Abstract
Escherichia coli is divided into four main phylogenetic groups, which each exhibit ecological specialization. To understand the population structure of E. coli in its primary habitat, we directly assessed the relative proportions of these phylogroups from the stools of 100 healthy human subjects using a new real-time PCR method, which allows a large number of samples to be studied. The detection threshold for our technique was 0.1% of the E. coli population, i.e., 105 CFU/g of feces; in other methods based on individual colony analysis, the threshold is 10%. One, two, three, or four phylogenetic groups were simultaneously found in 21%, 48%, 21%, and 8% of the subjects, respectively. Phylogroups present at a threshold of less than 10% of the population were found in 40% of the subjects, revealing high within-individual diversity. Phylogroups A and B2 were detected in 74% and 70% of the subjects, respectively; phylogroups B1 and D were detected in 36% and 32%, respectively. When phylogroup B2 was dominant, it tended not to cooccur with other phylogroups. In contrast, other phylogroups were present when phylogroup A was dominant. These data indicate a complex pattern of interactions between the members of a single species within the human gut and identify a reservoir of clones that are present at a low frequency. The presence of these minor clones could explain the fluctuation in the composition of the E. coli microbiota within single individuals that may be seen over time. They could also constitute reservoirs of virulent and/or resistant strains.
INTRODUCTION
Escherichia coli is the most common commensal aerobic bacterium in the human gut microbiota (1); it is also the Gram-negative bacillus most frequently implicated in human extraintestinal infections (2). This apparent paradox, which characterizes opportunistic pathogens, is classically associated with host deficiencies (for example, as a result of immune-compromising illness, surgery, or catheterization). However, there are still questions concerning the role of the intrinsic properties of the parasite and the conditions that may cause commensal intestinal E. coli strains to become extraintestinal pathogens.
Four main phylogenetic groups named A, B1, B2, and D have been distinguished among E. coli strains (3, 4), and distribution within these groups appears to correlate with the origin of the strains. Phylogroup B2 has been shown to include extraintestinal virulent strains (extraintestinal pathogenic E. coli [ExPEC]), which express numerous virulence factors (5, 6), whereas phylogroups A and B1 contain mostly human and animal commensal strains, respectively (7, 8, 9). However, more recently, an increase in B2 phylogroup strains was observed in human commensal strains originating from industrialized countries (10, 11, 12, 13).
Little is known about the diversity, transmission, and persistence of E. coli commensal strains within human populations (1). An individual can be colonized by more than one distinct strain at any given time (14, 15, 16). However, the few available studies that describe strain diversity are based on the arbitrary selection of colonies from a plate inoculated with fecal samples. The typing methods used included O typing (14, 17); biotyping (17); multilocus enzyme electrophoresis (18, 19, 20); and genotyping methods, such as pulsed-field gel electrophoresis (21), enterobacterial repetitive intergenic consensus (ERIC)-PCR (22, 23), random amplified polymorphic DNA (RAPD) alone (24) or combined with biochemical fingerprinting (25), ribotyping (26), and triplex phylogrouping PCR (27). The studies included a small number of subjects and/or selected a relatively small number of colonies on which to base their investigations of E. coli population structure. The ability to accurately characterize E. coli strain diversity in human subjects, and the likelihood of identifying a particular E. coli strain, are directly related to the number of colonies sampled and the underlying prevalence of the strain (21, 26). For example, for a 90% likelihood of identifying a strain present in ≥10% of colonies, 22 colonies must be sampled (21). New approaches are therefore needed to improve the sensitivity of detection. The detection of fecal strains present at low frequencies is necessary to understand within-individual fluctuations in strain composition that can be observed over time (18, 20, 28, 29).
To obtain insights into the population structure of E. coli in its primary habitat, we assessed the relative proportions of the main E. coli phylogenetic groups in the stools of 100 healthy human subjects; their intestinal microbiota had undergone minimal pathological and medical perturbations. A new, rapid, and sensitive real-time PCR strategy, using the same targets as triplex phylogrouping PCR (4), was applied to samples from the subjects.
MATERIALS AND METHODS
Subjects.
From May 2009 to December 2011, we recruited 100 healthy human subjects from the region of Ile-de-France (Paris, France, and its suburban area). All participants lived in the community and volunteered to self-collect a fecal swab sample. The subjects had no history of gastrointestinal disease and no symptoms of immunodepression, had not received antibiotic therapy in the previous month, and had not been hospitalized in the 3 months preceding inclusion. Written informed consent was obtained from each participant, and the study was approved by the ethics evaluation committee of Institut National de la Santé et de la Recherche Médicale (INSERM) (CCTIRS no. 09.243, CNIL no. 909277, and CQI no. 01-014).
Samples and characterization of isolated strains.
Fecal samples were self-collected by the subjects using Amies transport medium swabs (Venturi Transystem; Copan, Brescia, Italy) and sent by mail to the Avicenne hospital laboratory (Bobigny, France). The swabs were discharged in glycerol stock solution (Cryobank; Biovalley, Marne la Vallée, France) and stored at −80°C until they were used. The quantity of feces in 0.2 ml of solution was estimated in preliminary experiments to be 10 mg: E. coli counts from 10 swabs were compared to those from corresponding fresh feces, which were chosen for a variety of consistencies (data not shown). The stool-containing suspensions were plated onto Drigalski agar plates (Bio-Rad, Life Science, Marnes-la-Coquette, France). After 24 h of incubation at 37°C, one colony was randomly picked, identified as E. coli using API 20E (bioMérieux, Marcy l'Etoile, France), and stored in glycerol stock solution. Twenty to 30 E. coli colonies were randomly picked from each of 20 original plates and stored in glycerol stock solution at −80°C to serve as controls. The phylogenetic groups (A, B1, B2, and D) of the randomly picked E. coli strains were determined by the triplex PCR method (4) and by the recently reported quadriplex method, which also detects Escherichia clade strains (13, 30). The strains were tested for their antibiotic susceptibilities using the disk diffusion method according to the 2012 recommendations of the Comité de l'Antibiogramme de la Société Française de Microbiologie (Antimicrobial Committee of the French Society for Microbiology; http://www.sfm-microbiologie.org). The following antimicrobial agents were tested: amoxicillin, amoxicillin-clavulanic acid, cefoxitin, ceftriaxone, amikacin, ofloxacin, and sulfamethoxazole-trimethoprim.
DNA extraction from feces.
