Abstract
Water is a major route for infection of humans by exotoxin-producing bacteria, including Shiga toxin-producing Escherichia coli (STEC). While STEC has the potential to be present in nearly every type of water source, its distribution is sporadic, and an understanding of factors that govern its emergence and persistence within water is lacking. In this study, we examined the influence of microbe content on STEC persistence in freshwater. We found that depletion of microbes in the water leads to a considerable increase in the persistence of STEC, an effect that can be mitigated by adding grazing protists to the water. STEC strains appear to be more resistant to the impact of grazing protists than E. coli strains that lack the Shiga toxin (stx) gene. Our results demonstrate that the microcosm can dramatically influence the persistence of STEC in aquatic ecosystems and that the overall impact by microbes on STEC strains is fundamentally different from that of non-STEC strains of bacteria. Overall, these results provide insight into why STEC and possibly other exotoxin-producing bacterial pathogens display such variability in abundance, distribution, and persistence in aquatic ecosystems.
INTRODUCTION
Bacteria capable of producing exotoxins are common in the environment. These toxins kill host cells by disrupting their normal metabolic activities. Some examples of bacteria that contain such toxins include but are not limited to Clostridium, Vibrio, Escherichia, Staphylococcus, Shigella, and Bacillus species (1). The exotoxins produced by these bacteria are some of the most potent and lethal naturally occurring substances known to humankind. The United States Centers for Disease Control and Prevention lists over a dozen different diseases caused by exotoxins that afflict millions of people annually.
The majority of disease-causing exotoxins are classified as type III toxins. Many of these toxins are polypeptides organized in an A-B structural arrangement. Bacteria containing the genes for these types of exotoxins are responsible for diseases such as cholera, traveler's diarrhea, pertussis, shigellosis, and diphtheria (2). In these toxins, the A component is enzymatically active and alters the host cells' metabolic activity. The B component is responsible for receptor recognition at the host cell surface (3).
Shiga toxin (Stx) is one of the most intensively studied type III toxins. The B subunit of this polypeptide binds to the globotriaosylceramide-3 receptor on target cells, which allows the entry of the toxin into the host. Once inside the cell, the A subunit inhibits translation by inactivating the ribosome, ultimately causing cell death (4). There are two immunologically distinct forms of Stx, Stx1 and Stx2. These are further subcategorized on the basis of their biological activity and association with disease (5). Over 250,000 occurrences of Stx infection with over 60 human deaths are estimated to occur annually in the United States alone (6).
Transmission of this toxin to humans occurs through ingestion of food or water contaminated with Stx-producing bacteria. Although several bacterial types, including Shigella and Escherichia coli, harbor stx genes and/or produce Stx, Shiga toxin-producing E. coli (STEC) serotype O157:H7 is the most common bacterial type associated with human illness. In North America, E. coli O157:H7 is responsible for an average of over 75,000 illnesses annually, including an outbreak in 2001 due to a contaminated drinking water supply in Walkerton, Ontario, Canada, that resulted in approximately 2,500 illnesses and seven deaths (7, 8).
Drinking water is not the only aquatic source for transmitting STEC to humans. In fact, nearly every imaginable type of water system is capable of harboring E. coli O157:H7 and other STEC serotypes, including swimming pools, lagoons, wastewater, lakes, oceans, and rivers (9–14). Similar to the case in Walkerton, Ontario, exposure of humans to STEC in some of these aquatic ecosystems has led to human illness and death (15).
It is clear that water is a major reservoir of STEC and a significant source for transmission of STEC to humans in the environment. However, the factors that lead to the appearance, distribution, abundance, and persistence of this bacterial pathogen in aquatic ecosystems remain largely unknown. Significantly, studies examining the presence of E. coli O157:H7 genes or the Shiga toxin gene (stx) in the environment have found no correlation with the presence of traditional fecal indicator bacteria (FIB) that are used as a measure of feces-based contamination and water quality (16, 17). Together, these studies suggest that factors that govern the presence and survivability of STEC in aquatic environments are different from those that govern the presence and survivability of the more common classes of fecal coliforms.
Laboratory-based studies showed that STEC can kill grazing protists and is much more resistant to grazing by the protist Tetrahymena thermophila than non-STEC bacteria (18, 19). Additionally, studies have demonstrated an impaired ability of grazing protists to process ingested STEC (20). Together, these findings suggest that the higher resistance of STEC, in comparison to other bacterial types, to protist grazing could contribute to enhanced STEC persistence in aquatic environments.
In this study, we utilized quantitative PCR (qPCR) targeting E. coli to directly examine the role that grazing protists and other microbes have in mediating STEC survivability in recreational water. The qPCR approach has been shown to provide results that relate to E. coli colony counts enumerated on growth plates in water, can be used for rapid detection, and allows testing for other bacterial indicator genes in the sample (21). The results demonstrate the importance of the microcosm in influencing STEC persistence in aquatic environments, providing novel information on how STEC reacts to microbes in comparison to non-STEC bacterial strains. The extension of these studies to other exotoxin-producing bacteria and the implications that these findings have in the fields of microbial ecology, water quality management, and public health are considered.
MATERIALS AND METHODS
Detection probes.
