Abstract
IncA/C plasmids are a class of plasmids from the Enterobacteriaceae that are relatively large (49 to >180 kbp), that are readily transferred by conjugation, and that carry multiple antimicrobial resistance genes. Reconstruction of the phylogeny of these plasmids has been difficult because of the high rate of remodeling by recombination-mediated horizontal gene transfer (HGT). We hypothesized that evaluation of nucleotide polymorphisms relative to the rate of HGT would help to develop a clock to show whether anthropic practices have had significant influences on the lineages of the plasmid. A system was developed to rapidly sequence up to 191 known open reading frames from each of 39 recently isolated IncA/C plasmids from a diverse panel of Salmonella enterica and Escherichia coli strains. With these data plus sequences from GenBank, we were able to distinguish six distinct lineages that had extremely low numbers of polymorphisms within each lineage, especially among the largest group designated as group 1. Two regions, each about half the plasmid in size, could be distinguished with a separate lineal pattern. The distribution of group 1 showed that it has migrated extremely rapidly with fewer polymorphisms than can be expected in 2,000 years. Remodeling by frequent HGT was evident, with a pattern that appeared to have the highest rate just upstream of the putative conjugation origin of transfer (oriT). It seems likely that when an IncA/C plasmid is transferred by conjugation there is an opportunity for plasmid remodeling adjacent to the oriT, which was also adjacent to a multiple antimicrobial resistance gene cassette.
INTRODUCTION
Plasmids are often mobile units that can carry accessory genes to other strains (lineages) of bacteria within the species, and some will pass between many species of bacteria. Plasmids of the IncA/C incompatibility group have been found in many species of the Gammaproteobacteria. In a survey of veterinary isolates of Salmonella enterica from throughout the United States, Lindsey et al. (1) found that approximately half of the isolates were resistant to ampicillin and/or tetracycline and that, of these, 26% were positive for IncA/C plasmids. The defining characteristic of an IncA/C plasmid is the presence of the repA gene (2), a gene that should never be found on a bacterial genome (3). It is believed that all IncA/C plasmids were derived from a single common ancestor, with lateral gene transfer bringing in several important genes such as several antimicrobial resistance genes (4, 5). However, multiple plasmid sequence analyses have not produced clear lineages of IncA/C plasmids (6). It is clear that IncA/C plasmids have undergone considerable remodeling by horizontal gene transfer events, many of which appear to be transposon mediated. Total plasmid sequences have ranged in size from 49 to 182 kbp (4, 5).
Up to this point it has been difficult to tell whether the resistance genes have been gradually added to the backbone or whether there has been an ancestor that acquired a large piece of DNA with the resistance genes that has been eroded. Phylogenetic studies on the plasmid have not been done in a way that can appropriately clock the events that went into constructing the plasmid. We hypothesized that comparisons of reconstructed phylogenies of each individual open reading frame (ORF) would show how the IncA/C family of plasmids was constructed. Thus, we sequenced 39 IncA/C plasmids and compared their ORFs, along with 26 IncA/C plasmid sequences obtained from GenBank (National Center for Biotechnology Information).
For producing sequence assemblies, we relied on an “ORF-supervised analysis,” an ORF-centric approach that is analogous to “taxonomy-supervised analysis” of massively parallel sequencing of populations of small subunit ribosomal genes (7). ORF sequences from a known reference plasmid were used to search databases made with the raw sequence output files from a Roche 454 GS FLX sequencer (Roche, Nutley, NJ), which proved to be a rapid method for creating ORF assemblies. The same reference ORFs were used to search published (GenBank) whole plasmid sequences to extract ORF sequences without risk of introducing anomalies to the analyses due to sequencing errors that result in frameshifts or due to differences in ORF predicting software. Alignments were made of each ORF for all of the plasmids that had part or all of the given ORF. Downstream analyses were made of each ORF, as well as for concatenations of all the alignments, to give fine draft sequences for a total of 65 plasmids. We were able to determine that the plasmids had two major sequence regions with separate lineages and that together there are at least six lineages of IncA/C plasmids. The plasmid has disseminated very rapidly with many insertion/deletion events, but we could estimate that the most prevalent lineages were between 1,500 and 57,000 years old.
MATERIALS AND METHODS
Plasmid population.
Newly isolated plasmids were selected for sequencing based on the presence of a marker within the repA gene that defines an IncA/C plasmid (1, 8). The plasmids from Salmonella enterica were a subset of the plasmids previously used for microarray analysis for IncA/C plasmids (1). Plasmids from Escherichia coli were obtained from isolates described by Glenn et al. (9) and Lindsey et al. (10). Some of the bacterial isolates had several other plasmids, as well as the IncA/C plasmid that overwhelmed the sequencing strategy, which was overcome by transforming the plasmids into E. coli DH10B (11) and selecting resistant isolates that were subsequently shown by PCR to be positive for IncA/C (10). The population was augmented with complete sequences of plasmids that were found in GenBank using the repA gene from pAR060302 (12) in a BLASTn (13) search. A list of all of the plasmids is presented in Table S1 in the supplemental material.
