Abstract
The genus Arcobacter is composed of 17 species which have been isolated from various sources. Of particular interest are A. butzleri, A. cryaerophilus, and A. skirrowii, as these have been associated with human cases of diarrhea, the probable transmission routes being through the ingestion of contaminated drinking water and food. To date, only limited studies of virulence traits in this genus have been undertaken. The present study used 60 Arcobacter strains isolated from different sources, representing 16 of the 17 species of the genus, to investigate their ability to adhere to and invade the human intestinal cell line Caco-2. In addition, the presence of five putative virulence genes (ciaB, cadF, cj1349, hecA, and irgA) was screened for in these strains by PCR. All Arcobacter species except A. bivalviorum and Arcobacter sp. strain W63 adhered to Caco-2 cells, and most species (10/16) were invasive. The most invasive species were A. skirrowii, A. cryaerophilus, A. butzleri, and A. defluvii. All invasive strains were positive for ciaB (encoding a putative invasion protein). Other putative virulence genes were present in other species, i.e., A. butzleri (cadF, cj1349, irgA, and hecA), A. trophiarum (cj1349), A. ellisii (cj1349), and A. defluvii (irgA). No virulence genes were detected in strains which showed little or no invasion of Caco-2 cells. These results indicate that many Arcobacter species are potential pathogens of humans and animals.
INTRODUCTION
The genus Arcobacter was created in 1991 (1) and is considered an atypical group within the class Epsilonproteobacteria because its species have been isolated from many habitats and hosts (2). Currently, the genus is composed of 17 species (3–6). The perceived pathogenicity of some species, such as Arcobacter butzleri and A. cryaerophilus, is due to their recovery from stools of patients with diarrhea and occasionally from cases of bacteremia, endocarditis, and peritonitis (3). Clinical cases are probably underestimated due to the absence of specific protocols for their adequate detection and identification (3).
In an 8-year study, Vandenberg et al. (7) reported that the species A. butzleri was the fourth most common Campylobacter-like organism isolated from 67,599 human stools. This species was associated with cases of persistent and watery diarrhea and was less often associated with bloody diarrhea than was Campylobacter jejuni. Other Arcobacter species such as A. cryaerophilus, A. skirrowii, and A. thereius have also been isolated from the intestinal tracts and feces of asymptomatic farm animals, as well as being associated with diarrhea, abortions, and mastitis (3, 8).
The pathogenicity and virulence mechanisms of Arcobacter spp. are still poorly understood (3). Their adhesion, invasion, and cytotoxicity capacity have been studied in only 4 Arcobacter species (A. butzleri, A. cryaerophilus, A. skirrowii, and A. cibarius) using various cell lines, i.e., HEp-2, HeLa, INT407, CHO, and Caco-2 (reference 3 and references therein). These studies showed considerable variation in adhesion, invasion, and toxicity, depending upon the origin of strains and the cell lines studied (3, 9). The publication of the A. butzleri RM 4018 genome (10) reported the presence of several putative virulence genes in the organism, such as ciaB, cj1349, and cadF. These are homologous to genes associated with pathogenicity in other closely related organisms. The ciaB gene in Campylobacter spp. encodes an invasion protein injected directly into the cytoplasm of the host cells through a secretion system (11). The cj1349 gene in C. jejuni encodes proteins that enable adhesion to host cells by binding specifically to fibronectin (11), and the CadF protein also induces the internalization of bacterial cells by the activation of GTPases (11). In addition, there are homologs to the irgA gene, encoding an iron-regulated outer membrane protein in Vibrio cholerae, and the hecA gene, which encodes a filamentous hemagglutinin in uropathogenic Escherichia coli (10). In 2012, Douidah et al. (12) developed primers for these virulence genes and demonstrated their presence in strains of A. butzleri, A. cryaerophilus, and A. skirrowii. More recently, Karadas et al. (13) also determined the presence of these genes by PCR in 52 strains of A. butzleri and the capacity for adhesion to and invasion of HT-29 and Caco-2 cells in six strains. In that study, no correlation between virulence gene patterns and adhesive or invasive capabilities was observed. However, the incidence of these genes and their potential correlation with the capacity for adhesion to or invasion of human cell lines have not been studied for all Arcobacter spp., and this is the aim of the present study.
