Abstract
During a 3-year study, Clostridium perfringens was investigated in defined fecal sources from a temperate alluvial backwater area of a large river system. The results reveal that using C. perfringens as a conservative water quality indicator for total fecal pollution monitoring is no longer justified but suggest that it can be used as a tracer for excreta from nonherbivorous wildlife and human sewage.
TEXT
The reliable detection of fecal pollution in water is of high importance, as fecal material often contains a significant number of intestinal pathogens. The standard fecal indicator bacteria (SFIB), Escherichia coli and intestinal enterococci, have been used to monitor total fecal pollution for over a century. However, the suitability of the SFIB as general indicators has been increasingly questioned because these bacteria can establish “naturalized” populations in nonintestinal environments (1, 2). Genetic marker assays for total fecal pollution have also recently been developed (3), but there is still debate about their specificity, and there is still potential for further methodical improvements (4, 5). Clostridium perfringens has been suggested as an alternative to SFIB almost since the advent of the indicator concept (6). It has frequently been applied to various water resources on several continents (7–14). Despite the widespread application of C. perfringens as a general fecal indicator, recent studies of its basic molecular biology actually contradict the idea that it universally occurs in fecal pollution sources from different hosts. This contradiction is based on a genomic analysis that uncovered C. perfringens as an anaerobic, fastidious, pathogenic, “flesh-eating” organism, with the essential requirement of various amino acids (15). Based on this knowledge, C. perfringens is expected to occur in the fecal excreta from mixed-diet and carnivorous organisms, where its particular nutritional requirements are met by the food selection of the respective host.
The aim of this research was to investigate the quantitative occurrence of C. perfringens over a 3-year study period in well-defined wildlife and human-associated fecal sources that appeared in a typical backwater area of a large river system in the temperate zone. The hypothesis was that C. perfringens does not frequently occur in ruminant wildlife but is mainly associated with nonherbivorous animal excreta and human sewage. To date, there have been extremely limited systematic investigations on the quantitative occurrence of C. perfringens in wildlife fecal excrements (16–18). This paucity of data is surprising given the importance of C. perfringens in veterinary issues. However, there are numerous investigations available on the various clinically important C. perfringens pathotypes. These studies focus almost exclusively on qualitative characteristics (see, e.g., references 19 and 20). To the best of our knowledge, this report represents the first rigorous effort to quantitatively test the occurrence of C. perfringens in herbivorous and nonherbivorous animal excreta and in human sewage. To generate a robust data set, a multiyear longitudinal study was designed, and a defined, postmortem fecal sampling strategy was applied.
Investigation area, fecal sources, sampling, and bacteriological analysis.
The Porous Groundwater Well Aquifer (PGWA) backwater area is a riverine wetland located on the north side of the Danube River at the southeastern borderline of Vienna, Austria. The PGWA is a national park and an important water resource. Throughout an area of approximately 12 km2, fecal samples from freshly hunted animals (n = 73), which represent the dominant animal fecal emission sources in the area, were collected between 2010 and 2012. The hunted herbivorous ruminants included Cervus elaphus (red deer), Capreolus capreolus (roe deer), Ovis orientalis musimon (European mouflon), and Dama dama dama (European fallow deer). Additionally, Sus scrofa (wild boar) was included as a hunted, mixed-diet animal. The numbers of the hunted species included in this study reflect the relative abundances of the respective populations in this area. During the officially declared hunting season, fecal material was retrieved from the rectum immediately after the animals were shot. In addition to postmortem sampling, avian fecal droppings (n = 25) from the carnivorous Phalacrocorax carbo sinensis (great cormorant) and other mixed-diet avian species were collected (hunting of avian populations was not allowed). For domesticated animals (n = 20), feces from dogs and cats, which may occasionally enter the area, were collected nearby. Two representative wastewater treatment plants (WWTP1 and WWTP2) that discharged into the Danube River were also analyzed during this time period (n = 2 × 25). Direct sewage transfer from the Danube River to the investigated backwater happens frequently during flood events (21).