Plasmid extraction and sequencing.
Plasmid DNA was extracted using the PowerPrep HP plasmid midiprep system (OriGene Technologies, Inc., Rockville, MD). Purified plasmid preps were visualized by agarose electrophoresis for plasmid size and to determine whether the prep was contaminated with genomic DNA and/or RNA. Plasmid preps free from contamination were used to make shotgun genomic libraries using the GS Titanium general library preparation kit according to the manufacturer's protocol (Roche) and run on a Roche 454 genome sequencer FLX system.
Sequence analysis.
Sequence assemblies, preliminary manipulations, alignments, and preliminary analyses were all done from within the Geneious software package (Geneious version 6.0.4 created by Biomatters [http://www.geneious.com/]) unless otherwise noted. The sequencer output (.sff) files were imported into Geneious. If multiple sequencing runs were performed for a plasmid, the output files were grouped into a single list. The raw sequences were made into a BLAST database that was then searched by BLASTn (13) with the ORF sequences from the reference plasmid pAR060302 (4) using the following settings: an E value set to 1e−5; only return query-centric alignments, turn off the low-complexity filter, and maximum hits set to 10,000; the rest of the settings were left at the default. The resulting blast hits were then aligned with the Geneious alignment algorithm using the default settings. The alignments were visually inspected and edited as appropriate (mostly to trim off bases that extended beyond the start or end of the reference sequence). The consensus sequence of this build was extracted and considered the ORF sequence for the plasmid.
In order to identify ORFs from each of the completed plasmid sequences obtained from GenBank, each sequence was converted into a BLAST database and searched with the reference ORFs as described above. This ensured that the ORFs were interpreted in a uniform way for all of the plasmids. The annotation of the presence or absence of each gene for each plasmid was imported into Cluster 3.0 (M. Eisen, http://bonsai.hgc.jp/∼mdehoon/software/cluster/software.htm) for preliminary evaluation of relationships of the plasmids.
Copy number approximations.
For the new sequences, the number of copies of sequences in a given plasmid was estimated based on the number of raw sequence fragments that aligned to the reference sequence. The number of fragments was divided by the length of the reference ORF and then multiplied by 1,000 to give the number of fragments per thousand base-pairs of sequence. To normalize the data for all of the plasmids, this number was divided by the value derived for ORF 0003 (the repA gene, which is found in all IncA/C plasmids). The variation was visualized, and a 2.5-fold increase (the 95th percentile) over the value for the repA gene was set as the criterion for concluding the ORF was multicopy in that plasmid. One plasmid, Ec270, had an excessive variance with no outlier values; thus, no multicopy ORFs were inferred for that plasmid. The copy number of ORFs in GenBank acquired plasmid sequences was derived by counting the number of BLASTn hits that exceeded the default of 100 bases for each ORF.
All of the sequences for each ORF from all of the plasmids that were positive for that ORF were grouped together and aligned using the Geneious alignment algorithm. These alignments were also visually inspected but no editing was deemed necessary. The ORF alignments were concatenated in the same order that they are found in the pAR060302 reference to yield the complete alignment visualized in Fig. 1. The annotations for pAR060302 in GenBank file NC_012692 were used for all of the nomenclature used here.
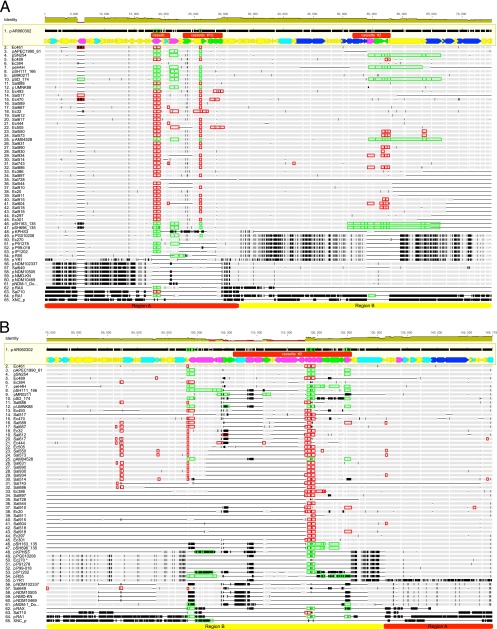
Fig 1.
Alignment map of ORF reconstructions of IncA/C plasmids concatenated in order found in the reference plasmid pAR060302. The reference sequence is at the top with an ORF map just below. The color codes for the ORFs are as follows: yellow = unknown function (hypothetical proteins), light blue = metabolic functions, dark blue = conjugation genes (tra), magenta = transposon functions, and green = antimicrobial resistance genes (including mercury resistance). The red bars represent putative cassette regions. Gray regions in alignments represent sequence identical to the reference. Vertical black hash marks represent single nucleotide polymorphisms (at the magnification shown, the alignment becomes a black bar due to saturation at ca. 10% difference from the reference). For comparison, plasmid XNC1_p averages ca. 11% difference from pAR060302. Gaps with horizontal lines that may or may not be dark, are missing sequence relative to any of the other sequences. Thus, there are some gaps in the reference sequence as well. Red boxes enclosing parts of the sequences represent regions that were deduced to be multicopy because of excesses of raw sequence fragments in that region. The genes were oriented (forward or reverse) in the same direction that they occur in pAR060302 so that the gaps would be closer to reality if they started in midgene. Green boxes enclosing parts of the sequences represent regions that were found to be multicopy when GenBank sequences were searched. Red and yellow bars at the bottom indicate region A and region B, respectively.
