Abstract
Blood samples from Apodemus agrarius from Poland yielded PCR amplicons of Bartonella species. These included B. grahamii, B. taylorii, and B. birtlesii, as is typical of European Apodemus, as well as B. elizabethae-like forms and a recombinant strain of B. taylorii, most closely related to an American isolate from Tamiasciurus hudsonicus.
TEXT
The alphaproteobacterium Bartonella is associated with emerging infection in humans. Between 30 and 40 species have been described (1), primarily from rodents; of 1,400 deposited citrate synthase (gltA) sequences, over 530 are from rodents (2). In Europe, studies on Bartonella in rodents have focused on Apodemus flavicollis and Apodemus sylvaticus and the Myodes and Microtus voles (2–5). These have a characteristic Bartonella microbiota, dominated by B. grahamii, several clades of B. taylorii, B. doshiae, and B. birtlesii. Apodemus agrarius, also a common rodent in this region, has been almost ignored. This mouse recently migrated into Europe (6) from Central Asia and can therefore give insights into the evolution and phylogeography of zoonotic pathogens within Eurasia.
Apodemus agrarius mice were captured at Wroclaw Mokry Dwor (WMD; 51°04′57″N, 17°06′14″E) and Ruda Milicka (RM; 51°31′56″N, 17°20′12″E) during spring and autumn of 2009 to 2011, and whole blood collected into 0.001 M EDTA. Infection with Bartonella spp. was detected by PCR amplification of gltA (7). Other housekeeping genes, including ribC, groEL, and the virB5 locus, were amplified as described previously (2, 8). PCR products were sequenced, and phylogenetic analysis carried out using MEGA 5.10 (9). Of 94 A. agrarius mice (77 from RM and 17 from WMD), 36 (38.3%) were infected with Bartonella. The prevalence was higher in WMD than in RM (52.9% and 35.1%, respectively).
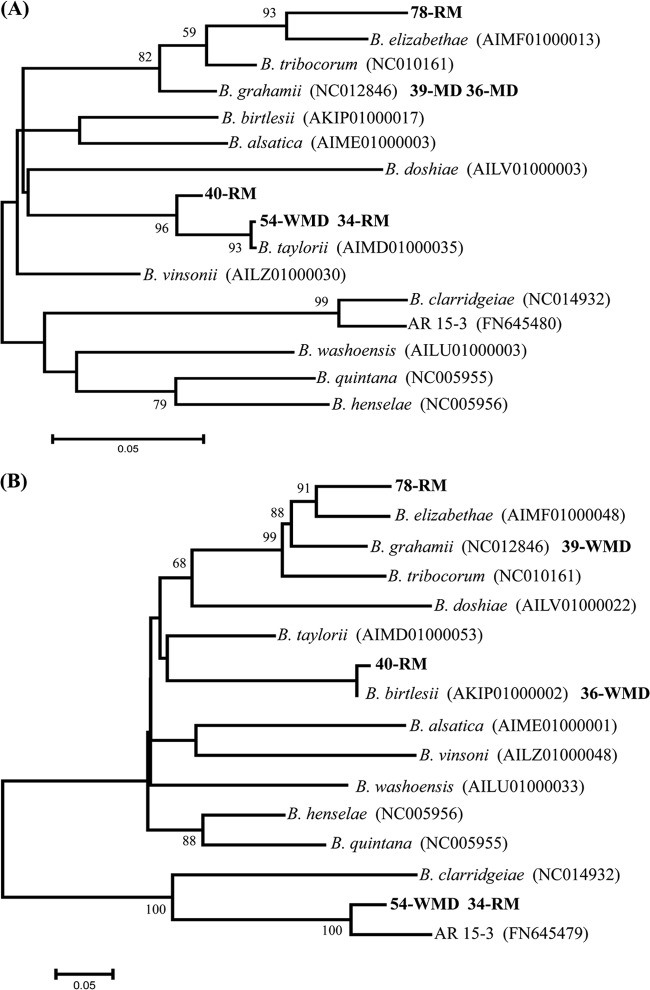
Six Bartonella sequences were analyzed in detail (Fig. 1). Based on gltA (Fig. 1A), 2 were identical to the B. grahamii-type isolate (accession number Z70016) and were not studied further. Three belonged to B. taylorii clades: 54-WMD and 34-RM showed 100% homology to sequences with GenBank accession numbers GU338950 and JQ694004, and 40-RM to sequences with accession numbers JQ694005 and GU338962. One (78-RM) was identical to Bartonella from A. agrarius in Slovakia (accession number DQ155392) and created a sister clade to Bartonella elizabethae, which normally infects rats.