All samples were aseptically collected in sterile 50-ml plastic vials (feces) or 1,000-ml glass bottles (WWTP effluent) and stored at 5 ± 3°C in the dark until analysis. Bacteriological analysis included C. perfringens, presumptive C. perfringens, E. coli, and intestinal enterococci, according to established ISO standards (for a detailed description of the microbiological methods and definition of parameters, refer to the supplemental material). To ensure direct comparability with the standardized water quality testing procedure, samples for clostridia analysis were not pasteurized. The European Drinking Water Directive stipulates this approach for C. perfringens enumeration, which includes vegetative cells and spores (22). However, a preliminary comparison of C. perfringens concentrations from pasteurized versus unpasteurized samples did not reveal significantly different results (data not shown), which supported our previous investigations on the pasteurization effect from samples of human origin (23). Selected WWTP influent and effluent samples were also used to demonstrate the differential persistence between C. perfringens and E. coli in microcosm experiments, spanning a minimum of 163 h. The data analysis was performed with SigmaPlot, version 11.0 (Systat Software, Inc., San Jose, CA), applying the nonparametric Mann-Whitney U test to analyze for pairwise differences. For further details on area and fecal sources, please refer to the supplemental material.
Prevalence and abundance of C. perfringens.
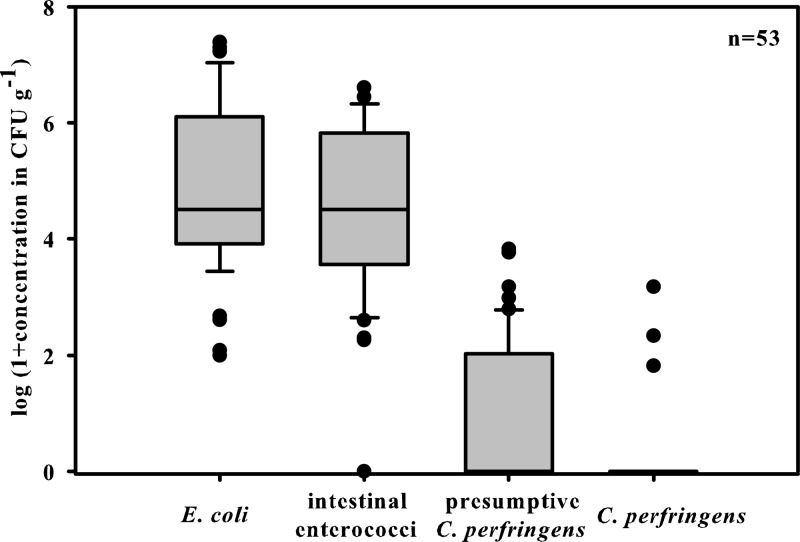
C. perfringens showed very low prevalence and abundance in fecal samples from ruminant herbivores (Table 1). C. perfringens could be detected in only 8% of the ruminant samples (n = 53) and at a low average concentration of log10 2.7 CFU per g feces for the positive fecal samples (Table 1). Consistently low prevalence and abundance of C. perfringens in ruminant wildlife excreta were also evident in comparisons of the results from the investigated years (see Table S1 in the supplemental material). The mixed-diet fauna, including wild boars and birds, had a prevalence rate of 54%, with a significantly increased average abundance of log10 5.2 CFU per g feces, compared to the samples from the herbivores (Table 1, P < 0.001). Remarkably, the highest abundance of C. perfringens in avian excreta was found in the piscivorous cormorants, reaching concentrations of up to log10 7.7 CFU per g feces. The examination of dog and cat feces showed C. perfringens prevalence rates of 67% and 63%, respectively, and the average concentrations were more than 4 orders of magnitude higher than those observed in the herbivore fecal samples. The unbalanced distribution of C. perfringens also became clear when the concentrations were compared to the abundances of E. coli and enterococci in the feces of herbivorous wildlife in the backwater area (Fig. 1). The recovered results are in agreement with the preliminary findings at an alpine watershed, where C. perfringens rarely occurred in herbivorous fecal sources (16). These results relate to a very specific environment (e.g., high altitude, little vegetation and food availability, long periods of cold temperatures). It was not possible to draw any substantial conclusions on the occurrence of C. perfringens in the fecal excreta from nonalpine areas based on this investigation (16). The recovered data also appear to be in agreement with those from a study of an Australian watershed, where a set of native and feral wildlife fecal samples (n = 72) was investigated (17). Unfortunately, the investigations were limited to two single sampling events and did not give information on the temporal variability of C. perfringens or the consistency of the results for the investigated sources.