The QIAamp DNA Stool Minikit (Qiagen, Courtabœuf, France) was used to extract DNA from 0.2 ml of frozen stool sample according to the manufacturer's recommendations with modifications. An internal DNA control corresponding to the R′ 1 region of the hepatitis delta virus (HDV) genome prepared as previously described (31) was added, before extraction, at 105 copies per sample. The DNA was eluted in a final volume of 200 μl and stored at −80°C.
Quantitative-PCR assay. (i) Design of primers and phylogroup-specific probes.
The sequences used in the triplex PCR phylogrouping method (4) (TspE4.C2, chuA, and yjaA) were obtained from GenBank, EMBL, and the Broad Institute (http://www.broadinstitute.org) databases (from the whole-genome sequences of Escherichia strains) and aligned using the Clustal W program (provided by the European Bioinformatics Institute). The strains were assigned in vitro or in silico to their corresponding phylogroups by the triplex PCR method and/or by multilocus sequence typing (MLST). Primers were designed to anneal to conserved sequences, whereas probes were designed to target unique sites, allowing specific detection of each phylogroup with the Primer Express 3.0 software (Applied Biosystems, Villebon Sur Yvette, France) and the Primer3 Plus online interface (32) (provided by the Whitehead Institute). A list of strains tested in silico is given in Table S1 in the supplemental material. Two strains belonging to phylogroup D (TA255 and TA280) have a phylogroup F chuA sequence (probably resulting from horizontal gene transfer) and were identified as B2 by our method. Primers and probes were manufactured by Sigma-Aldrich (Lyon, France) and Applied Biosystems, respectively. All primers and probes used are listed in Table 1.
Table 1.
Primers and phylogroup-specific probes
| Target organism | Primers and probe | Sequence (5′–3′)a | Target gene | Reference or source |
|---|---|---|---|---|
| E. coli | ECFW | CATGCCGCGTGTATGAAGAA | 16S rRNA | 33 |
| ECRV | CGGGTAACGTCAATGAGCAAA | |||
| 16S rRNA gene | FAM-TATTAACTTTACTCCCTTCCTCCCCGCTGAA-TAMRA | |||
| E. coli phylogroup B1 | TspFW | CAACGCTACCTTGGCGTTATC | TspE4.C2 | This study |
| TspRV | CCGCTCTCCAGGCAACATC | |||
| pTspE4.C2B1 | FAM-ATCAGCGAGGCCGCGCGA-TAMRA | |||
| E. coli phylogroup B2 | chuAFW | ATTAACCCGGATACCGTTACCA | chuA | This study |
| chuARV | CTGCTGCTGATATGTGTTGAGC | |||
| pchuAB2 | FAM-CGTACCGACCCAACCA-MGB | |||
| E. coli phylogroup D | chuAFW | ATTAACCCGGATACCGTTACCA | chuA | This study |
| chuARV | CTGCTGCTGATATGTGTTGAGC | |||
| pchuAD | FAM-CGTACCAACCCACCCAA-MGB | |||
| E. coli phylogroups A1 and B2 | yjaAFW | CGCCTGTTAATCGCCAATTT | yjaA | This study |
| yjaARV | AAAAGAATGCCAGGTTGAACG | |||
| pyjaAA1/B2 | FAM-AAGTTCTGCAAGATCTTGTTCTGCAACTCCA-TAMRA |
FAM, 6-carboxyfluorescein; TAMRA, 6-carboxytetramethylrhodamine.
(ii) Phylogroup-specific real-time PCR.
Purified DNA (20 μl) was added to 30 μl of PCR mixture containing 25 μl of TaqMan Universal PCR master mix II (2×; Applied Biosystems), 300 nM each primer, 100 nM fluorescent probe, and bovine serum albumin at a final concentration of 0.1 μg/μl (New England BioLabs, Evry, France). However, for the B1 phylogenetic group, primers (TspFW and TspRV) and probe (pTspE4.C2B1) were used at 900 nM and 250 nM, respectively. The reaction mixture was heated to 50°C for 2 min (initiation step) and then 95°C for 10 min. This was followed by 45 cycles of amplification that each consisted of 15 s at 95°C and 1 min at 60°C. A no-template control and a positive-control sample, which had been quantified previously, were included in each run. The run was considered valid if the interassay coefficient of variation (CV) for the control sample was less than 30%. Each assay was performed in duplicate in the same run. The DNA extraction was considered to be valid if the difference between the cycle threshold (CT) of the internal-control HDV and the average CT (calculated as the average of the HDV CT values for all samples in the series) was less than the standard deviation. The reactions, data acquisition, and analyses were performed using the ABI Prism 7000 sequence detection system (Applied Biosystems). Standard curves made from known concentrations of DNA for each set of primers and probes were used for quantification.
(iii) Preparation of phylogroup-specific PCR standards.
Quantification controls were obtained using one representative strain for each phylogroup: K-12 (phylogroup A), IAI1 (phylogroup B1), ED1a (phylogroup B2), and UMN026 (phylogroup D). The Genomic DNA Buffer Set (Qiagen) and Qiagen genomic tips 100 G (Qiagen) were used to extract DNA from bacterial cultures according to the manufacturer's recommendations. Bacterial counts were obtained by plating, the concentration of purified DNA was determined with a spectrophotometer, and the corresponding copy number was calculated according to genome length. The two methods of quantification gave similar results (data not shown). Serial 10-fold dilution series for 8 × 107 to 12 × 107 down to 0.8 × 101 to 1.2 × 101 target genomes were applied for real-time PCR.
(iv) E. coli real-time PCR quantification assay.
Quantitative PCR for E. coli 16S rRNA genes was performed as previously described (33) using 20 μl of DNA purified from stool samples.
Control strains.
The following bacteria were used to evaluate the specificity of PCR primers and probes: E. coli sensu stricto ED1a, CFT073, 536, S88, IAI1, 55989, IAI39, ECOR36, DAECT14, K-12 MG1655, HS, H10407, 042, and UMN026; Escherichia clade I (ROAR 185 and M863), clade II (ROAR 19), clade III (ROAR 291 and ROAR 438), clade IV (E243 and B49), and clade V (ROAR 292 and ROAR 129); Escherichia fergusonii ATCC 35469; Escherichia albertii CIP107988; Escherichia blattae CIP104942; Escherichia vulneris CIP103177; Escherichia hermannii CIP103176; and Salmonella enterica serovar Typhimurium, Klebsiella pneumoniae, Proteus mirabilis, Pseudomonas aeruginosa ATCC 27853, Enterococcus faecalis ATCC 33186, Enterococcus faecium, Streptococcus bovis, Bacteroides fragilis, Bacteroides thetaiotaomicron, Bacteroides vulgatus, Fusobacterium nucleatum, Prevotella melaninogenica, Clostridium difficile, Clostridium perfringens, Bifidobacterium adolescentis, Bifidobacterium breve, Eubacterium exiguum, Eubacterium lentum, Lactobacillus casei, and Lactobacillus cateniformis.
