, 17). KC808510-Scarlet Macaw/2009 and KC808511-Tree-Duck/2009 fusion proteins had virulent cleavage sites (112 R R Q K R F117), with multiple basic amino acids between positions 112 and 116 and a phenylalanine at position 117 (Table 2) (5, 17). However, the isolates obtained from poultry had virulent fusion protein cleavage sites (112 R R Q K R F117), except the isolate KC808501-Rooster/2010, which presented a lentogenic cleavage site (112 G R Q G R L117) (Table 3).
Table 3.
Characterization of isolates from poultry
| Isolate name | Fusion protein cleavage site | Virulence classification |
|---|---|---|
| Gamefowl/Mex(Mex)/616/2008 | R R Q K R F | High |
| Gamefowl/Mex(D.F)/619/2008 | R R Q K R F | High |
| Quail/Mex(Mex)/615/2009 | R R Q K R F | High |
| Chicken/Mexico/612/2009 | R R Q K R F | High |
| Chicken/Mexico/613/2009 | R R Q K R F | High |
| Chicken/Mex(PU)/634/2010 | R R Q K R F | High |
| Broiler/Mex(AG)/635/2010 | R R Q K R F | High |
| Rooster/Mex(HG)/676/2010 | G R Q G R L | Low |
| Chicken/Mex(QT)/678/2010 | R R Q K R F | High |
| Chicken/Mex(GT)/679/2010 | R R Q K R F | High |
| Chicken/Mex(HG)/682/2010 | R R Q K R F | High |
| Chicken/Mex(PU)/684/2010 | R R Q K R F | High |
| Chicken/Mex(AG)/685/2011 | R R Q K R F | High |
| Chicken/Mex(AG)/686/2011 | R R Q K R F | High |
Phylogenetic analysis.
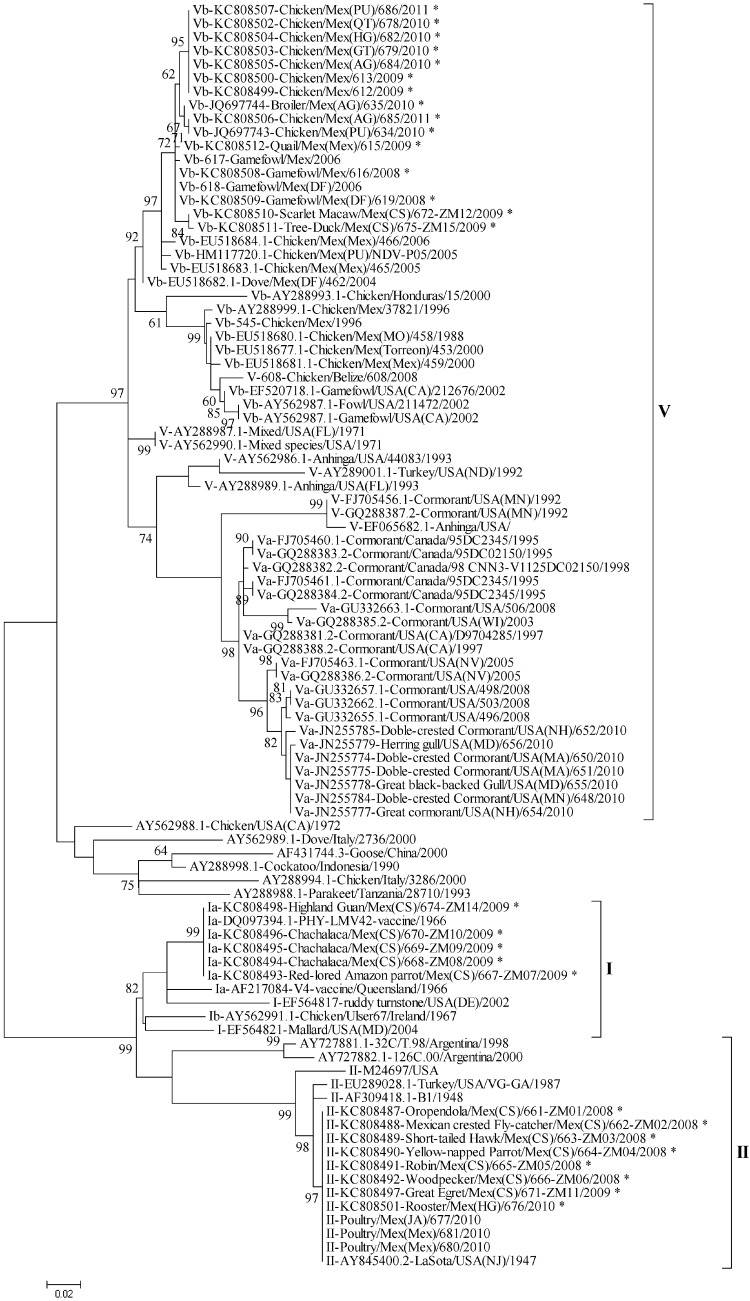
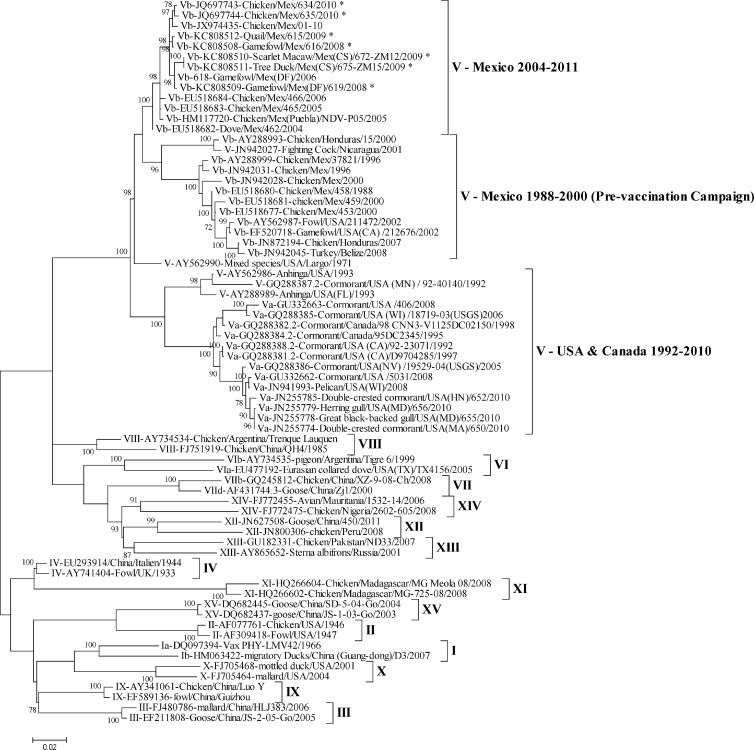
In order to localize the recent isolates into the phylogenetic trees and to compare them with older viruses isolated from Mexico in the past years, two analyses were performed. One tree was done using the 374-bp N-terminal region of the F gene, and the other was done with the full F gene sequence. These genomic regions have previously been used for NDV phylogenetic characterizations (10, 18). The 374-bp N-terminal region was used