Distance matrixes were determined for each ORF alignment using pairwise proportionate distances (P distance) after evaluation of a subset of the alignments with jModelTest (14). Dendrograms were constructed with the neighbor-joining algorithm from the distance matrixes. For estimations of the age of clones, the patristic distances from these trees were used.
Alignments for each ORF were exported in FastA files that were used in DnaSP (15) to calculate phylogenetic statistics. Codon analyses were done on the ORFs of selected representative plasmids using CodonW (16). This program classified ORFs by their relative synonymous codon usage (RSCU). The entire alignment in FastA format and in Geneious format is available at http://www.ars.usda.gov/Main/docs.htm?docid=23292 as a supplemental data file containing a table of ORF absence or presence and a table of the products of DnasSP analyses.
RESULTS
ORF-centric reference-based assemblies of high-throughput shotgun sequencing of IncA/C plasmids were rapidly created using the procedure described above. New sequences were developed from 13 independently isolated strains of Escherichia coli and 26 strains of Salmonella enterica. The data were augmented with completed sequences of 26 plasmids found in GenBank. Each ORF was aligned individually, and then the alignments were concatenated in the same order as the reference plasmid sequence (pAR060302). The concatenated sequence alignment is illustrated in Fig. 1. Polymorphic sites that differ from the reference sequence are represented by a vertical bar at the site. In the illustration, a difference of more than ca. 10% results in a saturation of the vertical bars and appears as a black block for the length of the section with that much difference. A map of the genes as annotated in pAR060302 (4) is shown at the top and indicates the direction of the gene transcription, as well as a color code for the function of the gene.
A total of 149,179 base positions were evaluated. Only the reference plasmid, pAR060302, had all 191 ORFs that were assessed, and the closest relative had 189 of the ORFs (see Table S2 in the supplemental material [http://www.ars.usda.gov/Main/docs.htm?docid=23292] for a list of the ORFs present in each plasmid). The plasmid with the smallest number of reference ORFs was pRAX, a plasmid collected in 1969 from a fish or turtle in Japan (5, 17), with 36 of the reference ORFs, and the next smallest is a contemporary plasmid isolated for the present study, Sal710, with 49 of the reference ORFs. By definition, all IncA/C plasmids have a homologous repA gene (pAR060302_0003), but only four other genes (0001, 0002, 0004, and 0008), all with unknown functions, were found in all of the analyzed plasmids, thus 186 ORFs participate in indels.
According to Call et al. (4) the synteny of IncA/C plasmids is well preserved. Our method of analysis does not allow evaluation of synteny, but many of the missing ORFs are most parsimoniously explained as creating gaps in syntenic arrangements. Of the 65 plasmids that were analyzed, Sal890 and Sal631 (Fig. 1, taxa numbers 26 and 27) were identical by both gene content and sequence and pSH 696_135 and pSH163_135 (Fig. 1, taxa numbers 46 and 47) had identical gene content but differed at 20 polymorphic sites. All of the remaining plasmids had differing patterns of insertions and deletions. The least common ORF was ORF 0136, a gene for a hypothetical protein only found in the reference plasmid, pAR060302. ORFs 140 through 144 represent a block of ORFs that are most commonly missing that can be considered the major indel in IncA/C plasmids. These ORFs have been annotated as members of a Tn21-like transposon (18). Of the 65 plasmids, we observed 45 variants of the major indel with 14 different right-hand (downstream) ends and 32 left-hand (upstream) ends. Figure 2 includes a display of the number of plasmids each ORF is found in. It can be seen that a second major indel occurs approximately between ORF 0035 and ORF 0049 that includes genes annotated as part of ISCR1 (19), but this block of ORFs was only absent in as many as 21 (32%) of the plasmids. ORF 0010, an IS1294-like element, was conspicuous in only being present in 8 (12%) of the plasmids. The order that the plasmids are presented in Fig. 1 is based on the results of a Cluster 3.0 (M. Eisen; available from http://rana.lbl.gov/EisenSoftware.htm) analysis of presence or absence of the ORFs.