The primers and probes targeting stx2 used in this study were ATTAACCACACCCCACCG (forward primer), GTCATGGAAACCGTTGTCAC (reverse primer), and CAGTTATTTTGCTGTGGATATACGAGGGCTTG (probe conjugated to a 6-carboxy-X-rhodamine on the 5′ end and black hole quencher 1 on the 3′ end). This primer, which recognizes the A subunit of the stx2 gene, detects several stx2 subtypes in water (22).
The primers and probes used to detect the uidA gene of E. coli were GTGTGATATCTACCCGCTTCGC (forward primer), AGAACGGTTTGTGGTTAATCAGGA (reverse primer), and TCGGCATCCGGTCAGTGGCAGT (probe conjugated to a 6-carboxy-X-rhodamine on the 5′ end and black hole quencher 1 on the 3′ end). These sequences specifically amplify the E. coli uidA gene and not genes from other closely related bacterial species and have also been used for E. coli detection in water (21, 23). All primers and probes were purchased from IDT Technologies.
Bacterial strains.
The STEC bacterial strains used in this study included the EDL933 strain obtained from the American Type Culture Collection (Manassas, VA), enterohemorrhagic E. coil (EHEC) O91:H21 isolate B2F1 (24), and EHEC O14:H4 isolate LB226692 (25). All STEC strains contain stx under the control of a phage lysogen. To test for phage induction, bacterial strains were grown to exponential phase and then treated with 1 μg ml−1 of mitomycin C or mock treated with water for 40 min. Following treatment, 1 ml of bacterial cells was centrifuged at 12,000 rpm for 5 min, and 200 μl of supernatant was subjected to DNase I treatment for 15 min at room temperature to remove free, nonencapsidated DNA, followed by protein inactivation by boiling for 5 min. Following boiling, 5 μl of the prepared sample was subject to qPCR targeting either the stx2 gene or the uidA gene. Serially diluted DNA from stock strain ATCC 43889 was used to create a standard curve that related the amplification signal of experimental samples to the amounts of the E. coli colonies on LB growth plates, which are represented as cell equivalents. Where needed, the resulting E. coli signal from the uidA primers was subtracted from the stx2 amplification signal to account for non-phage-encoding genes that were present in the sample, which yielded an stx2 concentration resulting from phage induction. The non-STEC bacterial strains used in this study included the common laboratory strain C600, the common laboratory strain MG1655 lysogenized with bacteriophage λ (λimm strain), and an EDL933Δstx strain, which is an EDL933 variant that bears deletions of the stx genes (26).
Water sample collection.
Water was obtained in sterilized 2-liter flasks from the beach areas of Presque Isle State Park (42°10′22.89″N, 80°05′29.46″W) or from Mill Creek stream (42°05′41.09″N, 80°04′20.71″W), located in Erie, PA. Collection and processing followed outlined procedures for collection and analysis of lake or stream freshwater (27). All water was maintained on ice and processed within a 2-h time frame.
Water sample treatment and analysis.
Water was separated into 50-ml aliquots and placed in separate 125-ml screw-top sterilized bottles (Corning). Depletion of microbes from some samples was achieved by filtration through a 20-nm-pore-size Anodisc alumina matrix filter (Whatman). To selectively deplete protists, water was filtered through a 5-μm-pore-size filter (Millipore). To recover protists, microbe-free water was used to resuspend protists from the 5-μm-pore-size filter through gentle agitation. This created a water sample that was reduced in viruses but retained protists. This approach to capture viruses depletes greater than 95% of all viruses, including all phages capable of infecting the indicator bacterial strain C600, which was determined by plaque assay (28) (results not shown). We estimated by cell counting that greater than 50% of the grazing protists were recovered off the filter by this method.
Bacterial addition to water was performed by centrifuging 1 ml of an exponentially growing bacterial culture grown in LB medium for 5 min at 5,000 rpm, removing the supernatant, and then resuspending the bacteria in the water to be used for the experiment to a final volume of 50 ml. The treatment of water with Tetrahymena thermophila occurred in a similar manner, with the exception that 20 ml of Tetrahymena cells grown in 2% proteose peptone was concentrated by centrifuging for 10 min at 3,000 rpm, followed by resuspension to a final concentration of 100 cells ml−1, a concentration which was determined by counting cells on a hemacytometer.
At various time points, 2 ml of water was filtered through a 0.45-μm-pore-size mixed cellulose ester filter (Fisher Scientific) to entrap bacteria. The filter was placed in 600 μl of 10 mM Tris, and bacterial DNA was released from the filter by boiling the sample for 7 min at 95°C. To determine the concentration of E. coli in each sample, qPCR targeting the uidA gene was performed on 5 μl of each sample following a previously outlined protocol (16). Serially diluted DNA from stock strain ATCC 43889 was used to create a standard curve that related the amplification signal of experimental samples to the amounts of the E. coli colonies on LB growth plates, which are represented as cell equivalents. Where indicated, statistical significance between the average of two treatment groups was determined by a P value of <0.05, obtained from a t test using Origin (version 6.0) software. All data presented represent typical results of experiments repeated two to three separate times from water samples obtained on different days.