due to the greater number of sequences available in GenBank for comparison; the full fusion region was used to confirm the classification according to the method proposed by Diel (3). These analyses helped to classify all the isolates presented here within the major class II genotypes V, I, and II (Fig. 1 and 2). Also, the full fusion region was used to confirm the identity between representative lentogenic isolates and the LaSota and PHY-LMV42 vaccine strains (Table 4).
Fig 1.
Molecular phylogenetic analysis by a maximum likelihood method using the 374-bp N-terminal region of the fusion gene. A total of 93 partial nucleotide sequences from genotypes I, II, and V were used in the analysis. There were a total of 374 positions in the final data set. Evolutionary analyses were conducted in MEGA5 (16). The new isolates are denoted with an asterisk. The evolutionary history was inferred by using the maximum likelihood method based on the general time-reversible model (23). The tree with the highest log likelihood (−3026.7428) is shown. The percentage of trees in which the associated taxa clustered together is shown next to the branches. The initial tree(s) for the heuristic search was obtained automatically by applying neighbor-joining and BioNJ algorithms to a matrix of pairwise distances estimated using the maximum composite likelihood (MCL) approach and then selecting the topology with a superior log likelihood value. A discrete gamma distribution was used to model evolutionary rate differences among sites (5 categories [+G, parameter = 0.7762]). The tree is drawn to scale, with branch lengths measured in the number of substitutions per site. The analysis involved 93 nucleotide sequences. Codon positions included were 1st, 2nd, 3rd, and noncoding. All positions containing gaps and missing data were eliminated.
Fig 2.