Table 1.
Prevalence and abundance of Clostridium perfringens (or presumptive C. perfringens) in ruminant and nonruminant fecal sources from an alluvial backwater system (2010 to 2012)a
| Fecal source | n | Prevalence (%) | Abundance (log10 CFU g−1 or log10 CFU 100 ml−1)b |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Median |
Mean |
Min |
Max |
||||||||
| CP | (p.CP) | CP | (p.CP) | CP | (p.CP) | CP | (p.CP) | CP | (p.CP) | ||
| Red deer | 40 | 8 | (33) | 2.3 | (2.6) | 2.2 | (3.1) | 1.8 | (2.0) | 2.3 | (3.8) |
| Roe deer | 8 | 0 | n. d. | n. d. | n. d. | n. d. | |||||
| Fallow deer | 3 | 33 | 3.2 | 3.2 | 3.2 | 3.2 | |||||
| Mouflon | 2 | 0 | n. d. | n. d. | n. d. | n. d. | |||||
| Σ Ruminants | 53 | 8 | (26) | 2.3 | (2.6) | 2.7 | (3.1) | 1.8 | (2.0) | 3.2 | (3.8) |
| Wild boar | 20 | 60 | (85) | 3.6 | (4.0) | 4.7 | 2.0 | 5.7 | |||
| Avianc | 19 | 47 | (58) | 2.8 | (3.3) | 5.5 | (6.1) | 1.8 | (2.1) | 6.4 | (7.1) |
| Σ Mixed diet | 39 | 54 | (72) | 3.4 | (3.7) | 5.2 | (5.7) | 1.8 | (2.0) | 6.4 | (7.1) |
| Cormorant | 6 | 100 | 7.2 | 7.3 | 3.5 | 7.7 | |||||
| Dog | 12 | 67 | (100) | 5.8 | 6.9 | (6.8) | 4.4 | (3.0) | 7.4 | ||
| Cat | 8 | 63 | (75) | 5.9 | (7.0) | 7.0 | (7.1) | 4.5 | 7.4 | ||
| Σ Carnivores | 26 | 73 | (92) | 5.9 | 7.1 | (7.0) | 3.5 | (3.0) | 7.7 | ||
| WWTP1 | 25 | 100 | 3.1 | 3.2 | 2.4 | (2.6) | 3.6 | ||||
| WWTP2 | 25 | 100 | 3.8 | 3.9 | 3.0 | 4.4 | |||||
| Σ WWTPs | 50 | 100 | 3.5 | 3.6 | (3.7) | 2.4 | (2.6) | 4.4 | |||
CP, Clostridium perfringens; (p.CP), presumptive C. perfringens; n, replicate number; WWTP, wastewater treatment plant. Presumptive C. perfringens results are shown explicitly only when they differ from those of C. perfringens.
Abundance data (i.e., median, mean, min, max) were calculated excluding nondetectable data and log transformed after the addition of a value of 1. CFU g−1 or 100 ml−1, CFU per g feces (wet weight) or per 100 ml of sewage effluent from WWTPs; Mean, arithmetic mean; Min, minimum; Max, maximum; n. d., not detectable. Detection limit = log10 1.7 to log10 2.0 CFU g−1 feces (except for 2 samples, log10 2.5 and log10 2.6 CFU g−1 feces) or log10 1.0 CFU 100 ml−1 sewage effluent (WWTPs).
Birds other than cormorants (see the supplemental material for more details).
Fig 1.
Observed concentrations of fecal indicators (Escherichia coli, intestinal enterococci, presumptive Clostridium perfringens, and C. perfringens) in fecal samples from herbivorous ruminants. CFU g−1, CFU per g feces (wet weight); boxes, 25th and 75th percentiles; lines within the boxes, medians; whiskers, 10th and 90th percentiles. Data are log transformed after the addition of a value of 1.