RESULTS
Recreational water samples were obtained from Mill Creek in Erie, PA, and inoculated with the ATCC EDL933 STEC strain or EDL933Δstx, a strain of EDL933 that bears a deletion of all stx genes. We measured the persistence of these bacterial strains in water by qPCR (Fig. 1). The concentration of the EDL933Δstx strain in water did not change significantly over the first 48 h postinoculation (Fig. 1). In contrast, the amount of EDL933 exhibited a statistically significant increase of over 3-fold between 0 and 36 h, before declining at 48 h to a concentration similar to that of the initial inoculum (Fig. 1). A statistically significant increase in the concentration of EDL933 but not that of the EDL933Δstx strain within a 20- to 36-h time frame was also observed when they were incubated in Mill Creek water samples analyzed on other days (see Fig. S1A in the supplemental material). Similar results were also obtained with Presque Isle State Park beach water samples (see Fig. S1B in the supplemental material). After 48 h, the concentrations of both bacterial strains declined sharply (Fig. 1).
Fig 1.
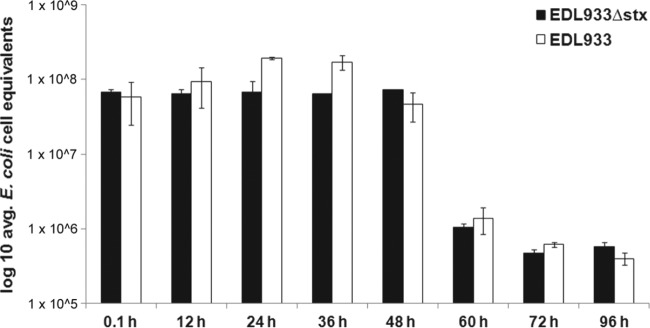
Bacterial strains were concentrated and added to recreational Mill Creek water as described in the text. Following incubation, E. coli cell equivalents in a fraction of the water were determined by qPCR at the indicated time points following incubation of the bacteria in the water. Error bars represent the standard error of the average of duplicate water samples treated in the same manner.
We compared the persistence of EDL933 in recreational water to that of two other STEC strains with inducible phage lysogens that contain and express stx genes (see Table S1 in the supplemental material). Similar to EDL933, these two other STEC strains persisted in this water for up to 20 h before beginning to decline. However, the persistence profiles of these STEC strains in this water differed from each other. Specifically, while the concentration of the EDL933 strain increased 2-fold within the first 20 h of incubation (Fig. 2), neither the B2F1 nor the LB226692 strain exhibited statistically significant growth. Additionally, we also found strain-specific differences in the rate at which different STEC strains declined in water. For example, the concentration of the EDL933 strain at 0 and 36 h postinoculation increased slightly, while the concentration of the LB226692 strain declined by >2-fold (Fig. 2). Regardless of these differences in persistence profiles within the first 36 h of incubation, the concentration of all STEC strains analyzed declined sharply after 60 h of incubation (Fig. 2).
Fig 2.
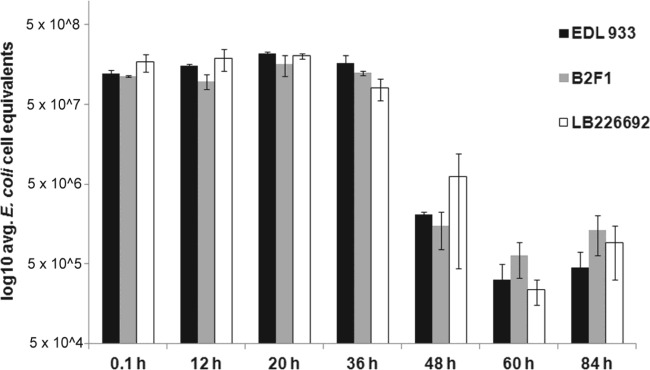
STEC strains were concentrated and added to 50 ml of recreational water obtained from Presque Isle State Park. At the indicated time points, 2 ml of the water sample was used to determine E. coli cell equivalents by qPCR. Error bars represent the standard error of average values of samples tested in duplicate for each STEC strain. Each STEC strain is listed in the key and described in Materials and Methods.
To substantially decrease total microbe levels in our water samples, we filtered the water through a 20-nm-pore-size filter. We used this microbe-depleted water to determine if the microcosm influences the persistence of STEC. We found that decreasing the total microbe level in freshwater facilitates a substantial increase in the persistence of STEC compared to that in unfiltered water (Fig. 3; compare the black bars and white bars). Specifically in unfiltered water, STEC levels decreased from an average of 7.7 × 107 E. coli cell equivalents immediately following inoculation to 5.5 × 105 E. coli cell equivalents 42 h later, a statistically significant difference between these two values. In contrast, over the same time period, the concentration of the same STEC strain incubated in water filtered to reduce the total microbe population was stable (Fig. 3). Interestingly, the concentration of STEC in microbe-depleted water did not exhibit any significant decrease in concentration until 110 h, or 5 days postinoculation. Even at 5 days postinoculation, the magnitude of the decrease in the STEC concentration was less than 2-fold compared to the concentration immediately postinoculation (Fig. 3). Similar results were obtained when microbes were inactivated in nonfiltered water following prolonged UV irradiation or were destroyed by autoclaving (see Fig. S2 in the supplemental material).
Fig 3.