Molecular phylogenetic analysis by a maximum likelihood method using the full F gene region. Two sequences representing each NDV genotype were used, and a total of 70 nucleotide sequences of the full fusion gene were involved in the analysis. There were a total of 1,651 positions in the final data set. Evolutionary analyses were conducted in MEGA5 (16). The new isolates are denoted with an asterisk. The evolutionary history was inferred by using the maximum likelihood method based on the general time-reversible model (23). The tree with the highest log likelihood (−15015.6528) is shown. The percentage of trees in which the associated taxa clustered together is shown next to the branches. The initial tree(s) for the heuristic search was obtained automatically by applying neighbor-joining and BioNJ algorithms to a matrix of pairwise distances estimated using the maximum composite likelihood (MCL) approach and then selecting the topology with a superior log likelihood value. A discrete gamma distribution was used to model evolutionary rate differences among sites (5 categories [+G, parameter = 0.6623]). The rate variation model allowed for some sites to be evolutionarily invariable ([+I], 37.2721% of sites). The tree is drawn to scale, with branch lengths measured in the number of substitutions per site. Codon positions included were 1st, 2nd, 3rd, and noncoding. All positions containing gaps and missing data were eliminated.
Table 4.
Estimates of evolutionary divergence between lentogenic full fusion sequences from genotypes I and IIa
| Isolate name | % divergence between each pair of isolates |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| PHY-LMV42 | KC808493 | KC808494 | KC808495 | KC808498 | KC808496 | LaSota | KC808487 | KC808489 | KC808490 | KC808497 | |
| Ia-PHY-LMV42 | 0.001 | 0.001 | 0.001 | 0.001 | 0.001 | 0.008 | 0.008 | 0.008 | 0.008 | 0.008 | |
| Ia-KC808493-Red-lored_Amazon_Parrot/2009 | 0.2 | 0.000 | 0.000 | 0.000 | 0.000 | 0.008 | 0.008 | 0.008 | 0.008 | 0.008 | |
| Ia-KC808494-Chachalaca/2009 | 0.2 | 0.0 | 0.000 | 0.000 | 0.000 | 0.008 | 0.008 | 0.008 | 0.008 | 0.008 | |
| Ia-KC808495-Chachalaca/2009 | 0.2 | 0.0 | 0.0 | 0.000 | 0.000 | 0.008 | 0.008 | 0.008 | 0.008 | 0.008 | |
| Ia-KC808498-Highland_Guan/2009 | 0.2 | 0.0 | 0.0 | 0.0 | 0.000 | 0.008 | 0.008 | 0.008 | 0.008 | 0.008 | |
| Ia-KC808496-Chachalaca/2009 | 0.2 | 0.0 | 0.0 | 0.0 | 0.0 | 0.008 | 0.008 | 0.008 | 0.008 | 0.008 | |
| II-LaSota | 10.5 | 10.4 | 10.4 | 10.4 | 10.4 | 10.4 | 0.001 | 0.001 | 0.001 | 0.001 | |
| II-KC808487-Oropendola/2008 | 10.4 | 10.3 | 10.3 | 10.3 | 10.3 | 10.3 | 0.2 | 0.000 | 0.000 | 0.000 | |
| II-KC808489-Short-tailed_Hawk/2008 | 10.4 | 10.3 | 10.3 | 10.3 | 10.3 | 10.3 | 0.2 | 0.0 | 0.000 | 0.000 | |
| II-KC808490-Yellow-naped_Parrot/2008 | 10.4 | 10.3 | 10.3 | 10.3 | 10.3 | 10.3 | 0.2 | 0.0 | 0.0 | 0.000 | |
| II-KC808497-Great_Egret/2009 | 10.4 | 10.3 | 10.3 | 10.3 | 10.3 | 10.3 | 0.2 | 0.0 | 0.0 | 0.0 | |
The analysis involved 11 nucleotide sequences of the full fusion gene from genotypes I and II (indicated at the start of each name). The percentages of divergence between sequences are shown. The divergences between the vaccine strains and their related isolates are in bold. The divergences between isolates from the same genotype are underlined. Standard error estimates are shown above the diagonal. Codon positions included were 1st, 2nd, 3rd, and noncoding. All positions containing gaps and missing data were eliminated. There were a total of 1,662 positions in the final data set. Evolutionary analyses were conducted in MEGA5 (16).