C. perfringens could be detected in 100% of the effluent WWTP samples, with concentrations in the expected range of log10 2.4 to log10 4.4 CFU per 100 ml wastewater (Table 1). Assuming a moderate dilution of human feces in the sewer channel (i.e., log10 1.0 dilution) and conservative treatment efficacy in the WWTP (i.e., log10 1.0 reduction), the C. perfringens concentrations would be approximately log10 4.4 to log10 6.4 CFU per g feces. These concentrations fit well with the previously reported median and average concentrations of log10 4.0 and log10 5.8 CFU per g feces from the investigated Viennese population (16).
Interestingly, the distinct patterns of C. perfringens concentrations between the considered fecal source groups were also reflected by the presumptive C. perfringens concentrations (Table 1, values in brackets). However, a clear trend toward higher presumptive C. perfringens concentrations was evident, most likely due to a broader taxonomic composition.
Implications of the results on the indicator capacity of C. perfringens.
The results of this longitudinal study strongly indicate that defining C. perfringens as a conservative indicator for total fecal pollution monitoring in water is no longer justified (24, 25). Instead, the results illustrate that C. perfringens is a conservative indicator for fecal excreta from nonherbivorous wildlife and human-associated sewage. In this respect, it is not surprising that C. perfringens proved to be an excellent indicator for point source emissions from wastewater treatment plants in rivers and other lotic systems (9, 11, 12) and as a tracer for sewage sludge pollution (13, 14). Although the feces of carnivorous wildlife can contain very high concentrations of C. perfringens, it seems unlikely that this type of feces plays a significant role in water pollution, as the abundance of predators is usually very low in comparison to that of the remaining animals. Depending on the situation, dogs and cats may play a substantial role in C. perfringens emission, especially in urban areas. As livestock are not found in the investigated backwater area, this type of fecal source was not investigated. However, a previous study in an alpine area indicated that herbivorous livestock may carry significant C. perfringens concentrations, which were most likely promoted by special feeding practices (16).
Recent studies indicated that the concentrations of C. perfringens are correlated with certain pathogens (Cryptosporidium spp.) and with infection risk from recreational activities (26, 27). Because of this finding, C. perfringens was suggested as a potential alternative indicator to SFIB for recreational water quality monitoring (28). The implicit rationale behind this suggestion is based on its wastewater-associated nature. It is important that C. perfringens is a spore-forming organism that can be extremely resistant to various environmental factors such as heat, low water availability, radiation, or disinfection procedures (8, 29, 30). C. perfringens does not reproduce in aquatic systems (31, 32). The spores may be detected long after a pollution event has occurred and far from the source. To demonstrate the persistent nature of C. perfringens, we performed microcosm experiments using various raw and treated wastewater effluents and the intrinsic C. perfringens and E. coli contaminants (see Fig. S1 in the supplemental material). Remarkable differential levels of persistence of C. perfringens and E. coli were evident, which highlighted the conservative nature of C. perfringens populations. Finally, as C. perfringens occurs at lower numbers than E. coli or enterococci (16, 17), larger sampling volumes have to be used for water quality monitoring and risk management.
Supplementary Material
ACKNOWLEDGMENTS
We acknowledge the support of the Forestry Administration Office (Vienna Municipal Department of Forestry and Urban Agriculture, MA 49) and the laboratory assistance of Sonja Knetsch and Andrea Lettl.
This paper was supported by the Austrian Science Fund (FWF) as part of the “Vienna Doctoral Program on Water Resource Systems” (W1219-N22) and the research project “Groundwater Resource Systems Vienna,” in cooperation with the Vienna Waterworks as part of the “(New) Danube-Lower Lobau Network Project” [Gewässervernetzung (Neue) Donau-Untere Lobau (Nationalpark Donau-Auen)] funded by the Government of Austria (Federal Ministry of Agriculture, Forestry, Environment & Water Management), the Government of Vienna, and the European Agricultural Fund for Rural Development (project LE 07-13). J.V. also acknowledges UTM (Universiti Technologi Malaysia) in cooperation with IWA (International Water Association) for providing and financing her attendance at a course on scientific publication in January 2013, where this work was significantly improved.