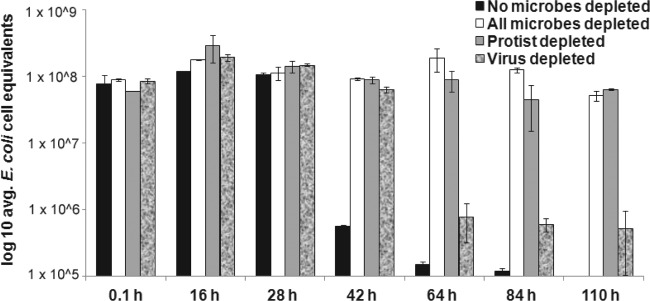
STEC strains were concentrated and added to 50 ml of recreational water obtained from Presque Isle State Park. Water samples were not filtered (black bars) or selectively filtered to greatly deplete all microbes (white bars), protists (gray bars), or viruses (textured bars). At the indicated time points, 2 ml of the water sample was used to determine E. coli cell equivalents by qPCR. Error bars represent the standard error of average values of samples tested in duplicate for each treatment group.
To further refine our analysis of which types of microbes might be influencing STEC persistence in freshwater, we employed selective filtration to reduce protist or virus levels in freshwater samples (Fig. 3). Our results show that depleting either protists or viruses from recreational freshwater increases STEC persistence. For example, at 42 h, the concentration of E. coli cell equivalents in water depleted of protists (Fig. 3, gray bars) or viruses (Fig. 3, shaded bars) was statistically significantly higher than the STEC concentration in water that contained these microbes (Fig. 3, black bars). Even after 110 h, the average levels of E. coli cell equivalents in the protist-depleted water sample were not statistically significantly different from the same value postinoculation (Fig. 3; compare gray bars postinoculation and at 110 h).
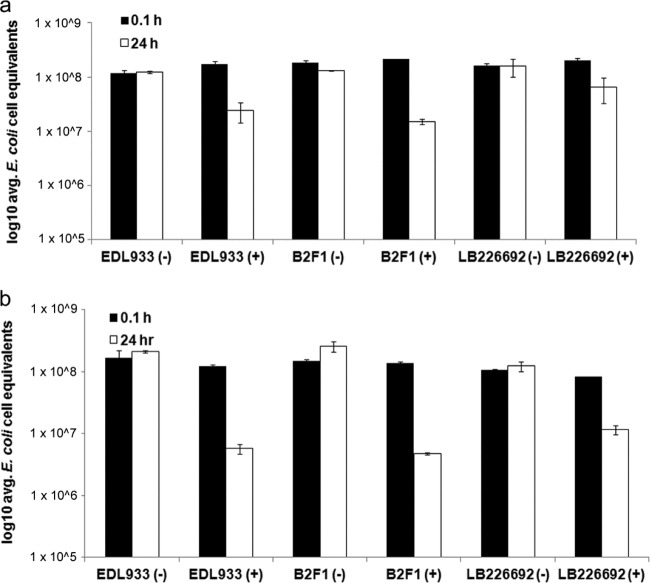
We directly tested the ability of the grazing protist Tetrahymena thermophila to reduce STEC levels in water by coincubating these two microbes with each other in freshwater samples (Fig. 4). In unfiltered water, all three STEC strains tested demonstrated statistically significant reductions in levels when Tetrahymena was added to the microcosm (Fig. 4a). In all cases, the presence of Tetrahymena decreased STEC levels by greater than 2-fold within a 24-h period compared to the levels in the same water samples that did not have Tetrahymena added to them (Fig. 4a). The ability of added Tetrahymena to decrease STEC persistence was greater in water samples that were filtered to reduce the endogenous microbe content (Fig. 4b). This effect was seen regardless of the STEC strain examined (Fig. 4b). However, the precise effect of Tetrahymena addition on bacterial persistence varied between the individual STEC strains. For example, in the water samples where the endogenous microbe content was kept intact, the level of the B2F1 strain was reduced by greater than 8-fold compared to that in water samples not treated with Tetrahymena, while under identical conditions, the concentration of the LB226692 strain was reduced by only approximately 2-fold, a statistically significant difference when these levels of reductions were compared (Fig. 4a). Similarly, in water samples that were prefiltered to remove native microbes, the concentration of the B2F1 STEC strain decreased by greater than 50-fold due to the presence of Tetrahymena within a 24-h period, while the concentration of the LB226692 strain decreased by only approximately 10-fold during the same time frame (Fig. 4b).
Fig 4.
STEC strains were concentrated and added to 50 ml of recreational water obtained from Presque Isle State Park. This water was left untreated to keep microbes from the water present (a) or filtered through a 20-nm-pore-size filter to greatly deplete all microbes (b). As indicated by the plus or minus signs in parentheses, some of the water samples were treated with the protist Tetrahymena thermophila, which, when present, was added at a concentration of 100 protists ml−1. At the indicated time points, 2 ml of the water sample was used to determine E. coli cell equivalents by qPCR. Error bars represent the standard error of average values of samples tested in duplicate for each STEC strain.