Fig 1.
Maximum likelihood trees of (A) gltA gene fragment (Tamura 3-parameter model with gamma distribution) and (B) virB5 gene (general time reversible model with gamma distribution) for analyzed strains of Bartonella (boldface). Accession numbers of reference sequences obtained from GenBank are shown.
Sequencing of virB5 revealed 5 variants (Fig. 1B). One (39-WMD) was typical for B. grahamii (8, 10), and two (40-RM and 36-WMD) were most similar to B. birtlesii (98.9% and 100% homology to the sequences with accession numbers AKIP01000002 and AIMC01000045). The 78-RM Bartonella sequence, most similar to B. elizabethae at gltA, also grouped with this species at virB5 but with low homology (88.3% to the sequences with accession numbers AIMF01000048 and AILW01000028). groEL was also most similar to B. elizabethae but again with low homology (92.7%). Sequences 54-WMD and 34-RM, which by gltA were B. taylorii, were identical at virB5, forming a sister group to AR 15-3 (GenBank accession number FN645479), isolated from an American squirrel, Tamiasciurus hudsonicus, imported into Japan (11). The ribC gene of these strains also grouped with AR 15-3.
The Bartonella species from A. agrarius therefore include B. taylorii and B. grahamii, the typical species infecting rodents in this region. However, strains resembling B. elizabethae were also collected, while two others had ribC and virB5 sequences that were closest to a Bartonella from the American squirrel Tamiasciurus hudsonicus. Bartonella elizabethae is a worldwide rat pathogen, and the presence of a B. elizabethae-like Bartonella from A. agrarius could be explained by cross-infection from a rat-infecting form. However, this genotype is 5% different from B. elizabethae at the loci sequenced, and an identical isolate has been collected elsewhere from A. agrarius (GenBank accession number DQ155392). We therefore hypothesize that this clade is specific to A. agrarius, brought into Europe by the westward expansion of the host. Similarly, sequences most similar to those from a Bartonella species infecting American squirrels can best be explained by the westward migration of A. agrarius, bringing Asian Bartonella clades into western Europe. These clades clearly recombine freely, and the form most similar to isolate AR 15-3 is strongly introgressed with B. taylorii, having an AR 15-3-like virB5 and ribC and a B. taylorii gltA sequence. Recombination has strongly influenced other collected strains. One with B. grahamii gltA had virB5 identical to that of B. birtlesii, while another with B. taylorii gltA had virB5 very similar and groEL identical to those of B. birtlesii. This combination has been noted previously from Apodemus flavicollis in Northeast Poland (2).
We would interpret these results as evidence that A. agrarius brought a central Asian Bartonella microbiota with it during its expansion into Western Europe and that traces of this microbiota can still be discerned, despite widespread introgression into B. taylorii and B. grahamii. Key questions are the extent of host specificity shown by Bartonella clades and the barriers which may exist to recombination and the generation of novel variants. Numerous studies (e.g., references 1, 2, and 12) have led to disparate conclusions. On one hand, a strong association between specific Bartonella strains and certain rodents occurs, including B. elizabethae in rats or B. washoensis in squirrels (13, 14). On the other hand, B. grahamii has been isolated from rodents that include Apodemus, Microtus, and Myodes species (15, 16). A key element in host specificity appears to be the virB5 infectivity gene, which tracks host identity much more closely than other housekeeping genes (8). It is interesting to note that the A. agrarius-infecting strains can best be differentiated by their distinct virB5 genes, while other housekeeping genes have become introgressed with those of B. grahamii and B. taylorii. A further point to note is that the greatest diversity of Bartonella clades and of recombinant strains can be found within open, regenerating grassland habitats, the preferred habitat of A. agrarius, rather than within forest (2).
The expansion of A. agrarius west from Central Asia represents one of the most dramatic range extensions of a small mammal without human assistance; the expansion is complex and may have been made up of several phases (17), but most European records are less than 10,000 years before present (BP) (18), and the species did not occur in Poland before ca. 1,000 years BP (6). The role of this rodent as a link between rodent pathogen communities of the east and west Palaearctic region has been ignored by epidemiologists and evolutionary biologists, despite the potential importance of this species in the appearance of novel recombinant Bartonella clades within Western Europe.
Nucleotide sequence accession numbers.
virB5 gene sequences obtained in this study were deposited in the GenBank database under accession numbers KF051532 (40-RM), KF051533 (54-WMD), and KF051534 (78-RM).