This work was a joint investigation of the Interuniversity Cooperation Centre for Water & Health (www.waterandhealth.at).
Footnotes
Published ahead of print 7 June 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.01396-13.
REFERENCES
- 1. Ishii S, Sadowsky MJ. 2008. Escherichia coli in the environment: implications for water quality and human health. Microbes Environ. 23:101–108 [DOI] [PubMed] [Google Scholar]
- 2. Byappanahalli MN, Nevers MB, Korajkic A, Staley ZR, Harwood VJ. 2012. Enterococci in the environment. Microbiol. Mol. Biol. Rev. 76:685–706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Wuertz S, Wang D, Reischer GH, Farnleitner AH. 2011. Library-independent bacterial source tracking methods, p 61–112 In Hagedorn C, Blanch AR, Harwood VJ. (ed), Microbial source tracking: methods, applications, and case studies. Springer, New York, NY [Google Scholar]
- 4. van der Wielen PWJJ, Medema G. 2010. Unsuitability of quantitative Bacteroidales 16S rRNA gene assays for discerning fecal contamination of drinking water. Appl. Environ. Microbiol. 76:4876–4881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Vierheilig J, Farnleitner AH, Kollanur D, Blöschl G, Reischer GH. 2012. High abundance of genetic Bacteroidetes markers for total fecal pollution in pristine alpine soils suggests lack in specificity for feces. J. Microbiol. Methods 88:433–435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Klein E. 1895. Ueber einen pathogenen aneroben Darmbacillus Bacillus enteritidis sporogenes. Centralbl. Bakteriol. Abt. I 18:737–743 [Google Scholar]
- 7. Bonde GJ. 1966. Bacteriological methods for estimation of water pollution. Health Lab. Sci. 3:124–128 [PubMed] [Google Scholar]
- 8. Bisson JW, Cabelli VJ. 1980. Clostridium perfringens as a water pollution indicator. J. Water Pollut. Control Fed. 52:241–248 [PubMed] [Google Scholar]
- 9. Fujioka RS, Shizumura LK. 1985. Clostridium perfringens, a reliable indicator of stream water quality. J. Water Pollut. Control Fed. 57:986–992 [Google Scholar]
- 10. Sartory DP. 1988. Faecal clostridia and indicator bacteria levels in an eutrophic impoundment. Water SA 14:115–117 [Google Scholar]
- 11. Sorensen DL, Eberl SG, Dicksa RA. 1989. Clostridium perfringens as a point source indicator in non-point polluted streams. Water Res. 23:191–197 [Google Scholar]
- 12. Byamukama D, Mach RL, Kansiime F, Manafi M, Farnleitner AH. 2005. Discrimination efficacy of fecal pollution detection in different aquatic habitats of a high-altitude tropical country, using presumptive coliforms, Escherichia coli, and Clostridium perfringens spores. Appl. Environ. Microbiol. 71:65–71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Hill RT, Straube WL, Palmisano AC, Gibson SL, Colwell RR. 1996. Distribution of sewage indicated by Clostridium perfringens at a deep-water disposal site after cessation of sewage disposal. Appl. Environ. Microbiol. 62:1741–1746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Skanavis C, Yanko WA. 2001. Clostridium perfringens as a potential indicator for the presence of sewage solids in marine sediments. Mar. Pollut. Bull. 42:31–35 [DOI] [PubMed] [Google Scholar]