Statistical analyses.
A factorial analysis of correspondence (FAC) (34) was used to describe associations among the data; a two-way table was analyzed using SPAD.N software (Cisia, Saint Mandé, France). The table had 98 rows (corresponding to the 98 subjects with E. coli identified in their stool samples) and 22 columns (corresponding to the 10 variables). The 10 variables were the phylogenetic group (A, B1, B2, or D) of the unique randomly selected strain, the dominant phylogenetic group, the intermediate phylogenetic group, and the minor phylogenetic group (defined below); the presence of a high quantitative level of E. coli in stools (more than 108 CFU per gram); subject gender; subject age (above or below 60 years old); and strain resistance to amoxicillin, ofloxacin, or sulfamethoxazole-trimethoprim. We used a binary code for each variable: present, 1; absent, 0. The FAC uses a covariance matrix based on χ2 distances. The computation determines a plane defined by two principal axes of the analysis. The first axis, F1, accounts for most of the variance, and the second axis, F2 (orthogonal to F1), accounts for the largest part of the variance not accounted for by F1 (34). The variables of the unique randomly selected strains were used as illustrative variables. All other variables were used to compute the plane.
RESULTS
Characteristics of the study population and of randomly selected E. coli commensal strains (one per individual).
The study included 46 males and 54 females between the ages of 26 and 86 years (median, 58.0 ± 11.7 years). All but two of the subjects carried E. coli. We studied one randomly selected clone per individual, which was considered to be the dominant clone. The resistance levels of these 98 E. coli dominant clones to amoxicillin, sulfamethoxazole-trimethoprim, amoxicillin-clavulanic acid, and ofloxacin were 25%, 14%, 7%, and 2%, respectively. No resistance to broad-spectrum cephalosporins or amikacin was detected.
The classical phylogroup triplex PCR method used on the dominant clones showed the prevalence of the different phylogroups to be as follows: phylogroup A, 31%; phylogroup B1, 13%; phylogroup B2, 33%; and phylogroup D, 21%.
Stool sample quantification of the four main E. coli phylogroups.
We developed a real-time PCR assay targeting the genes involved in the classical phylogrouping triplex PCR method (4) to directly quantify the four main E. coli phylogroups (A, B1, B2, and D) from stool samples. In this assay, the phylogroups B1 (chuA negative, yjaA negative, and TspE4.C2 positive), B2 (chuA positive, yjaA positive, and TspE4.C2 variable), and D (chuA positive, yjaA negative, and TspE4.C2 variable) were quantified by real-time PCR using probes specifically targeting B1 TspE4.C2, B2 chuA, and D chuA sequences, respectively (Table 2). The proportion of each phylogroup was calculated as a percentage of the total E. coli count obtained by quantitative PCR for 16S rRNA genes (33). The proportion of phylogroup A, which encompasses subgroups A0 (chuA, yjaA, and TspE4.C2 negative) and A1 (chuA negative, yjaA positive, and TspE4.C2 negative) (Table 2), was calculated by subtracting the proportions of phylogroups B1, B2, and D from the total E. coli count. The proportion of subgroup A1 was confirmed using an yjaA-specific probe (pyjaAA1/B2), which quantifies the A1 subgroup and the B2 phylogroup using the following formula: A1 = yjaApositive − B2. This pyjaAA1/B2 probe was used to estimate the lower limit of detection of the proportion of phylogroup A. The comparison between the proportion of phylogroup A (calculated by subtraction) and the proportion of subgroup A1 (quantified by the pyjaAA1/B2 probe) allowed us to estimate the threshold of phylogroup A detection by subtraction. Concordance was observed when the proportion of phylogroup A was at least 15% of the total E. coli count (data not shown). With this approach and using four probes (16S rRNA gene, pTspE4.C2B1, pchuAB2, and pchuAD), we estimated that phylogroup A was actually present at over 15% and that real-time PCR variations did not make a significant contribution to the results. This threshold value was further supported by comparison to PCR findings for several clones of plated fecal samples (see below).
Table 2.
E. coli phylogenetic group and subgroup determination using the results of PCR amplification of the chuA gene, yjaA gene, and DNA fragment TspE4.C2a
| E. coli phylogenetic group and subgroup | Presence of the sequenceb |
||
|---|---|---|---|
| chuA | yjaA | TspE4.C2 | |
| A0 | − | − | − |
| A1 | − | + | − |
| B1 | − | − | + |
| B22 | + | + | − |
| B23 | + | + | + |
| D1 | + | − | − |
| D2 | + | − | + |
As in reference 4.
+, present; −, absent.
Specificity, sensitivity, and reproducibility of the assay. (i) Assay specificity.
The specificity of the E. coli primers and probes was confirmed using an extensive set of control strains; this set (mainly gastrointestinal tract bacteria of non-Escherichia species [see Materials and Methods]) produced negative PCR results. The specificity of the 16S rRNA gene assay (33), as well as that of the phylogroup probes and primers, was tested using DNAs from various phylogenetic groups of E. coli sensu stricto and from closely related bacterial species of the genus Escherichia (including the recently described Escherichia clades [30, 35]). The 16S rRNA gene probe detects the phylogroups of E. coli sensu stricto, E. albertii, E. fergusonii, and the Escherichia clades, but not E. blattae, E. hermanii, and E. vulneris, in accordance with their taxonomic status (36, 37).
Each phylogroup-specific probe gave positive PCR results for the corresponding target bacteria and negative PCR results for nontarget microorganisms, with a small number of exceptions. The Escherichia clade III and IV strains, and some strains of clade I (ROAR 185), were recognized by the chuA B2 probe. The strains of the recently reported phylogroup F, defined by MLST (38), are considered D1 subgroup by the triplex PCR method (Table 2) but were split into B2 and D phylogroups by the real-time PCR method. This split corresponds to the different branches of the phylogroup F strains on the MLST tree (39). We did not detect a signal above the threshold for the nontemplate controls in any of the assays (CT > 40).