All the virulent isolates were clustered into the new clade that emerged with NDV isolated after the intensive vaccination campaigns were established in Mexico in 2002 (10). The lentogenic isolates KC808487-Oropendola/2008, KC808488-Flycatcher/2008, KC808489-Hawk/2008, KC808490-Yellow-naped Parrot/2008, KC808491-Robin/2008, KC808492-Woodpecker/2008, KC808497-Great-Egret/2009, and KC808501-Rooster/2010 were highly similar to the NDV LaSota vaccine strain (genotype II) and were clustered together. Moreover, the isolates KC808493-Red-lored Parrot/2009, KC808494-Chachalaca/2009, KC808495-Chachalaca/2009, KC808496-Chachalaca/2009, and KC808498-Highland-Guan/2009 were highly similar to the PHY-LMV42 vaccine strain (genotype I) (Fig. 1 and Table 4).
The full fusion gene sequence analysis confirmed that all the virulent isolates continue to be clustered within the subgenotype Vb together with other NDV isolates that have been obtained since 2004 in poultry. These isolates are clearly different from those vNDV that have been isolated since 1992 in the United States and Canada from poultry and free-living wild birds (Fig. 2).
DISCUSSION
Virus characterization tests and phylogenetic analysis of sequences have demonstrated that vNDV subgenotype Vb continues to circulate in Mexico (3, 8, 10). Our phylogenetic analysis shows that all the vNDV isolated from poultry and from captive wild birds from 2008 and 2011 continued to be part of subgenotype Vb, with slight divergences that indicate that these viruses continue evolving. After virulent outbreaks affected the country in 2000, intensive vaccination campaigns were established in 2002 as part of the efforts to control ND in Mexico (10). Presumably, this campaign worked, since no vNDV were reported in Mexico until 2004, when vNDV reappeared, forming a new clade within the subgenotype Vb group. These virulent viruses clustered into the new clade were only 93% to 94% similar to the virulent viruses isolated prior to 2001, demonstrating phylogenetic divergence between earlier (prior to 2001) and recent (after 2001) vNDV isolates (10) (Fig. 2).
Highly related vNDV are present in different geographic regions of Mexico. The isolates KC808510-Scarlet Macaw/2009 and KC808511-Tree-Duck/2009, obtained in Chiapas from captive wild birds in April 2009 and May 2009, respectively, were identical to each other. Also, those two isolates were closely related to the isolate KC808512-Quail/2009, which was isolated in the state of Mexico from poultry, but the geographical distance between the places of isolation made it unlikely that the outbreak in the zoo was related to the occurrence in the state of Mexico.
The origin of the vNDV isolated at the Chiapas zoo is unknown; however, the geographic and temporal proximity suggest that they are related to viruses present in poultry. In January 2009, ND was simultaneously identified in Tecpatan, Chiapas (lat 17.149305, long −93.418948), where clinical disease was detected in 10 out of 20 backyard birds (poultry), causing 8 birds to die, and in Cintalapa, Chiapas (lat 16.591961, long −93.869374), where 72 out of 90 backyard hens died; the circulating viruses from both municipalities were identified as vNDV, with ICPIs ranging from 1.64 to 1.70. By the end of March 2009, a new episode with high mortality in backyard hens and turkeys was reported at the north of Tuxtla Gutierrez, 9.76 km from the zoo. Unfortunately, it was not possible to further analyze those samples, but considering when the outbreaks occurred and the time when the viruses KC808510-Scarlet Macaw/2009 and KC808511-Tree-Duck/2009 were isolated, it seems to be more likely that the occurrence in the zoo may be related to those outbreak strains.