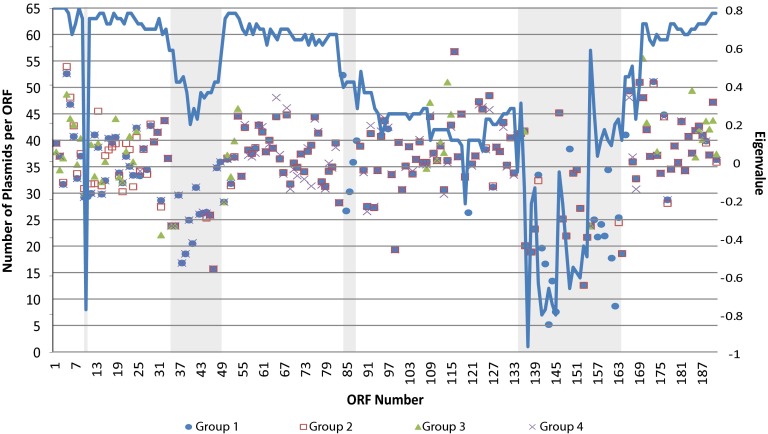
Fig 2.
Number of plasmids with each ORF (blue line) and first eigenvalue for principal component analysis of codon usage for representative plasmids in groups 1 through 4 (groups with more than one member). The gray-shaded regions represent regions that consist of transposon-associated genes.
Patterns of polymorphisms can also be seen in Fig. 1, which shows gray areas as sections where the aligned sequence is the same as the reference (pAR060302) and vertical lines where there is a difference from the reference sequence. In the graphic, the visualization of polymorphisms becomes saturated with roughly 10% difference in the sequences such that the aligned sequence appears as a horizontal bar. Based on visualizing the patterns of polymorphisms, two major regions of the plasmids can be discerned; we designated region A to include genes 0001 through 0054 at the left end and 0168 to 190 at the right end, and region B includes ORFs 0055 through 0167 (pAR060302 numbering, out of a total of 0190, see the red and yellow bars at the bottom of Fig. 1). By inspection of the polymorphism patterns in Fig. 1, we were able to distinguish six groups of plasmids. Plasmids 1 through 47, designated as group 1, (as numbered in Fig. 1) have a region A designated as type A1 and a region B designated as type B1. Plasmids 48 through 54, designated as group 2, have region A of type A1 and a second region B type designated type B2. Plasmids 56 through 61, designated group 3, have a second type of region A, designated type A2, with region B being mostly type B1. Plasmids 62 through 64, designated group 4, constitute a more loosely associated group that have regions A and B that both differ from the above types. Plasmid 55, pYR1, was distinctive with a unique region A and region B conforming to type B2. Plasmid 65, XNC_p, also had unique regions A and B that excluded it from the other groups. The vast majority—46 of 54—of the plasmids from Salmonella and E. coli were in group 1, with only one plasmid from another species found in that group (pMR0211) from Providencia stuartii (see Table S1 in the supplemental material). There appears to be little segregation within group 1 by either species or serovar (in the case of Salmonella enterica). The average patristic distances within groups 1, 2, and 3 were 0.00015354, 0.00424819, and 0.00011113 base changes per site, respectively.
Several different phylogenetic reconstructions were evaluated. A tree is presented in Fig. 3 that represents a reconstruction of the deduced phylogeny for ORF 0003, the repA gene that is present in all of the plasmids. In that tree, four of five of the groups distinguished above can be seen and XNC_p categorized independently. Group 1 and group 2 plasmids shared sequences for ORF 0003. Analysis of the alignment for ORF 0003 for recombination revealed that the sequence for pYR1 is apparently the product of a recombination of a fragment at the 3′ end from an ancestor of group 4 plasmids and 5′ end two-thirds was typical of ORF 0003 seen in groups 1 and 2. This could be visualized in Fig. 1 if it is substantially magnified. A phylogenic reconstruction based on the complete concatenated sequences is also presented in Fig. 3. The grouping seen in this tree is also congruent with the group assignments given above with the exception of the placement of pKPHS3; this is a reflection of the small size of that plasmid in which most of region B is missing. Group 1 is a large homogenous group of plasmids, all but two of our new sequences fell into this group, and when considered alone, the new sequences in this group had 293 parsimoniously informative sties and only 165 singleton sites.
Fig 3.
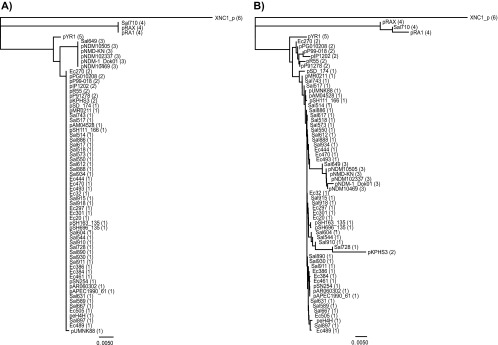
Phylogenic reconstruction based on sequences for ORF 0003 (A) and for the complete concatenated sequences (B). Plasmid identifiers are followed in parentheses by the group assignment for the plasmid.