Spiking experiments were carried out to test the ability of our assay to pick out a specific bacterial DNA from a complex DNA background. Defined amounts of DNA from each of the studied phylogenetic groups B1, B2, and D were quantified by real-time PCR and spiked with DNA belonging to other phylogenetic group strains. No significant difference was observed, indicating high specificity of the probes used and no evidence of cross-reactivity.
(ii) Assay sensitivity, linearity, and efficiency.
The detection limits of the real-time PCR assays were examined using 10-fold serial dilutions of target DNAs extracted from pure culture of E. coli A, B1, B2, and D. The sensitivity of the probe specific for pyjaAA1/B2, pchuAB2, pchuAD, and pTspE4.C2B1 was below 10 CFU per PCR, which corresponded to approximately 9 × 104 CFU/g feces. The cycle number at which product fluorescence exceeded a defined threshold varied linearly over a range of concentrations from 106 to 10 bacteria per PCR mixture (r2 = 0.99) (data not shown). The linear range of all assays was therefore 9 × 109 to 9 × 104 bacteria per g of feces. The efficiencies of the PCRs were around 95%, as the slopes of the standard curves were close to the optimal theoretical values (see Table S2 in the supplemental material).
(iii) Assay reproducibility.
Based on the CT values of 15 replicates, the intra-assay reproducibility was found to be high (CV of CT values, <1%, corresponding to a CV of copies of <20%) (see Table S2 in the supplemental material).
Comparison with PCR typing of individual clones.
To test the robustness of our real-time PCR results, we compared our phylogroup quantification to the triplex PCR typing of 20 to 30 clones from 20 subjects. From the number of randomly selected colonies, the probability of detecting a minor clone (defined as a clone constituting up to 10% of the E. coli population) is estimated to be >90% (20). We found a correlation between our quantitative-PCR assay and the phylogroup determination of individual clones. In all cases, the phylogroups detected by the classical PCR typing of individual clones were also detected by the quantitative-PCR assay; we found similar quantitative trends but some differences in the proportions of the phylogroups. Interestingly, the real-time PCR assay detected minor clones, not found by plating, in 40% of the cases (see Table S3 in the supplemental material).
Quantitative determination of E. coli phylogenetic group composition.
Preliminary global quantification of E. coli in the stools of the subjects was carried out using real-time PCR with the 16S rRNA gene E. coli probe and primers (33). Negative real-time PCR results were found for the two subjects for whom E. coli also failed to be detected by plating. Excluding these two subjects, we found an average of 7.84 ± 0.54 log CFU of E. coli per gram of stool. The respective proportions of the four phylogenetic groups A, B1, B2, and D were determined for the 98 subjects for whom E. coli was detected. To verify that the use of transportation swabs sent by regular mail did not bias E. coli quantification, we compared real-time PCR results obtained from eight fresh control fecal samples and from their corresponding swabs, which were collected using the same procedure described in Materials and Methods, and no significant difference was found (see Table S4 in the supplemental material), in accordance with recent data on storage conditions of samples (40). However, if possible, analysis of fresh feces is preferred.
The phylogenetic groups A, B1, B2, and D were detected in 74%, 36%, 70%, and 32% of the subjects, respectively; the groups A and B2 were more frequently detected than B1 and D. However, the proportions of the four phylogroups varied widely among the subjects. The within-subject prevalence of each phylogenetic group varied from 0 to 100%. One, two, three, or four phylogenetic groups were found in 21%, 48%, 21%, and 8% of the subjects, respectively. When only a single group was detected, it was most commonly phylogroup B2 (62%); phylogroup A was found in 19% of subjects and phylogroups B1 and D in 9.5% of subjects. When the proportions of the phylogroups were defined according to three categories, as suggested by Schlager et al. (20) (dominant phylogenetic group, >50% of the E. coli population; intermediate phylogenetic group, 10 to 50% of the E. coli population; and minor phylogenetic group, <10% of the E. coli population), the proportions of the three categories varied significantly according to the four phylogenetic groups (Table 3). Phylogroups B2 and A were equally prevalent as the dominant group and in a higher proportion of samples than phylogroup B1 or D. Phylogroup A was the most common intermediate phylogenetic group, and phylogroup D was the most common minor phylogenetic group. Minor phylogroups (less than 10% of the total E. coli population) were detected in 40% of the samples.
Table 3.
Distribution of phylogenetic groups according to their proportions determined by quantitative PCR of fecal samples from 100 healthy volunteers
| Phylogroup | Distribution (%)a |
||
|---|---|---|---|
| DPG | IPG | MPG | |
| A | 38 | 27 | 35 |
| B1 | 13 | 10 | 77 |
| B2 | 37 | 13 | 50 |
| D | 11 | 5 | 84 |
DPG, dominant phylogenetic group (>50%); IPG, intermediate phylogenetic group (10 to 50%); MPG, minor phylogenetic group (<10%) (20).
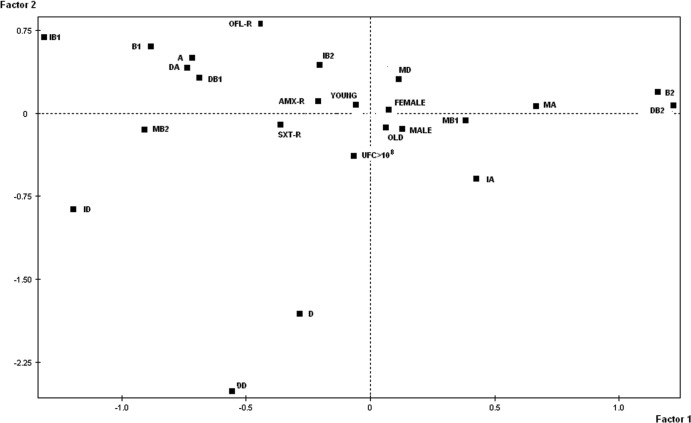
When the 12 combinations (three categories and four phylogenetic groups) were considered, the proportions of the categories varied according to the phylogenetic group considered. This complex variation is described by a FAC (Fig. 1). On the F1-F2 plane of the FAC, which accounted for 32.7% of the total variance, the dominant phylogenetic group B2 was distinguished by the positive values of the first factor, whereas the dominant phylogenetic groups B1, A, and D were distinguished by the negative values of this factor. The intermediate phylogenetic groups B1, B2, and D were projected on the negative values of the F1 axis and thus appeared to be associated with the dominant phylogenetic groups A and B1. The intermediate phylogenetic group A and the minor phylogenetic groups A, B1, and D were projected on the positive values of the F1 axis and thus are associated with the dominant phylogenetic group B2. This analysis allows us to distinguish between two types of structures within the E. coli intestinal commensal populations, according to the major dominant group B2 or A. The dominant phylogenetic group B2 was associated with the absence of the other groups, whereas the dominant phylogenetic group A was associated with variable proportions of other groups. The second axis, F2, characterized the dominant phylogenetic group D by its negative value. No link with the absolute quantity of E. coli bacteria was observed, as the variable >108 CFU was projected near the origin of the axes and thus was not related to phylogenetic repartition.