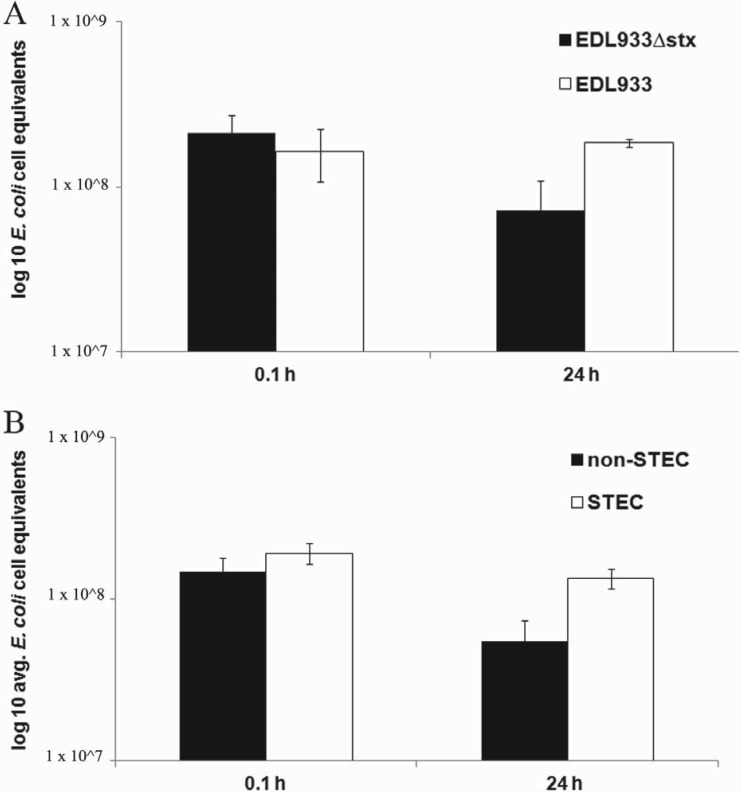
To determine the potential role of Stx expression on bacterial persistence in recreational freshwater, we next compared the persistence of STEC strains in the presence of Tetrahymena to that of non-STEC strains which do not contain or express Stx (Fig. 5). Our data demonstrate that the STEC strains utilized in this study are more resistant to protist grazing in recreational freshwater than the non-STEC strains (Fig. 5). For example, the concentration of the non-STEC strain EDL933Δstx decreased by nearly 3-fold within 24 h after the addition of Tetrahymena, while the concentration of the EDL933 STEC strain slightly increased during that same time frame when treated in the same way, a statistically significant difference when the different concentrations of these two strains of bacteria were compared (Fig. 5A). The same result was obtained when other STEC strains were compared to non-STEC strains. Specifically, the average amount of non-STEC strains of E. coli fell 2.5-fold, from 1.49 × 108 to 5.52 × 107 E. coli cell equivalents, in a 24-h period. In contrast, under identical conditions, the average amount of STEC in this water decreased only 1.4-fold during the same time frame (Fig. 5B). In all cases, the average levels of E. coli cell equivalents in control samples not treated with Tetrahymena were not statistically different between the postinoculation and 24-h readings (Fig. 4 and data not shown).
Fig 5.
Bacterial strains were concentrated and added to 50 ml of recreational water obtained from Presque Isle State Park. The protist Tetrahymena thermophila was added to the water sample at a concentration of 100 protists ml−1. At the indicated time points, 2 ml of the water sample was used to determine E. coli cell equivalents by qPCR. Error bars represent the standard error of average values of samples tested in duplicate for each STEC strain. A comparison of EDL933 to EDL933Δstx (A) or all three STEC strains to the non-STEC strains described in Materials and Methods (B) is shown.
DISCUSSION
Although STEC is present in many different aquatic ecosystems, studies that examine the factors regulating the persistence of this microbe in water environments are lacking. Here, we show that the EDL933 STEC strain can persist in a freshwater environment for up to 48 h before beginning to decline (Fig. 1). This persistence is marked by a statistically significant increase in the concentration of this microbe at between 20 and 36 h postinoculation. This phenomenon was not observed with the EDL933Δstx strain (Fig. 1). Since the only difference between these two strains is the ability to express Stx, we conclude that Stx must be acting to facilitate the survival of E. coli when initially introduced in freshwater.
Interestingly, we did not observe a statistically significant increase in the concentration of STEC strains B2F1 and LB226692 in freshwater between the 20- and 36-h-postinoculation time frame as we did with the EDL933 strain (Fig. 2). The E. coli EDL933 strain is an O157:H7 serotype strain, while the B2F1 strain is an O91:H21 serotype strain and the LB226692 strain is an O14:H4 serotype strain. Serotype-dependent differences in the presence or expression of cell surface markers may be contributing to the observed differences in freshwater survivability of these different STEC strains. Consistent with this suggestion, mutation of the ecf operon has a dramatic impact on the membrane composition and persistence of O157:H7 strains in drinking water (29). Additionally, other as yet undiscovered gene products besides stx that are expressed by STEC may act in concert with stx to influence predator grazing. For example, in another bacterial species, Vibrio cholerae, an extracellular protease protects this bacterium from grazing by predators, including Tetrahymena (30). The presence or differential expression of genes such as these in some STEC strains but not others could explain the difference in the persistence profiles of the STEC strains observed in this study.
Alternatively, the precise isoform(s) of the Shiga toxins produced by a particular bacterial strain could influence the ability of STEC strains to survive in freshwater. All three strains studied here contain stx2 genes. However, the EDL933 and LB226692 strains contain the stx2a subtype, while the B2F1 strain contains two copies of the stx2d subtype. Both subtypes display similar potencies in vitro, but their potency in protist killing has not yet been studied (31). Moreover, of the strains studied, only EDL933 contains an stx1 gene. While this Shiga toxin variant appears to be less frequently associated with hemolytic-uremic syndrome in humans, its potency in killing protists grazing has not yet been explored and could provide one potential explanation as to why the EDL933 strain demonstrated enhanced survival within the first 24 h in freshwater compared to the survival of the B2F1 or LB226692 strain (5).