In order to see whether any signature could be detected in the sequences that would suggest the origins of the sequences, codon usage analysis was performed with representative sequences from group 1 (pAR060302; Fig. 1, taxon number 1), group 2 (Ec270; Fig. 1, taxon number 50), group 3 (Sal649; Fig. 1, taxon number 57) and group 4 (Sal 710; Fig. 1, taxon number 63) using the program CodonW. The eigenvalue for the first dimension of the variance space was plotted against the position of each ORF (Fig. 2). An eigenvalue of 0 (labeled on the right side of the graphic) is a normative value. Points that cluster together are indicative of ORFs with similar codon usage. It can be seen (Fig. 2) that there is not a substantial or consistent difference in eigenvalues for each group. However, the eigenvalues drop in three regions that coincide with transposon sequences (shaded in gray in the figure).
As an alternative to internal signature for origin of sequences, we searched the NCBI nr (nonredundant) database by BLASTn (13) with each of the ORFs and scored each with how often neighboring ORFs were found on the same fragments in the database that were definitively not other IncA/C plasmids. This creates a score based on the frequency of co-occurrence of neighboring ORFs on fragments of the GenBank nr database. There were three segments of plasmid that are labeled in Fig. 1 with red bars putatively identified as cassettes (raw data are presented in Table S2 in the supplemental material [http://www.ars.usda.gov/Main/docs.htm?docid=23292]). The cassettes largely correlated with transposon sequences. The first and third of these cassettes had an elevated G+C content and the second contained a transposase with a decreased G+C content, the rest of the ORFs in both region A and region B had G+C content of ca. 50% (see Table S2 in the supplemental material [http://www.ars.usda.gov/Main/docs.htm?docid=23292]).
Without complete assembly of the plasmids we cannot know the real copy number of each ORF or the location of the copies. However, it is the multiple copies that lead to poor assembly results. The copy number can be inferred from the number of fragments in the raw sequence files that align to the ORF. The number of fragments per thousand bases of sequence for each ORF was divided by the number of fragments per thousand bases for ORF 0003 (repA) and plotted for comparison of each ORF (Fig. 4). The variance noted for each plasmid suggested a cutoff 2.5 times the ORF 0003 value for concluding probable multicopy ORFs. One plasmid (Ec270) had excessive variance that did not allow confident calling of multicopies. ORFs that were concluded to be multicopy are indicated in Fig. 1 by enclosure in red boxes. Sequences obtained from GenBank were analyzed for multicopies by BLASTn searches, and ORFs found to be multicopy in them are indicated by enclosure in green boxes. The most commonly multicopied ORFs were—using the designation in pAR060302 reference plasmid—ORFs 0034, 0035, 0154, and 0155, annotated as members of the insB family. As many as 10 copies of that ORF could be found in one GenBank sequence (pIP1202), and two newly sequenced plasmids (Ec461 and Ec470) had signal as much as 26-times signal for that ORF over the ORF 0003 reference.
Fig 4.
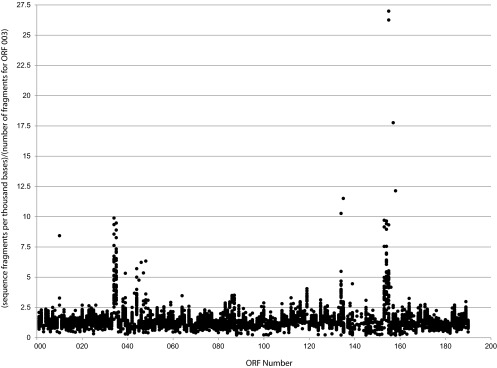
Copy number analysis. For newly sequenced plasmids, estimates of the number of copies of each ORF were made by calculating the number of raw sequence fragments per thousand bases of the ORF divided by the number calculated for ORF 003 (thus, the value for ORF 0003 = 1.0) plotted against the y axis and the position of the ORF in the reference sequence (pAR060302).
DISCUSSION
Since this investigation focused on the coevolution of different segments of IncA/C plasmids, an ORF-centric approach was used to maximize the information from draft-level sequences. pAR060302 was selected to be the reference sequence because it was the longest known IncA/C plasmid at the time of the initiation of this project. This strategy is unable to detect novel sequences (the IS26 to mph-mel insertion noted by Fernandez-Alarcon [12] was not included in the analysis) and does not allow for analysis of synteny. For genes that were annotated as multicopy in the reference sequence, it could be deduced that they were similarly multicopy in the newly sequenced plasmids by the presence of multiples of raw read fragments. For instance, a gene that was demonstrated to have four copies in pAR060302 might have about 300 fragments of raw sequences per thousand base-pair length of the gene, while the single-copy genes may have about 60 to 125 fragments per thousand base pairs. However, polymorphisms between the copies are ambiguous in the final assemblies.
It is difficult to construct whole assemblies of IncA/C plasmids with pyrosequencing data because the short fragments of data are not suited for identifying the multiple sequence redundancies, both short (<100 bp) and long (whole genes). Our method of ORF based assemblies provides all of the information that is delivered by hybridization array analyses; it also delivers sequence data that help to provide the relative age of the genes. Our attempts to assemble total plasmid sequences yielded very poor quality assemblies including apparent errors in synteny because of the high level of repeats sequenced in short fragments. Thus, it is reasonable to rely on the gene arrangements of total plasmid assemblies from high quality Sanger sequencing fragments as references as we have done. Assuming that neighboring genes are more likely to share phylogenies except when disrupted by lateral gene transfers, the gene order in the plasmid we used as the reference, pAR060302, is substantially supported by our analyses as discussed further below.