Fig 1.
FAC for the 98 subjects with E. coli found in their feces. Projections on the plane F1-F2 of the four phylogenetic groups (A, B1, B2, and D) of the unique randomly selected strains, of the dominant phylogenetic group (DA, DB1, DB2, or DD), of the intermediate phylogenetic group (IA, IB1, IB2, or ID), and of the minor phylogenetic group (MA, MB1, MB2, or MD) defined for each of the four phylogenetic groups are shown. The dominant, intermediate, and minor groups were defined from data from the quantitative-PCR assay. The abundance of E. coli in stools (CFU > 108); the gender of the subjects (male or female); the age of the subjects (older or younger than 60 years); and the resistance of the unique randomly selected strains to amoxicillin (AMX-R), ofloxacin (OFL-R), and sulfamethoxazole-trimethoprim (SXT-R) were also projected on the plane.
E. coli phylogenetic group composition: relevance of subject characteristics and the phylogroup and antibiotic resistance of a randomly selected clone.
There is no significant difference in the phylogenetic group composition according to the age or gender of the subjects: these variables were projected near the origin of the axes on the FAC (Fig. 1).
In 78% of the cases, the dominant phylogenetic group determined by the real-time PCR was identical to the one determined by the method of Clermont et al. (4) on one randomly selected clone. In 22% of the cases, the phylogenetic group of the randomly selected clone was present in the sample, but as an intermediate or minor phylogenetic group. There was a discrepancy in six cases, which corresponded to the selected clone being identified as phylogroup D whereas the quantitative PCR identified it as belonging to phylogroup B2. We have shown that in all six of these cases, the randomly selected clone belonged to phylogroup F (13): this is supported by the fact that the pchuAB2 probe that we designed is positive for some F group strains (see Table S3 in the supplemental material). The good correlation between the quantitative-PCR assay and classical phylogroup determination (based on one randomly selected clone) is shown in Table 3, where the proportions of the dominant phylogenetic groups (A, 38%; B1, 13%; B2, 37%; D, 11%) are close to the proportions detected in the randomly selected clones (A, 31%; B1, 13%; B2, 33%; D, 21%), as well as on the FAC (Fig. 1). The phylogenetic group of the randomly selected clone (considered an illustrative variable) is projected close to the dominant phylogenetic group on the plane, with the exception of the D phylogroup variable on the negative values of F2, which is attracted by the dominant phylogroup B2 due to the six cases of discrepancy. The resistance levels of the randomly selected clones to amoxicillin, sulfamethoxazole-trimethoprim, or ofloxacin were also considered illustrative variables. They were associated with the dominant A phylogroup for negative values of F1 and distantly related to the dominant B2 phylogroup on the FAC (Fig. 1).
DISCUSSION
We have developed an original quantitative-PCR assay, based on the classical phylotyping method of Clermont and colleagues (4), to determine the relative proportions of the four main E. coli phylogroups, A, B1, B2, and D. On average, 7.84 ± 0.54 log CFU of E. coli were found per gram of feces, in accordance with previous studies (33, 41, 42). When assessing the population structure of intestinal commensal bacteria in humans, it is necessary to study subjects whose microbiotas are not perturbed by external factors, such as antibiotic consumption, hospitalization, or intestinal disease. We were able to study real commensal E. coli populations, as our 100 healthy subjects conformed to strict inclusion criteria; we excluded any potential subject with a physiopathological condition that could disturb the intestinal microbiota equilibrium. The level of acquired resistance to antibiotics in our study population was much lower than that reported in community-acquired E. coli urinary tract infections in France (ONERBA 2008 and AFORCOPI-BIO network 2007 [http://www.onerba.org]): amoxicillin, 25% versus 44%; sulfamethoxazole-trimethoprim, 14% versus 20%; amoxicillin-clavulanic acid, 7% versus 24%; and ofloxacin, 2% versus 11%. This supports our belief that the study populations are commensal and in the same range of resistance found in a recent study in Paris on fecal strains (43).
The specificity of our assay is satisfactory, although it has some limitations. Some strains of the recently reported Escherichia clades (30, 35) gave a signal with the pchuAB2 probe, which can falsely increase the proportion of phylogroup B2. However, as these clade strains were yjaA negative, the opposite of the E. coli B2 strains, a strong B2 signal with a weak or negative yjaA signal should indicate the presence of clade I, III, or IV strains. The prevalence of these Escherichia clades in human commensal populations is very low (around 2 to 3%) (35) and thus will not significantly affect the results of our assay. We did not detect any Escherichia clade among the 100 randomly selected clones using the new Clermont phylotyping method (13) (see Table S3 in the supplemental material). Some strains classified as phylogroup D by the classical triplex PCR method and belonging to the recently reported phylogroup F (identified by MLST [38]) appeared to belong to phylogroup B2 due to a positive signal with the pchuAB2 probe. Phylogroup F is closely related to phylogroup B2 (1). This probe cross-reactivity was responsible for all six discrepancies observed between our quantitative-PCR assay and the classical triplex PCR phylogrouping of the randomly selected clones (see Table S3 in the supplemental material). However, it may not significantly alter the results, as phylogroup F strains have been found to represent less than 8% of the population in a large cohort of fecal strains from Australia and France (13).
We have shown that our assay is highly reproducible, with a detection threshold of 105 CFU/g of feces; this corresponds to 0.1% of the E. coli population, much lower than the 10% threshold obtained by other methods (20, 21, 23, 26, 29). Previous studies picked 20 to 30 isolates from an agar plate per individual and used a semiquantitative method of assessment (20, 23, 25). We found good correlation between data obtained with such an approach and that obtained using our quantitative method (see Table S3 in the supplemental material). However, real-time PCR may be faster, easier to perform, and more suitable for large sample sizes. In 40% of cases (8/20), real-time PCR detected phylogroups that were not found by plating. Our specific probes were able to detect groups with very low prevalence thanks to a low detection threshold. To reach a level of detection (0.1%) similar that in our assay by the alternative semiquantitative method, a large number of isolates would have to be studied from each fecal sample. The classical qualitative method appeared to underestimate the presence of minor groups. This explains the apparent differences in fecal carriage of the E. coli phylogroups (e.g., the prevalence of the B2 phylogroup in the studied population is apparently 33% if only major clones are considered but 70% if all clones, including minor clones, are considered). We acknowledge that, in giving a global estimation of the structure of the commensal strains, our method does not allow us to study the E. coli clones separately.