Footnotes
Published ahead of print 7 June 2013
REFERENCES
- 1. Kosoy M, Hayman DT, Chan KS. 2012. Bartonella bacteria in nature: where does population variability end and a species start? Infect. Genet. Evol. 12:894–904 [DOI] [PubMed] [Google Scholar]
- 2. Paziewska A, Harris PD, Zwolińska L, Bajer A, Siński E. 2011. Recombination within and between species of the alpha proteobacterium Bartonella infecting rodents. Microb. Ecol. 61:134–145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Holmberg M, Mills JN, McGill S, Benjamin G, Ellis BA. 2003. Bartonella infection in sylvatic small mammals of central Sweden. Epidemiol. Infect. 130:149–157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Knap N, Duh D, Birtles R, Trilar T, Petrovec M, Avsic-Zupanc T. 2007. Molecular detection of Bartonella species infecting rodents in Slovenia. FEMS Immunol. Med. Microbiol. 50:45–50 [DOI] [PubMed] [Google Scholar]
- 5. Welc-Faleciak R, Paziewska A, Bajer A, Behnke JM, Siński E. 2008. Bartonella spp. infection in rodents from different habitats in the Mazury Lake District, Northeast Poland. Vector Borne Zoonotic Dis. 8:467–474 [DOI] [PubMed] [Google Scholar]
- 6. Kowalski K. 2001. Pleistocene rodents of Europe. Folia Quat. 72:3–389 [Google Scholar]
- 7. Norman AF, Regnery R, Jameson P, Greene C, Krause DC. 1995. Differentiation of Bartonella-like isolates at the species level by PCR-restriction fragment length polymorphism in the citrate synthase gene. J. Clin. Microbiol. 33:1797–1803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Paziewska A, Siński E, Harris PD. 2012. Recombination, diversity and allele sharing of infectivity proteins between Bartonella species from rodents. Microb. Ecol. 64:525–536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Berglund EC, Frank AC, Calteau A, Vinnere Pettersson O, Granberg F, Eriksson AS, Näslund K, Holmberg M, Lindroos H, Andersson SG. 2009. Run-off replication of host-adaptability genes is associated with gene transfer agents in the genome of mouse-infecting Bartonella grahamii. PLoS Genet. 5:e1000546. 10.1371/journal.pgen.1000546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. 2011. MEGA5: Molecular Evolutionary Genetics Analysis using maximum likelihood, evolutionary distance and maximum parsimony methods. Mol. Biol. Evol. 28:2731–2739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Inoue K, Maruyama S, Kabeya H, Hagiya K, Izumi Y, Une Y, Yoshikawa Y. 2009. Exotic small mammals as potential reservoirs of zoonotic Bartonella spp. Emerg. Infect. Dis. 15:526–532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Chomel BB, Boulouis HJ, Breitschwerdt EB, Kasten RW, Vayssier-Taussat M, Birtles RJ, Koehler JE, Dehio C. 2009. Ecological fitness and strategies of adaptation of Bartonella species to their hosts and vectors. Vet. Res. 40:29. 10.1051/vetres/2009011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Ying B, Kosoy MY, Maupin GO, Tsuchiya KR, Gage KL. 2002. Genetic and ecologic characteristics of Bartonella communities in rodents in southern China. Am. J. Trop. Med. Hyg. 66:622–627 [DOI] [PubMed] [Google Scholar]
- 14. Kosoy M, Murray M, Gilmore RD, Jr, Bai Y, Gage KL. 2003. Bartonella strains from ground squirrels are identical to Bartonella washoensis isolated from a human patient. J. Clin. Microbiol. 41:645–650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Birtles RJ, Harrison TG, Molyneux DH. 1994. Grahamella in small woodland mammals in the U.K.: isolation, prevalence and host specificity. Ann. Trop. Med. Parasitol. 88:317–327 [DOI] [PubMed] [Google Scholar]
- 16. Inoue K, Kabeya H, Kosoy MY, Bai Y, Smirnov G, McColl D, Artsob H, Maruyama S. 2009. Evolutional and geographical relationships of Bartonella grahamii isolates from wild rodents by multi-locus sequencing analysis. Microb. Ecol. 57:534–541 [DOI] [PubMed] [Google Scholar]
- 17. Aguilar JP, Pélissié T, Sigé B, Michaux J. 2008. Occurrence of the stripe field mouse lineage (Apodemus agrarius, Pallas 1771; Rodentia; Mammalia) in the late Pleistocene of southwestern France. C. R. Palevol. 7:217–225 [Google Scholar]
- 18. Toškan B, Kryštufek B. 2006. Noteworthy rodent records from the upper Pleistocene and Holocene of Slovenia. Mammalia 70:98–105 [Google Scholar]