Both nonvaccinated free-living wild birds and captive wild birds were carrying ND vaccine viruses. The phylogenetic analysis of the 374-bp N-terminal region demonstrated high similarity between the isolates KC808487-Oropendola/2008, KC808488-Flycatcher/2008, KC808489-Hawk/2008, KC808490-Yellow-naped Parrot/2008, KC808491-Robin/2008, KC808492-Woodpecker/2008, KC808497-Great-Egret/2009, and KC808501-Rooster/2010 and the LaSota vaccine strain (Fig. 1). In addition, when the sequence of the full fusion gene from representative isolates (KC808487-Oropendola/2008, KC808489-Hawk/2008, KC808490-Yellow-naped Parrot/2008, and KC808497-Great-Egret/2009) was analyzed, the identities between the LaSota vaccine strain and those isolates were 99.8% (Table 4). Moreover, the identities between KC808493-Red-lored Parrot/2009, KC808494-Chachalaca/2009, KC808495-Chachalaca/2009, KC808496-Chachalaca/2009, and KC808498-Highland-Guan/2009 and the PHY-LMV42 vaccine strain were 99.8% (Fig. 1 and Table 4). The identities between isolates of the same genotype were equal to 100% no matter the ICPI value for both LaSota-like and PHY-LMV42-like isolates (Table 4), suggesting that the observed differences in ICPI may be due to the existence of additional changes elsewhere in the genome. This identity and the known ability of NDV to rapidly evolve (19, 20) suggest that these isolates are not lentogenic viruses that have been circulating in free-living birds since 1947 (for the viruses similar to the LaSota vaccine) or 1966 (for the viruses similar to the vaccine PHY-LMV42). Most likely, these isolates are spillover from recent infections with live vaccines utilized in poultry vaccination programs. Our findings suggest that vaccine viruses may be escaping from poultry and being carried by free-living wild birds, which may be playing a role in their dissemination. A similar situation was observed in Luxembourg, where lentogenic viruses highly similar to the LaSota strain were isolated from waterfowl (21). In a previous study conducted in our lab to evaluate the presence of lentogenic NDV in waterfowl in the United States, we have found no evidence of spillover from vaccinated animals into wild birds; however, an isolate similar to viruses of genotype Ia from wild birds was identified in a commercial turkey along with several cases of NDV genotypes from wild birds identified in live bird markets (22), suggesting that viruses from wild birds may spill over into poultry.
The results presented here may be explained by differences in biosecurity practices between Mexico and the United States, widespread presence of backyard chickens in the proximity of poultry farms in Mexico, presence of other avian species that are more susceptible to infection in Mexico, or a combination of these and other factors. Since vaccination of poultry with live NDV is the accepted standard against ND, with billons of doses used worldwide, there is a need to perform additional epidemiological surveillance in free-living wild birds in the vicinity of poultry farms to determine the role of wild birds in the spread of NDV and to identify possible weaknesses in the poultry industry that may allow this escape to happen. As new live NDV recombinant vaccines enter the market around the world, it is important to identify the risks of spillover of lentogenic and virulent NDV from poultry into the environment. In addition, since previous studies performed by our lab and others have demonstrated that vaccines homologous to the circulating virulent viruses reduce viral shedding and may potentially reduce transmission and spillover from poultry into the environment, the development of vaccines that prevent virus replication should be explored (7, 12).
ACKNOWLEDGMENTS
We acknowledge the excellent work and technical collaboration of Dawn Williams-Coplin and Tim Olivier.
This research was funded by USDA, ARS, CRIS project number 66612-32000-064.
Footnotes
Published ahead of print 14 June 2013
REFERENCES
- 1. Alexander DJ, Senne DA. 2008. Newcastle disease, other avian paramyxoviruses and pneumovirus infections, p 75–100 In Saif YM, Fadly AM, Glisson JR, McDougald LR, Nolan LK, Swayne DE. (ed), Diseases of poultry, 12th ed Blackwell Publishing, Ames, IA [Google Scholar]
- 2. Mayo MA. 2002. A summary of taxonomic changes recently approved by ICTV. Arch. Virol. 147:1655–1663 [DOI] [PubMed] [Google Scholar]
- 3. Diel DG, da Silva LH, Liu H, Wang Z, Miller PJ, Afonso CL. 2012. Genetic diversity of avian paramyxovirus type 1: proposal for a unified nomenclature and classification system of Newcastle disease virus genotypes. Infect. Genet. Evol. 12:1770–1779 [DOI] [PubMed] [Google Scholar]