Each person commonly carries a dominant strain of E. coli that constitutes more than half of the total colonies isolated (other strains are present at various levels) (14, 15, 16, 18, 44). Depending on the study, between 92% and 100% of human samples contained a dominant clone representing more than 50% of the E. coli population (17, 20, 21, 23). When we compare the results of the quantitative PCR with those based on a qualitative approach in which a single clone was randomly selected per individual, we obtained 78% correlation if the prevalence of a given phylogenetic group is 50% (see Table S3 in the supplemental material). Our method ensures correct identification of E. coli dominant-strain phylogroups.
The prevalence of E. coli phylogroups in the stools of humans has been shown to depend on sex, age, year of sampling, country of origin, and diet (1, 12, 45). In our population and using our approach, phylogroups A and B2 were detected in 74% and 70% of cases, respectively, whereas phylogroups B1 and D were detected less often. Phylogroups A and B2 were overrepresented among the dominant phylogroups (Table 3). These results are in accordance with previous data (reviewed in reference 1) and confirm the prominent role played now by phylogroup B2, as well as phylogroup A, in commensal microbiotas of French subjects. The B2 phylogroup strains appear to be opposed to the B1 phylogroup strains, which are more animal specific (9). We found no correlations to be associated with gender or age in our samples.
High intrahost diversity was found, as two, three, and four phylogenetic groups were simultaneously detected in 48%, 21%, and 8% of subjects, respectively. There was a nonrandom, complex level of phylogroup combinations within single subjects. As illustrated in the FAC analysis (Fig. 1), there is antagonism between phylogroups B1 and B2. This could correspond to different ecological niches defined, for example, by different diets. Among the most prevalent phylogroups (A and B2), there appear to be distinct population structures. A dominant phylogroup B2 was associated with an absence of other groups, whereas the presence of the other phylogroups was detected with a dominant phylogroup A (Fig. 1). These data support the notion that extraintestinal virulence may be a by-product of commensalism: numerous “extraintestinal virulence genes” (coding for adhesins, iron capture systems, toxins, and protectins) exhibited by phylogroup B2 strains (6, 46) may have evolved primarily to allow the bacteria to be good intestinal colonizers (24, 28, 47–49). When phylogroup B2 is dominant, it does not cooccur with the residence of other phylogroup strains, possibly due to higher fitness (50, 51). Alternatively, dominance of phylogroup B2 may reflect the presence of a specific gut environment or parasitism by viruses or bacteriocins that favor it. As previously reported (52), an association between the non-B2 phylogroup strains, especially phylogroup A, is observed with antibiotic resistance (Fig. 1). This is of particular interest, as the intestinal microbiota plays a critical role in the emergence of antibiotic resistance (53, 54, 55). Further work is needed to investigate the characteristics of the hosts corresponding to the two patterns of dominant phylogroups. This would help us to identify the ecological forces shaping population structure and to better understand the impact of exposure to opportunistic E. coli extraintestinal infections and the emergence of antibiotic resistance.
We have developed a rapid and reliable real-time PCR assay that quantifies E. coli phylogroups directly from stool samples and detects, with high frequency, the presence of subdominant clones. The presence of minor clones could explain the fluctuation in the composition of the E. coli microbiota within single individuals that may be seen over time. They could also constitute reservoirs of virulent and/or resistant strains. Long-term studies of subject cohorts, using our approach, will allow these notions to be tested.
Supplementary Material
ACKNOWLEDGMENTS
We are grateful to Hervé Jacquier for providing some of the control strains.
The Coliville Group is composed of the following medical practitioners who were involved in the recruitment of subjects for the study: Monique Allouche, Jean-Pierre Aubert, Isabelle Aubin, Ghislaine Audran, Dan Baruch, Philippe Birembaux, Max Budowski, Emilie Chemla, Alain Eddi, Marc Frarier, Eric Galam, Julien Gelly, Serge Joly, Jean-François Millet, Michel Nougairede, Nadja Pillon, Guy Septavaux, Catherine Szwebel, Philippe Vellard, Raymond Wakim, Xavier Watelet, and Philippe Zerr.
Footnotes
Published ahead of print 14 June 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.01423-13.
REFERENCES
- 1. Tenaillon O, Skurnik D, Picard B, Denamur E. 2010. The population genetics of commensal Escherichia coli. Nat. Rev. Microbiol. 8:207–217 [DOI] [PubMed] [Google Scholar]
- 2. Russo TA, Johnson JR. 2003. Medical and economic impact of extraintestinal infections due to Escherichia coli: focus on an increasingly important endemic problem. Microbes Infect. 5:449–456 [DOI] [PubMed] [Google Scholar]
- 3. Herzer PJ, Inouye S, Inouye M, Whittam TS. 1990. Phylogenetic distribution of branched RNA-linked multicopy single-stranded DNA among natural isolates of Escherichia coli. J. Bacteriol. 172:6175–6181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Clermont O, Bonacorsi S, Bingen E. 2000. Rapid and simple determination of the Escherichia coli phylogenetic group. Appl. Environ. Microbiol. 66:4555–4558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Bingen E, Picard B, Brahimi N, Mathy S, Desjardins P, Elion J, Denamur E. 1998. Phylogenetic analysis of Escherichia coli strains causing neonatal meningitis suggests horizontal gene transfer from a predominant pool of highly virulent B2 group strains. J. Infect. Dis. 177:642–650 [DOI] [PubMed] [Google Scholar]