Additionally, the degree to which these stx genes are induced in these strains may also play a role in STEC persistence in freshwater. In all three strains, at least one copy of the stx-encoding genes is on a prophage, meaning that stx is produced upon induction of that prophage (4) (see Table S1 in the supplemental material). The stx2a genes in EDL933 and LB226692 are located on inducible prophages, whereas only one of the two stx2a genes, the stx2ad1 allele, in the B2F1 strain is located on an inducible phage. While the general arrangement of these genes on the different prophages is similar, these phages differ in their ability to be induced (and, consequently, the amounts of Stx produced differ) (see Table S1 in the supplemental material). Under controlled conditions in the laboratory, the EDL933 and LB226692 strains induced and expressed stx at similar levels, which were higher than the level observed for the B2F1 strain (see Table S1 in the supplemental material). These expression profiles do not fully explain why the EDL933 strain exhibited greater survival in freshwater at early time frames than the other two STEC strains analyzed. However, phage induction can be influenced by both the type and the concentration of the inducing factor (32). For example, the Stx1- and Stx2-encoding phages in EDL933 respond differently to various inducing agents (33, 34). These inducing factors could also be present in the aquatic environment, such as UV sunlight and chemicals, or present within organisms, such as nutrients and reactive oxygen species (18, 35). Thus, the inducibility, the level of Stx expression, and the type of Stx expressed by the different STEC strains may all contribute to the differences in STEC survival in aquatic ecosystems. Regardless of the mechanism, our results demonstrate that the genetic background impacts the survivability of STEC in freshwater.
Although the genetic composition impacts the precise freshwater survivability of the STEC strains utilized in this study, all STEC strains were susceptible to grazing by protists (Fig. 3 and Fig. 4). However, the extent to which protists graze upon STEC is significantly reduced compared to that for non-STEC bacterial strains (Fig. 5). The non-STEC strains tested included the common laboratory strain C600, which does not contain a phage lysogen, and the common laboratory strain MG1655 lysogenized with bacteriophage λ. Both strains exhibited markedly lower survival within the first 24 h of incubation in freshwater than any of the STEC strains (Fig. 5 and data not shown). Additionally, a direct comparison of the EDL933 strain to the EDL933Δstx strain, which differs from EDL933 only in the absence of the stx genes, demonstrated a survival advantage of the STEC strain over that of the identical strain unable to produce Stx (Fig. 1). Together, the results of these studies indicate that the presence of stx genes in E. coli and not the induction of phage itself offers the bacteria a competitive advantage for survival in freshwater compared to the situation for bacteria that do not contain or express stx genes. These data are consistent with those from laboratory-based studies that have demonstrated that STEC strains can escape from grazing and kill protists due to the production of Stx (18–20). Thus, our results support the notion that the production of this exotoxin by bacteria provides a defense against protist grazing in freshwater (9). These data are, to our knowledge, the first evidence that the presence of stx genes can increase bacterial survival in an aquatic context.
The mechanisms by which bacterially produced exotoxins kill their host cells are distinct from one another. While Stx acts by inhibiting protein synthesis through modification of eukaryotic rRNA, diphtheria toxin inactivates protein synthesis by inactivating the eukaryotic elongation factor-2 protein (1). Other exotoxins, such as cholera toxin, heat-labile toxin, and pertussis toxin, act on eukaryotic cells through alterations made to signaling cascades (1). While the specific cellular targets differ, all of these exotoxins have the ability to influence the abundance, distribution, and function of grazing protists or other eukaryotic cells in the environment. Thus, similar to Stx, other exotoxins produced by bacteria may offer a competitive survival advantage to bacteria in water. We are currently testing the extent to which these and other exotoxins can act in such a way, and that work will test the generality of our findings that we report here for STEC.
Exotoxin-producing bacteria, including STEC, can be highly abundant in aquatic ecosystems (9). In the case of STEC, abundance and distribution patterns in water are difficult to predict since STEC levels do not appear to correlate with general E. coli levels or those of other fecal indicator bacteria that are used as a measure of water quality (16, 17). Our results reported here demonstrate that STEC interacts with the microcosm in freshwater in a fundamentally different way than non-STEC bacteria. This finding provides a possible explanation as to why STEC abundance in aquatic environments cannot be easily predicted by traditional bacteriological testing protocols. Specifically, we propose that STEC levels do not correlate with the overall levels of E. coli or the levels of other fecal indicator bacteria because of the enhanced ability of STEC to resist predation by grazing protists. In direct support of this, we show that the addition of Tetrahymena to freshwater decreased the concentration of E. coli without stx genes >3-fold in a 24-h period, while it did not significantly alter the concentration of STEC bacteria during the same time frame (Fig. 5). Additionally, we found a greater increase in the STEC concentration in Mill Creek water over a 20- to 36-h period than in Presque Isle State Park beach waters (Fig. 1; see Fig. S1 in the supplemental material). In addition to having a bacterial composition different from that of Presque Isle State Park beach water, Mill Creek water is characterized by a much higher concentration of grazing protists than Presque Isle State Park beach water (36) (data not shown). Our work here provides a starting point to clarify how naturally occurring grazing protist concentrations relate to the presence and abundance of STEC in water. This work should lead to a better understanding of how STEC relates to non-STEC and other fecal indicator bacteria in water, work which will inevitably influence how water management decisions are made.