- 15. Shimizu T, Ohtani K, Hirakawa H, Ohshima K, Yamashita A, Shiba T, Ogasawara N, Hattori M, Kuhara S, Hayashi H. 2002. Complete genome sequence of Clostridium perfringens, an anaerobic flesh-eater. Proc. Natl. Acad. Sci. U. S. A. 99:996–1001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Farnleitner AH, Ryzinska-Paier G, Reischer GH, Burtscher MM, Knetsch S, Kirschner AKT, Dirnböck T, Kuschnig G, Mach RL, Sommer R. 2010. Escherichia coli and enterococci are sensitive and reliable indicators for human, livestock and wildlife faecal pollution in alpine mountainous water resources. J. Appl. Microbiol. 109:1599–1608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Cox P, Griffith M, Angles M, Deere D, Ferguson C. 2005. Concentrations of pathogens and indicators in animal feces in the Sydney watershed. Appl. Environ. Microbiol. 71:5929–5934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Ferguson CM, Charles K, Deere DA. 2009. Quantification of microbial sources in drinking-water catchments. Crit. Rev. Environ. Sci. Tech. 39:1–40 [Google Scholar]
- 19. Popoff MR, Bouvet P. 2009. Clostridial toxins. Future Microbiol. 4:1021–1064 [DOI] [PubMed] [Google Scholar]
- 20. Songer JG. 2010. Clostridia as agents of zoonotic disease. Vet. Microbiol. 140:399–404 [DOI] [PubMed] [Google Scholar]
- 21. Derx J, Blaschke AP, Farnleitner AH, Pang L, Blöschl G, Schijven JF. 2013. Effects of fluctuations in river water level on virus removal by bank filtration and aquifer passage—a scenario analysis. J. Contam. Hydrol. 147:34–44 [DOI] [PubMed] [Google Scholar]
- 22. Council of the EU 1998. Council directive 98/83/EC of 3 November 1998 on the quality of water intended for human consumption. Official Journal of the European Community L 330, p 32–54 http://eur-lex.europa.eu/LexUriServ/LexUriServ.do?uri=CELEX:31998L0083:en:NOT
- 23. Sherpa AM, Byamukama D, Shrestha RR, Haberl R, Mach RL, Farnleitner AH. 2009. Use of faecal pollution indicators to estimate pathogen die off conditions in source separated faeces in Kathmandu Valley, Nepal. J. Water Health 7:97–107 [DOI] [PubMed] [Google Scholar]
- 24. Toranzos GA, McFeters GA, Borrego JJ, Savill M. 2007. Detection of microorganisms in environmental freshwaters and drinking waters, p 249–264 In Hurst CJ, Crawford RL, Garland JL, Lipson DA, Mills AL, Stetzenbach LD. (ed), Manual of environmental microbiology, 3rd ed ASM Press, Washington, DC [Google Scholar]
- 25. Yates MV. 2007. Classical indicators in the 21st century—far and beyond the coliform. Water Environ. Res. 79:279–286 [DOI] [PubMed] [Google Scholar]
- 26. Brookes JD, Hipsey MR, Burch MD, Regel RH, Linden LG, Ferguson CM, Antenucci JP. 2005. Relative value of surrogate indicators for detecting pathogens in lakes and reservoirs. Environ. Sci. Technol. 39:8614–8621 [DOI] [PubMed] [Google Scholar]
- 27. Viau EJ, Lee D, Boehm AB. 2011. Swimmer risk of gastrointestinal illness from exposure to tropical coastal waters impacted by terrestrial dry-weather runoff. Environ. Sci. Technol. 45:7158–7165 [DOI] [PubMed] [Google Scholar]
- 28. Boehm AB, Ashbolt NJ, Colford JM, Jr, Dunbar LE, Fleming LE, Gold MA, Hansel JA, Hunter PR, Ichida AM, McGee CD, Soller JA, Weisberg SB. 2009. A sea change ahead for recreational water quality criteria. J. Water Health 7:9–20 [DOI] [PubMed] [Google Scholar]
- 29. Tyrrell SA, Rippey SR, Watkins WD. 1995. Inactivation of bacterial and viral indicators in secondary sewage effluents, using chlorine and ozone. Water Res. 29:2483–2490 [Google Scholar]
- 30. Payment P. 1999. Poor efficacy of residual chlorine disinfectant in drinking water to inactivate waterborne pathogens in distribution systems. Can. J. Microbiol. 45:709–715 [PubMed] [Google Scholar]
- 31. Wright RC. 1989. The survival patterns of selected faecal bacteria in tropical fresh waters. Epidemiol. Infect. 103:603–611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Desmarais TR, Solo-Gabriele HM, Palmer CJ. 2002. Influence of soil on fecal indicator organisms in a tidally influenced subtropical environment. Appl. Environ. Microbiol. 68:1165–1172 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.