Based on hybridization array analyses, we had previously (10) concluded that IncA/C plasmids occurred in two major lineages that differed mainly on the presence or absence of genes corresponding to ORF numbers 0088 through 0165 (see Fig. 2). However, the sequence analyses reported here do not support that criterion for defining lineages. Likewise, clustering based on PCR tests for the presence or absence of ORFs as described by Welch et al. (20) is not congruent with the ancestry revealed by sequence analyses for IncA/C plasmids. At least six lineage groups of IncA/C plasmids can be identified based on the patterns of polymorphisms.
Current technologies for massively parallel sequencing (“next generation sequencing”) are known to be plagued with a relatively high error rate, one as high as 1% (21). However, the IncA/C ORFs that were in group 1 are more remarkable for their sequence homogeneity than for the diversity that was expected. The ratio of parsimoniously informative sites to singleton polymorphic sites in our new sequence, 293:165, would argue that our analyses are not substantially impacted by sequencing errors, although we were not without some ambiguous base calls. At the same time, the diversity imparted by insertions and/or deletions was remarkable; only two pairs of plasmids had the same ORF content.
With some small exceptions, the constituent parts of each lineage probably arose from a single common ancestor in an evolutionarily recent period. However, in the longer term, the polymorphism map clearly shows that there are two major homology regions in the plasmid: for the sake of discussion, we will define region A to include genes 0001 through 0054 at the left end and 0168 to 190 at the right end (pAR060302 numbering, out of a total of 0190, with 0001 directly following 0190), and region B includes genes in between (0055 to 0167). Although the major differences between the lineages can be accounted for by recombination, the accumulation of polymorphisms by point mutations in nonrecombinant ORFs is congruent with the groupings for most genes. For instance, five of the six lineages can be seen in the clustering seen for the repA gene (pAR060302, Fig. 3).
Region A has at least four lineages, and region B has at least three lineages, but phylogenetic reconstruction of region B is confounded by a high rate of insertion/deletion events (no part of region B was shared by all 65 plasmids). The lineages combine with each other in six ways making the six lineage groups. The regions were bounded by insertion sequence elements: region B begins a few genes after the sul2 containing element and ends a few genes after the Tn21-like element.
Within a lineage, the major influence on plasmid remodeling is by insertion or deletions. The bulk of the indels were in region B. One plasmid, Sal728, had only a single complete ORF in region B, insB3, a transposon-associated gene, and portions of the flanking ORFs at both ends of that gene. Almost all of the indels within region A occur in and around the sul2-containing insertion element. Other than ORF 0135.2 that was only present in the reference sequence, ORFs 0140 (aacC) and 0144 (insF) (pAR060302 annotation) were the most commonly absent and were part of a larger indel that had 52 variations in its start, fragments present within the indel, and end. Figure 2 shows the number of plasmids containing each ORF, and it can be seen that the indel containing ORF 0140 has more start positions than end positions. It is difficult to time indel events or to determine how many events a given indel represents. Our data can be most parsimoniously interpreted to show that plasmids in lineage 1 and 4 gained region B (ORFs 55 through 167) in its entirety. The decay in the number of plasmids per ORF as seen in left-hand side and moving to the right (as displayed in Fig. 2) of the major indel in region B bears the signature of random interruptions of plasmid transfer during conjugation. Transfer of plasmids by conjugation begins at the oriT (origin of transfer) and involves the transfer of a single strand of the plasmid in a 5′-to-3′ direction (22). In order for conjugation to be successful, the oriT crosses to the new host, followed by enough of the plasmid to that ensure genes needed for replication are also transferred. Becker and Meyer (22) have presented a model for this kind of plasmid remodeling and also include the possibility that the 3′ end of the transferred plasmid DNA is degraded and thus shortened by exonuclease I before the plasmid can recircularize in the new host strain. Thus, it is likely that the oriT is close to the beginning of the downstream section of region A (near ORF 0168); however, the oriT has not been identified for IncA/C, and we were unable to find any candidate sequences in our analyses. The location of the transposon at the downstream end of region B may be a coincidence or the transfer of the transposon sequences may be favored by the relaxation of the DNA near the oriT during conjugation (23). The low level of polymorphisms in the remaining ORFs indicates that the losses have been an extremely rapid process.
Transposon-associated ORFs tended to be absent more often, and they tended to have RSCU scores that deviated from the norm. It is noteworthy that the bulk of the plasmid region B, ORFs from number 87 up to 134, are not known transposon-associated genes, are frequently absent from IncA/C plasmids, and yet have RSCU scores that are more reflective of the ORFs that are neither transposon-associated or frequently absent. This supports the hypothesis that region B was acquired as a whole and degraded as described above.