- 6. Picard B, Garcia JS, Gouriou S, Duriez P, Brahimi N, Bingen E, Elion J, Denamur E. 1999. The link between phylogeny and virulence in Escherichia coli extraintestinal infection. Infect. Immun. 67:546–553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Duriez P, Clermont O, Bonacorsi S, Bingen E, Chaventré A, Elion J, Picard B, Denamur E. 2001. Commensal Escherichia coli isolates are phylogenetically distributed among geographically distinct human populations. Microbiology 147:1671–1676 [DOI] [PubMed] [Google Scholar]
- 8. Gordon DM, Cowling A. 2003. The distribution and genetic structure of Escherichia coli in Australian vertebrates: host and geographic effects. Microbiology 149:3575–3586 [DOI] [PubMed] [Google Scholar]
- 9. Escobar-Páramo P, Le Menac'h A, Le Gall T, Amorin C, Gouriou S, Picard B, Skurnik D, Denamur E. 2006. Identification of forces shaping the commensal Escherichia coli genetic structure by comparing animal and human isolates. Environ. Microbiol. 8:1975–1984 [DOI] [PubMed] [Google Scholar]
- 10. Obata-Yasuoka M, Ba-Thein W, Tsukamoto T, Yoshikawa H, Hayashi H. 2002. Vaginal Escherichia coli share common virulence factor profiles, serotypes and phylogeny with other extraintestinal E. coli. Microbiology 148:2745–2752 [DOI] [PubMed] [Google Scholar]
- 11. Zhang L, Foxman B, Marrs C. 2002. Both urinary and rectal Escherichia coli isolates are dominated by strains of phylogenetic group B2. J. Clin. Microbiol. 40:3951–3955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Gordon DM, Stern SE, Collignon PJ. 2005. Influence of the age and sex of human hosts on the distribution of Escherichia coli ECOR groups and virulence traits. Microbiology 151:15–23 [DOI] [PubMed] [Google Scholar]
- 13. Clermont O, Christenson JK, Denamur E, Gordon DM. 2013. The Clermont Escherichia coli phylo-typing method revisited: improvement of specificity and detection of new phylo-groups. Environ. Microbiol. Rep. 5:58–65 [DOI] [PubMed] [Google Scholar]
- 14. Sears HJ, Brownlee I, Uchiyama JK. 1950. Persistence of individual strains of Escherichia coli in the intestinal tract of man. J. Bacteriol. 59:293–301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Sears HJ, Brownlee I. 1952. Further observations on the persistence of individual strains of Escherichia coli in the intestinal tract of man. J. Bacteriol. 63:47–57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Sears HJ, Janes H, Saloum R, Brownlee I, Lamoreaux LF. 1956. Persistence of individual strains of Escherichia coli in man and dog under varying conditions. J. Bacteriol. 71:370–372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Lidin-Janson G, Kaijser B, Lincoln K, Olling S, Wedel H. 1978. The homogeneity of the faecal coliform flora of normal school-girls, characterized by serological and biochemical properties. Med. Microbiol. Immunol. 164:247–253 [DOI] [PubMed] [Google Scholar]
- 18. Caugant DA, Levin BR, Selander RK. 1981. Genetic diversity and temporal variation in the E. coli population of a human host. Genetics 98:467–490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Caugant DA, Levin BR, Selander RK. 1984. Distribution of multilocus genotypes of Escherichia coli within and between host families. J. Hyg. (London) 92:377–384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Schlager TA, Hendley JO, Bell AL, Whittam TS. 2002. Clonal diversity of Escherichia coli colonizing stools and urinary tracts of young girls. Infect. Immun. 70:1225–1229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Lautenbach E, Bilker WB, Tolomeo P, Maslow JN. 2008. Impact of diversity of colonizing strains on strategies for sampling Escherichia coli from fecal specimens. J. Clin. Microbiol. 46:3094–3096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Moreno E, Andreu A, Pigrau C, Kuskowski MA, Johnson JR, Prats G. 2008. Relationship between Escherichia coli strains causing acute cystitis in women and the fecal E. coli population of the host. J. Clin. Microbiol. 46:2529–2534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Moreno E, Johnson JR, Pérez T, Prats G, Kuskowski MA, Andreu A. 2009. Structure and urovirulence characteristics of the fecal Escherichia coli population among healthy women. Microbes Infect. 11:274–280 [DOI] [PubMed] [Google Scholar]
- 24. Nowrouzian FL, Wold AE, Adlerberth I. 2005. Escherichia coli strains belonging to phylogenetic group B2 have superior capacity to persist in the intestinal microflora of infants. J. Infect. Dis. 191:1078–1083 [DOI] [PubMed] [Google Scholar]
- 25. Vollmerhausen TL, Ramos NL, Gündogdu A, Robinson W, Brauner A, Katouli M. 2011. Population structure and uropathogenic virulence-associated genes of faecal Escherichia coli from healthy young and elderly adults. J. Med. Microbiol. 60:574–581 [DOI] [PubMed] [Google Scholar]
- 26. Anderson MA, Whitlock JE, Harwood VJ. 2006. Diversity and distribution of Escherichia coli genotypes and antibiotic resistance phenotypes in feces of humans, cattle, and horses. Appl. Environ. Microbiol. 72:6914–6922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Escobar-Páramo P, Grenet K, Le Menac'h A, Rode L, Salgado E, Amorin C, Gouriou S, Picard B, Rahimy MC, Andremont A, Denamur E, Ruimy R. 2004. Large-scale population structure of human commensal Escherichia coli isolates. Appl. Environ. Microbiol. 70:5698–5700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Nowrouzian FL, Adlerberth I, Wold AE. 2006. Enhanced persistence in the colonic microbiota of Escherichia coli strains belonging to phylogenetic group B2: role of virulence factors and adherence to colonic cells. Microbes Infect. 8:834–840 [DOI] [PubMed] [Google Scholar]
- 29. De Muinck EJ, Oslashien T, Storrø O, Johnsen R, Stenseth NC, Rønningen KS, Rudi K. 2011. Diversity, transmission and persistence of Escherichia coli in a cohort of mothers and their infants. Environ. Microbiol. Rep. 3:352–359 [DOI] [PubMed] [Google Scholar]