Currently, nearly all recreational water managers at locations in the United States and worldwide utilize a type of fecal indicator bacteria plating approach to make decisions regarding the suitability of water for human occupancy, a process which takes 24 to 48 h or longer to complete. During this time, our data indicate that STEC isolates that would have been present in the water on the day of testing would begin to be significantly reduced by the time that the results of testing are available to make management decisions, making the procedure of little value to protect the public from exposure to this human pathogen. Thus, even if we do not yet know the exact role that protists have in mediating STEC persistence in all water types, our results provide an impetus for developing faster and more specific protocols to warn the public of unsafe swimming conditions due to the presence of STEC in the environment.
Supplementary Material
ACKNOWLEDGMENTS
This work was funded by grants from the National Science Foundation to G.B.K. (MCB-0956454) and S.A.M. (DBI-0922718) and from Mercyhurst University.
We also acknowledge and thank members of the G. B. Koudelka and S. A. Mauro laboratories for providing input on this work prior to publication.
Footnotes
Published ahead of print 7 June 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.01281-13.
REFERENCES
- 1. Burns BL, Barbieri JT, Iglewski BH, Rappuoli R. (ed). 2003. Bacterial protein toxins. ASM Press, Washington, DC [Google Scholar]
- 2. Stedman TL. 2006. Stedman's medical dictionary, 28th ed Lippincott Williams & Wilkins, Baltimore, MD [Google Scholar]
- 3. Saelinger CB. 2003. Receptors for bacterial toxins, p 131–148 In Burns BL, Barbieri JT, Iglewski BH, Rappuoli R. (ed), Bacterial protein toxins. ASM Press, Washington, DC [Google Scholar]
- 4. Tyler JS, Livny J, Friedman DI. 2005. Lambdoid phages and Shiga toxin, p 131–164 In Waldor MK, Friedman DI, Adhya SL. (ed), Phages: their role in bacterial pathogenesis and biotechnology. ASM Press, Washington, DC [Google Scholar]
- 5. Gyles CL. 2007. Shiga toxin-producing Escherichia coli: an overview. J. Anim. Sci. 85:E45–E62 [DOI] [PubMed] [Google Scholar]
- 6. Heiman K, Fullerton K, Mody R, Strockbine N. 2012. National enteric disease surveillance: STEC surveillance overview. CDC, U.S. Department of Health and Human Services, Atlanta, GA [Google Scholar]
- 7. Rangel JM, Sparling PH, Crowe C, Griffin PM, Swerdlow DL. 2012. Epidemiology of Escherichia coli O157:H7 outbreaks, United States, 1982–2002. Emerg. Infect. Dis. 11:603–609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Anonymous 2012. Waterborne disease outbreak. epiTRENDS 17(6):1–4 [Google Scholar]
- 9. Mauro SA, Koudelka GB. 2011. Shiga toxin: expression, distribution, and its role in the environment. Toxins 3:608–625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. McCarthy TA, Barrett NL, Hadler JL, Salsbury B, Howard RT, Dingman DW, Brinkman CD, Bibb WF, Cartter ML. 2001. Hemolytic-uremic syndrome and Escherichia coli O121 at a lake in Connecticut. Pediatrics 108:E59. 10.1542/peds.108.4.e59 [DOI] [PubMed] [Google Scholar]
- 11. Casas V, Miyake J, Balsley H, Roark J, Telles S, Leeds S, Zurita I, Breitbart M, Bartlett D, Azam F, Rohwer F. 2006. Widespread occurrence of phage-encoded exotoxin genes in terrestrial and aquatic environments in Southern California. FEMS Microbiol. Lett. 261:141–149 [DOI] [PubMed] [Google Scholar]
- 12. Blanch AR, Garcia-Aljaro C, Muniesa M, Jofre J. 2003. Detection, enumeration and isolation of strains carrying the stx2 gene from urban sewage. Water Sci. Technol. 47:109–116 [PubMed] [Google Scholar]
- 13. Chern EC, Tsai YL, Olson BH. 2004. Occurrence of genes associated with enterotoxigenic and enterohemorrhagic Escherichia coli in agricultural waste lagoons. Appl. Environ. Microbiol. 70:356–362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Muniesa M, Jofre J. 2000. Occurrence of phages infecting Escherichia coli O157:H7 carrying the Stx2 gene in sewage from different countries. FEMS Microbiol. Lett. 183:197–200 [DOI] [PubMed] [Google Scholar]
- 15. Muniesa M, Jofre J, García-Aljaro C, Blanch AR. 2006. Occurrence of Escherichia coli O157:H7 and other enterohemorrhagic Escherichia coli in the environment. Environ. Sci. Technol. 40:7141–7149 [DOI] [PubMed] [Google Scholar]
- 16. Smith CJ, Olszewski AM, Mauro SA. 2009. Correlation of Shiga toxin gene frequency with commonly used microbial indicators of recreational water quality. Appl. Environ. Microbiol. 75:316–321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Walters SP, Thebo AL, Boehm AB. 2011. Impact of urbanization and agriculture on the occurrence of bacterial pathogens and stx genes in coastal waterbodies of central California. Water Res. 45:1752–1762 [DOI] [PubMed] [Google Scholar]