Identification of the gene cassettes was a summary of GenBank blast searches that attempts to find out how often the genes occur in places other than IncA/C. The GenBank database is a poor surrogate for what is probably out in nature, but it is the only thing we have. Not only are there sampling biases, but it is difficult to determine whether the fragments are actually parts of other IncA/C plasmids, not to mention redundancies with other fragments or completed sequences. However, there were 10,345 hits altogether, and care was taken to exclude IncA/C plasmids. Although the genes in the three identified cassettes appear to be common outside of IncA/C plasmids and the cassettes in part or completely participated in insertion/deletions, these genes were not likely to be involved in the horizontal gene transfers that separated the different lineages of IncA/C plasmids.
There was no apparent segregation of the lineages in relation to the species or serotype of the carrying bacteria or the host from which the original isolation was made (see Table S1 in the supplemental material). This means that the migration of the most prevalent group we found has been extremely rapid, resulting in wide distribution of a lineage with a very small number of polymorphisms and yet displaying a substantial number of insertion or deletion events. This is also consistent with the conjugation process playing a role in the indel remodeling of the plasmids.
In the study by Lindsey et al. (1), we reported the prevalence of IncA/C and noted that pSal710, the new plasmid that was the most different from any other and is most similar to the older IncA/C sequences from GenBank (especially pRAX), came from a member of a PFGE defined “epidemic clone” that had 5 members with only 2 being positive for IncA/C. The lineage based on PFGE would not support the conclusion that other members of the clone lost the plasmid. Unfortunately, we do not have the sequence for the other IncA/C plasmid from a member of that clone. The only antimicrobial resistance gene on pSal710 was sul. The parent strain was resistant to tetracycline (TET) and sulfamethoxazole (SUL) only; therefore, TET resistance has to be on some other genetic element. Thus, we have found an older IncA/C plasmid with no known selective advantage in some members of a newer clone of Salmonella. A reasonable hypothesis is that when the plasmid was introduced into the lineage of bacteria it also had the TET gene that was transferred to another element within the bacteria, perhaps chromosomal.
Our earlier studies (1, 10, 24) suggested that the transfer of antimicrobial resistance determinants via complete IncA/C plasmids is relatively slow compared to the more frequent exchange orchestrated by transposable elements. We now believe that IncA/C plasmids are frequently transferred between bacterial lineages and remodeled in the process of transferring. We continue to believe that transposons contribute to the rapid remodeling of the plasmids, but we cannot distinguish whether the conjugation process accelerates the mobilization of the transposons. The donors of the transposons remain unknown but may be other plasmids or conjugative units that are more transient in the bacterial population. Using the mutation rate estimate of 2.45 × 10−10 per generation (25) and the estimate that E. coli averages 300 generations per year (26), based on the average patristic distances, we placed the group 1 plasmid lineage at about 2,000 years old, the group 2 lineage at about 1,500 years old, and the group 3 lineage at about 57,000 years old. There were not enough members of the other groups to make estimates of lineage ages. The plasmids we label as group 1 appear to be a young lineage, and the current prevalence of the lineage indicates that it has rapidly expanded. We also found members of older lineages, meaning that they are still extant as well. Thus, the newest lineages have arisen in approximately the last two thousand years, the same period that has seen great increase in human impacts on environmental microbiota.
Supplementary Material
Footnotes
Published ahead of print 7 June 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.01259-13.
REFERENCES
- 1. Lindsey RL, Fedorka-Cray PJ, Frye JG, Meinersmann RJ. 2009. Inc A/C plasmids are prevalent in multidrug-resistant Salmonella enterica isolates. Appl. Environ. Microbiol. 75:1908–1915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Llanes C, Gabant P, Couturier M, Bayer L, Plesiat P. 1996. Molecular analysis of the replication elements of the broad-host-range RepA/C replicon. Plasmid 36:26–35 [DOI] [PubMed] [Google Scholar]
- 3. Thomas CM. 2004. Evolution and population genetics of bacterial plasmids, p 509–528 In Funnell BE, Phillips GJ. (ed), Plasmid biology. ASM Press, Washington, DC [Google Scholar]