- 30. Walk ST, Alm EW, Gordon DM, Ram JL, Toranzos GA, Tiedje JM, Whittam TS. 2009. Cryptic lineages of the genus Escherichia. Appl. Environ. Microbiol. 75:6534–6544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Le Gal F, Gordien E, Affolabi D, Hanslik T, Alloui C, Dény P, Gault E. 2005. Quantification of hepatitis delta virus RNA in serum by consensus real-time PCR indicates different patterns of virological response to interferon therapy in chronically infected patients. J. Clin. Microbiol. 43:2363–2369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Untergasser A, Nijveen H, Rao X, Bisseling T, Geurts R, Leunissen JAM. 2007. Primer3Plus, an enhanced web interface to Primer3. Nucleic Acids Res. 35:W71–W74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Penders J, Thijs C, Vink C, Stelma FF, Snijders B, Kummeling I, van den Brandt PA, Stobberingh EE. 2006. Factors influencing the composition of the intestinal microbiota in early infancy. Pediatrics 118:511–521 [DOI] [PubMed] [Google Scholar]
- 34. Greenacre M. 1992. Correspondence analysis in medical research. Stat. Methods Med. Res. 1:97–117 [DOI] [PubMed] [Google Scholar]
- 35. Clermont O, Gordon DM, Brisse S, Walk ST, Denamur E. 2011. Characterization of the cryptic Escherichia lineages: rapid identification and prevalence. Environ. Microbiol. 13:2468–2477 [DOI] [PubMed] [Google Scholar]
- 36. Hartl DL. 1992. Population genetics of microbial organisms. Curr. Opin. Genet. Dev. 2:937–942 [DOI] [PubMed] [Google Scholar]
- 37. Priest FG, Barker M. 2010. Gram-negative bacteria associated with brewery yeasts: reclassification of Obesumbacterium proteus biogroup 2 as Shimwellia pseudoproteus gen. nov., sp. nov., and transfer of Escherichia blattae to Shimwellia blattae comb. nov. Int. J. Syst. Evol. Microbiol. 60:828–833 [DOI] [PubMed] [Google Scholar]
- 38. Jaureguy F, Landraud L, Passet V, Diancourt L, Frapy E, Guigon G, Carbonnelle E, Lortholary O, Clermont O, Denamur E, Picard B, Nassif X, Brisse S. 2008. Phylogenetic and genomic diversity of human bacteremic Escherichia coli strains. BMC Genomics 9:560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Clermont O, Olier M, Hoede C, Diancourt L, Brisse S, Keroudean M, Glodt J, Picard B, Oswald E, Denamur E. 2011. Animal and human pathogenic Escherichia coli strains share common genetic backgrounds. Infect. Genet. Evol. 11:654–662 [DOI] [PubMed] [Google Scholar]
- 40. Lauber CL, Zhou N, Gordon JI, Knight R, Fierer N. 2010. Effect of storage conditions on the assessment of bacterial community structure in soil and human-associated samples. FEMS Microbiol. Lett. 307:80–86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Mitsuoka T, Hayakawa K. 1972. The fecal flora of man. I. Communication: the composition of the fecal flora of different age groups. Zentralbl. Bakteriol. Orig. A 223:333–342 [PubMed] [Google Scholar]
- 42. Furet J-P, Firmesse O, Gourmelon M, Bridonneau C, Tap J, Mondot S, Doré J, Corthier G. 2009. Comparative assessment of human and farm animal faecal microbiota using real-time quantitative PCR. FEMS Microbiol. Ecol. 68:351–362 [DOI] [PubMed] [Google Scholar]
- 43. Nicolas-Chanoine M-H, Gruson C, Bialek-Davenet S, Bertrand X, Thomas-Jean F, Bert F, Moyat M, Meiller E, Marcon E, Danchin N, Noussair L, Moreau R, Leflon-Guibout V. 2013. 10-Fold increase (2006–11) in the rate of healthy subjects with extended-spectrum β-lactamase-producing Escherichia coli faecal carriage in a Parisian check-up centre. J. Antimicrob. Chemother. 68:562–568 [DOI] [PubMed] [Google Scholar]
- 44. Bettelheim KA, Faiers M, Shooter RA. 1972. Serotypes of Escherichia coli in normal stools. Lancet ii:1223–1224 [PubMed] [Google Scholar]
- 45. Skurnik D, Bonnet D, Bernède-Bauduin C, Michel R, Guette C, Becker Balaire J-MC, Chau F, Mohler J, Jarlier V, Boutin Moreau J-PB, Guillemot D, Denamur E, Andremont A, Ruimy R. 2008. Characteristics of human intestinal Escherichia coli with changing environments. Environ. Microbiol. 10:2132–2137 [DOI] [PubMed] [Google Scholar]
- 46. Escobar-Páramo P, Clermont O, Blanc-Potard H, Bui A-B, Le Bouguénec C, Denamur E. 2004. A specific genetic background is required for acquisition and expression of virulence factors in Escherichia coli. Mol. Biol. Evol. 21:1085–1094 [DOI] [PubMed] [Google Scholar]
- 47. Le Gall T, Clermont O, Gouriou S, Picard B, Nassif X, Denamur E, Tenaillon O. 2007. Extraintestinal virulence is a coincidental by-product of commensalism in B2 phylogenetic group Escherichia coli strains. Mol. Biol. Evol. 24:2373–2384 [DOI] [PubMed] [Google Scholar]
- 48. Ostblom A, Adlerberth I, Wold AE, Nowrouzian FL. 2011. Pathogenicity island markers, virulence determinants malX and usp, and the capacity of Escherichia coli to persist in infants' commensal microbiotas. Appl. Environ. Microbiol. 77:2303–2308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Nowrouzian FL, Oswald E. 2012. Escherichia coli strains with the capacity for long-term persistence in the bowel microbiota carry the potentially genotoxic pks island. Microb. Pathog. 53:180–182 [DOI] [PubMed] [Google Scholar]
- 50. Adiba S, Nizak C, van Baalen M, Denamur E, Depaulis F. 2010. From grazing resistance to pathogenesis: the coincidental evolution of virulence factors. PLoS One 5:e11882. 10.1371/journal.pone.0011882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Diard M, Garry L, Selva M, Mosser T, Denamur E, Matic I. 2010. Pathogenicity-associated islands in extraintestinal pathogenic Escherichia coli are fitness elements involved in intestinal colonization. J. Bacteriol. 192:4885–4893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Mammeri H, Eb F, Berkani A, Nordmann P. 2008. Molecular characterization of AmpC-producing Escherichia coli clinical isolates recovered in a French hospital. J. Antimicrob. Chemother. 61:498–503 [DOI] [PubMed] [Google Scholar]
- 53. Scanvic-Hameg A, Chachaty E, Rey J, Pousson C, Ozoux ML, Brunel E, Andremont A. 2002. Impact of quinupristin/dalfopristin (RP59500) on the faecal microflora in healthy volunteers. J. Antimicrob. Chemother. 49:135–139 [DOI] [PubMed] [Google Scholar]
- 54. Andremont A. 2003. Commensal flora may play key role in spreading antibiotic resistance. ASM News 63:601–607 [Google Scholar]
- 55. Grenet K, Guillemot D, Jarlier V, Moreau B, Dubourdieu S, Ruimy R, Armand-Lefevre L, Bau P, Andremont A. 2004. Antibacterial resistance, Wayampis Amerindians, French Guyana. Emerg. Infect. Dis. 10:1150–1153 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.