- 18. Lainhart W, Stolfa G, Koudelka GB. 2009. Shiga toxin as a bacterial defense against a eukaryotic predator, Tetrahymena thermophila. J. Bacteriol. 191:5116–5122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Stolfa G, Koudelka GB. 2013. Entry and killing of Tetrahymena thermophila by bacterially produced Shiga toxin. mBio 4(1):e00416–12. 10.1128/mBio.00416-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Gourabathini P, Brandl MT, Redding KS, Gunderson JH, Berk SG. 2008. Interaction between food-borne pathogens and protozoa isolated from lettuce and spinach. Appl. Environ. Microbiol. 74:2518–2525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Noble RT, Blackwood AD, Griffith JF, McGee CD, Weisberg SB. 2010. Comparison of rapid quantitative PCR-based and conventional culture-based methods for enumeration of Enterococcus spp. and Escherichia coli in recreational waters. Appl. Environ. Microbiol. 76:7437–7443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Ibekwe MA, Watt PM, Grieve CM, Sharma VK, Lyons SR. 2002. Multiplex fluorogenic real-time PCR for detection and quantification of Escherichia coli O157:H7 in dairy wastewater wetlands. Appl. Environ. Microbiol. 68:4853–4862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Frahm E, Obst U. 2003. Application of the fluorogenic probe technique (TaqMan PCR) to the detection of Enterococcus spp. and Escherichia coli in water samples. J. Microbiol. Methods 52:123–131 [DOI] [PubMed] [Google Scholar]
- 24. Melton-Celsa AR, Darnell SC, O'Brien AD. 1996. Activation of Shiga-like toxins by mouse and human intestinal mucus correlates with virulence of enterohemorrhagic Escherichia coli O91:H21 isolates in orally infected, streptomycin-treated mice. Infect. Immun. 64:1569–1576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Mellmann A, Harmsen D, Cummings CA, Zentz EB, Leopold SR, Rico A, Prior K, Szczepanowski R, Ji Y, Zhang W, McLaughlin SF, Henkhaus JK, Leopold B, Bielaszewska M, Prager R, Brzoska PM, Moore RL, Guenther S, Rothberg JM, Karch H. 2011. Prospective genomic characterization of the German enterohemorrhagic Escherichia coli O104:H4 outbreak by rapid next generation sequencing technology. PLoS One 6:e22751. 10.1371/journal.pone.0022751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Gobert AP, Vareille M, Glasser AL, Hindre T, de Sablet T, Martin C. 2007. Shiga toxin produced by enterohemorrhagic Escherichia coli inhibits PI3K/NF-κB signaling pathway in globotriaosylceramide-3-negative human intestinal epithelial cells. J. Immunol. 178:8168–8174 [DOI] [PubMed] [Google Scholar]
- 27. Clescerl LS, Greenberg AE, Eaton AD. 1985. Standard methods for examination of water and wastewater. American Public Health Association, Washington, DC [Google Scholar]
- 28. Olszewski A, Smith CJ, Mauro SA. 2010. Seasonal variation of viral abundance in Lake Erie beach waters of Presque Isle State Park. J. Penn. Acad. Sci. 84:55–59 [Google Scholar]
- 29. Yoon JW, Lim JY, Park YH, Hovde CJ. 2005. Involvement of the Escherichia coli O157:H7(pO157) ecf operon and lipid A myristoyl transferase activity in bacterial survival in the bovine gastrointestinal tract and bacterial persistence in farm water troughs. Infect. Immun. 73:2367–2378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Vaitkevicius K, Lindmark B, Ou G, Song T, Toma C, Iwanaga M, Zhu J, Andersson A, Hammarstrom ML, Tuck S, Wai SN. 2006. A Vibrio cholerae protease needed for killing of Caenorhabditis elegans has a role in protection from natural predator grazing. Proc. Natl. Acad. Sci. U. S. A. 103:9280–9285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Fuller CA, Pellino CA, Flagler MJ, Strasser JE, Weiss AA. 2011. Shiga toxin subtypes display dramatic differences in potency. Infect. Immun. 79:1329–1337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Corogeanu D, Willmes R, Wolke M, Plum G, Utermohlen O, Kronke M. 2012. Therapeutic concentrations of antibiotics inhibit Shiga toxin release from enterohemorrhagic E. coli O104:H4 from the 2011 German outbreak. BMC Microbiol. 12:160. 10.1186/1471-2180-12-160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Imamovic L, Muniesa M. 2012. Characterizing RecA-independent induction of Shiga toxin 2-encoding phages by EDTA treatment. PLoS One 7:e32393. 10.1371/journal.pone.0032393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. O'Brien AD, Newland JW, Miller SF, Holmes RK, Smith HW, Formal SB. 1984. Shiga-like toxin-converting phages from Escherichia coli strains that cause hemorrhagic colitis or infantile diarrhea. Science 226:694–696 [DOI] [PubMed] [Google Scholar]
- 35. McDaniel L, Paul JH. 2005. Effect of nutrient addition and environmental factors on prophage induction in natural populations of marine Synechococcus species. Appl. Environ. Microbiol. 71:842–850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Lybrook RP. 2006. Operation creek sweep—surface water E. coli assessment of major tributaries to the western portion of the Lake Erie watershed, Erie County. Pennsylvania Department of Environmental Protection, Northwest Regional Office, Meadville, PA [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.