- 4. Courtney SC, Susta L, Gomez D, Hines NL, Pedersen JC, Brown CC, Miller PJ, Afonso CL. 2013. Highly divergent virulent isolates of Newcastle disease virus from the Dominican Republic are members of a new genotype that may have evolved unnoticed for over 2 decades. J. Clin. Microbiol. 51:508–517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. World Organization for Animal Health (OIE) Manual of diagnostic tests and vaccines for terrestrial animals, chapter 2.3.14. OIE, Paris, France: http://www.oie.int/fileadmin/Home/eng/Health_standards/tahm/2.03.14_NEWCASTLE_DIS.pdf Accessed November 2012 [Google Scholar]
- 6. Goldhaft TM. 1980. Historical note on the origin of the LaSota strain of Newcastle disease virus. Avian Dis. 24:297–301 [PubMed] [Google Scholar]
- 7. Miller PJ, King DJ, Afonso CL, Suarez DL. 2007. Antigenic differences among Newcastle disease virus strains of different genotypes used in vaccine formulation affect viral shedding after a virulent challenge. Vaccine 25:7238–7246 [DOI] [PubMed] [Google Scholar]
- 8. Merino R, Villegas H, Quintana JA, Calderon N. 2009. Characterization of Newcastle disease viruses isolated from chicken, gamefowl, pigeon and quail in Mexico. Vet. Res. Commun. 33:1023–1030 [DOI] [PubMed] [Google Scholar]
- 9. Marquez MA. 1978. Historia de la enfermedad de Newcastle en Mexico, p 1–18 III Convencion Nacional ANECA. ANECA, Mazatlan, Sinaloa, Mexico [Google Scholar]
- 10. Perozo F, Merino R, Afonso CL, Villegas P, Calderon N. 2008. Biological and phylogenetic characterization of virulent Newcastle disease virus circulating in Mexico. Avian Dis. 52:472–479 [DOI] [PubMed] [Google Scholar]
- 11. Absalon AE, Mariano-Matias A, Vasquez-Marquez A, Morales-Garzon A, Cortes-Espinosa DV, Ortega-Garcia R, Lucio-Decanini E. 2012. Complete genome sequence of a velogenic Newcastle disease virus isolated in Mexico. Virus Genes 45:304–310 [DOI] [PubMed] [Google Scholar]
- 12. Miller PJ, Estevez C, Yu Q, Suarez DL, King DJ. 2009. Comparison of viral shedding following vaccination with inactivated and live Newcastle disease vaccines formulated with wild-type and recombinant viruses. Avian Dis. 53:39–49 [DOI] [PubMed] [Google Scholar]
- 13. Alexander DJ, Senne DA. 2008. Newcastle disease and other paramyxoviruses, p 135–141 In Swayne DE, Glisson JR, Pearson JE, Reed WM, Jackwood MW, Woolcock PR. (ed), A laboratory manual for the isolation identification and characterization of avian pathogens, 5th ed OmniPress, Inc, Madison, WI [Google Scholar]
- 14. Alexander DJ, Parsons G. 1986. Pathogenicity for chickens of avian paramyxovirus type 1 isolates obtained from pigeons in Great Britain during 1983-85. Avian Pathol. 15:487–493 [DOI] [PubMed] [Google Scholar]
- 15. Perozo F, Villegas P, Estevez C, Alvarado I, Purvis LB. 2006. Use of FTA filter paper for the molecular detection of Newcastle disease virus. Avian Pathol. 35:93–98 [DOI] [PubMed] [Google Scholar]
- 16. Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. 2011. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 28:2731–2739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Panda A, Huang Z, Elankumaran S, Rockemann DD, Samal SK. 2004. Role of fusion protein cleavage site in the virulence of Newcastle disease virus. Microb. Pathog. 36:1–10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Aldous EW, Mynn JK, Banks J, Alexander DJ. 2003. A molecular epidemiological study of avian paramyxovirus type 1 (Newcastle disease virus) isolates by phylogenetic analysis of a partial nucleotide sequence of the fusion protein gene. Avian Pathol. 32:239–256 [DOI] [PubMed] [Google Scholar]
- 19. Miller PJ, Decanini EL, Afonso CL. 2010. Newcastle disease: evolution of genotypes and the related diagnostic challenges. Infect. Genet. Evol. 10:26–35 [DOI] [PubMed] [Google Scholar]
- 20. Miller PJ, Kim LM, Ip HS, Afonso CL. 2009. Evolutionary dynamics of Newcastle disease virus. Virology 391:64–72 [DOI] [PubMed] [Google Scholar]
- 21. Snoeck CJ, Marinelli M, Charpentier E, Sausy A, Conzemius T, Losch S, Muller CP. 2013. Characterization of Newcastle disease viruses in wild and domestic birds in Luxembourg from 2006 to 2008. Appl. Environ. Microbiol. 79:639–645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Kim LM, King DJ, Curry PE, Suarez DL, Swayne DE, Stallknecht DE, Slemons RD, Pedersen JC, Senne DA, Winker K, Afonso CL. 2007. Phylogenetic diversity among low-virulence Newcastle disease viruses from waterfowl and shorebirds and comparison of genotype distributions to those of poultry-origin isolates. J. Virol. 81:12641–12653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Nei M, Kumar S. 2000. Molecular evolution and phylogenetics. Oxford University Press, New York, NY [Google Scholar]