- 4. Call DR, Singer RS, Meng D, Broschat SL, Orfe LH, Anderson JM, Herndon DR, Kappmeyer LS, Daniels JB, Besser TE. 2010. blaCMY-2-positive IncA/C plasmids from Escherichia coli and Salmonella enterica are a distinct component of a larger lineage of plasmids. Antimicrob. Agents Chemother. 54:590–596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Fricke WF, Welch TJ, McDermott PF, Mammel MK, LeClerc JE, White DG, Cebula TA, Ravel J. 2009. Comparative genomics of the IncA/C multidrug resistance plasmid family. J. Bacteriol. 191:4750–4757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Del Castillo CS, Hikima J, Jang HB, Nho SW, Jung TS, Wongtavatchai J, Kondo H, Hirono I, Takeyama H, Aoki T. 2013. Comparative sequence analysis of a multidrug-resistant plasmid from Aeromonas hydrophila. Antimicrob. Agents Chemother. 57:120–129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Sul WJ, Cole JR, da Jesus CE, Wang Q, Farris RJ, Fish JA, Tiedje JM. 2011. Bacterial community comparisons by taxonomy-supervised analysis independent of sequence alignment and clustering. Proc. Natl. Acad. Sci. 108:14637–14642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Carattoli A, Bertini A, Villa L, Falbo V, Hopkins KL, Threlfall EJ. 2005. Identification of plasmids by PCR-based replicon typing. J. Microbiol. Methods 63:219–228 [DOI] [PubMed] [Google Scholar]
- 9. Glenn LM, Englen MD, Lindsey RL, Frank JF, Turpin JE, Berrang ME, Meinersmann RJ, Fedorka-Cray PJ, Frye JG. 2012. Analysis of antimicrobial resistance genes detected in multiple-drug-resistant Escherichia coli isolates from broiler chicken carcasses. Microb. Drug Resist. 18:453–463 [DOI] [PubMed] [Google Scholar]
- 10. Lindsey RL, Frye JG, Fedorka-Cray PJ, Meinersmann RJ. 2011. Microarray-based analysis of IncA/C plasmid-associated genes from multidrug-resistant Salmonella enterica. Appl. Environ. Microbiol. 77:6991–6999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Durfee T, Nelson R, Baldwin S, GPlunkett, Burland V, Mau B, Petrosino JF, Qin X, Muzny DM, Ayele M, Gibbs RA, Csörgo B, Pósfai G, Weinstock GM, Blattner FR. 2008. The complete genome sequence of Escherichia coli DH10B: insights into the biology of a laboratory workhorse. J. Bacteriol. 190:2597–2606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Fernandez-Alarcon C, Singer RS, Johnson TJ. 2011. Comparative genomics of multidrug resistance-encoding IncA/C plasmids from commensal and pathogenic Escherichia coli from multiple animal sources. PLoS One 6:E23415. 10.1371/journal.pone.0023415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403–410 [DOI] [PubMed] [Google Scholar]
- 14. Posada D. 2009. Selection of models of DNA evolution with jModelTest. Methods Mol. Biol. 537:93–112. 10.1007/978-1-59745-251-9_5 [DOI] [PubMed] [Google Scholar]
- 15. Librado P, Rozas J. 2009. DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics 25:1451–1452 [DOI] [PubMed] [Google Scholar]
- 16. Peden JF. 2005. CodonW. http://sourceforge.net/projects/codonw/
- 17. Aoki T, Egusa S, Ogata Y, Watanabe T. 1971. Detection of resistance factors in fish pathogen Aeromonas liquefaciens. J. Gen. Microbiol. 65:343–349 [DOI] [PubMed] [Google Scholar]
- 18. Liebert CA, Hall RM, Summers AO. 1999. Transposon Tn21, flagship of the floating genome. Microbiol. Mol. Biol. Rev. 63:507–522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Toleman MA, Bennett PM, Walsh TR. 2006. ISCR elements: novel gene-capturing systems of the 21st century? Microbiol. Mol. Biol. Rev. 70:296–316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Welch TJ, Fricke WF, McDermott PF, White DG, Rosso ML, Rasko DA, Mammel MK, Eppinger M, Rosovitz MJ, Wagner D, Rahalison L, Leclerc JE, Hinshaw JM, Lindler LE, Cebula TA, Carniel E, Ravel J. 2007. Multiple antimicrobial resistance in plague: an emerging public health risk. PLoS One 2:e309. 10.1371/journal.pone.0000309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kirsch S, Klein CA. 2012. Sequence error storms and the landscape of mutations in cancer. Proc. Natl. Acad. Sci. U. S. A. 109:14289–14290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Becker EC, Meyer R. 2012. Origin and fate of the 3′ ends of single-stranded DNA generated by conjugal transfer of plasmid R1162. J. Bacteriol. 194:5368–5376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Jandle S, Meyer R. 2006. Stringent and relaxed recognition of oriT by related systems for plasmid mobilization: implications for horizontal gene transfer. J. Bacteriol. 188:499–506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Lindsey RL, Frye JG, Thitaram SN, Meinersmann RJ, Fedorka-Cray PJ, Englen MD. 2011. Characterization of multidrug-resistant Escherichia coli by antimicrobial resistance profiles, plasmid replicon typing, and pulsed-field gel electrophoresis. Microb. Drug Resist. 17:157–163 [DOI] [PubMed] [Google Scholar]
- 25. Lee H, Popodi E, Tang H, Foster LF. 2012. Rate and molecular spectrum of spontaneous mutations in the bacterium Escherichia coli as determined by whole-genome sequencing. Proc. Natl. Acad. Sci. U. S. A. 109:E2774–2783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Ochman H, Elwyn S, Moran NA. 1999. Calibrating bacterial evolution. Proc. Natl. Acad. Sci. U. S. A. 96:12638–12643 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.