MATERIALS AND METHODS
A total of 60 Arcobacter strains belonging to 15 of the 17 accepted species were studied (Table 1). The species A. halophilus and A. marinus, both so far known only by the type strains, were not included in the study because they are halophiles and do not grow in the standard media used for the cultivation of other Arcobacter spp. or for tissue culture studies (14, 15). Strain W63, which represented a new Arcobacter species (under proposal) on the basis of the 16S rRNA gene (data not shown), was included.
Table 1.
Virulence genotypes of 60 Arcobacter strains and their relative capacities for adhesion to and invasion of Caco-2 cellse
| Species and strain | Origin | Presence of virulence gene: |
Viable count (log10 CFU ml−1) |
|||||
|---|---|---|---|---|---|---|---|---|
| ciaB | irgA | hecA | cj1349 | cadF | Adhesion | Invasion | ||
| Controls | ||||||||
| Salmonella enterica NCTC 3046 | ND | ND | ND | ND | ND | 6.46 | 4.53 | |
| Escherichia coli K-12 | ND | ND | ND | ND | ND | 6.54 | NI | |
| A. butzleri LMG 10828T | Human blood | + | + | + | + | + | ND | ND |
| Tested strains | ||||||||
| A. butzleri | ||||||||
| F1 | Mussels | +a | + | +a | +a | +a | 4.62 | 2.24 |
| F15 | Turkey meat | + | − | − | + | + | 3.98 | 1.57 |
| F27 | Duck meat | + | − | − | + | + | 4.69 | 2.73 |
| F46b | Pig meat | + | − | − | + | + | 4.73 | 1.96 |
| F47 | Chicken meat | + | − | − | + | + | 4.21 | 2.35 |
| F49 | Pig meat | + | − | − | + | + | 4.53 | 2.29 |
| F50 | Beef | + | − | − | + | + | 4.69 | 3.17 |
| F63 | Clams | + | − | − | + | + | 5.13 | 3.10 |
| F71-1 | Clams | + | − | − | + | + | 6.12 | 2.72 |
| F87 | Mussels | + | +a | − | + | + | 4.23 | NI |
| SW21 | Sewage | + | − | − | + | + | 2.95 | 1.99 |
| SW28-5 | Sewage | + | − | − | − | − | 6.57 | 1.70 |
| A. cryaerophilus | ||||||||
| F55 | Clams | +a | − | − | − | − | 5.57 | 2.73 |
| F93-1 | Clams | + | − | − | − | − | 5.52 | 2.39 |
| FE4 | Chicken feces | + | − | − | − | − | 6.31 | 3.06 |
| FE5 | Chicken feces | + | − | − | − | − | 5.37 | 2.31 |
| FE14 | Sheep feces | + | − | − | − | − | 5.99 | 2.94 |
| A. cibarius | ||||||||
| NC81b | Piggery effluent | + | − | − | − | − | 4.35 | NI |
| NC88b | Piggery effluent | − | − | − | − | − | 5.71 | 2.51 |
| H743b | Poultry meat | + | − | − | − | − | 5.64 | 2.12 |
| H746b | Poultry meat | + | − | − | − | − | 5.09 | 2.12 |
| CECT 7203Tb | Poultry meat | +a | − | − | − | − | 4.96 | NI |
| A. skirrowii | ||||||||
| S7-1b | Sludge WWTP | +a | − | +a | +a | +a | 7.53 | 5.00c |
| F28 | Pig meat | + | − | − | + | − | 3.13 | NI |
| A. nitrofigilis | ||||||||
| CECT 7204Tb | Roots of Spartina alterniflora | +a | − | − | − | − | NA | NI |
| F72b | Mussels | + | − | − | − | − | 5.12 | 2.69 |
| F2176b | Mussels | + | − | − | − | − | 5.39 | 2.91 |
| A. thereius | ||||||||
| LMG 24486Tb | Porcine abortion | − | − | − | − | − | 4.68 | NI |
| LMG 24487b | Porcine abortion | − | − | − | − | − | 5.08 | 1.94 |
| F61-1b | Pig meat | − | − | − | − | − | 4.40 | 1.55 |
| F93-4b | Mussels | − | − | − | − | − | 5.19 | 2.18 |
| SW24b | Sewage | − | − | − | − | − | 4.27 | 1.58 |
| A. mytili | ||||||||
| T234b,d | Brackish water | − | − | − | − | − | 4.15 | NI |
| CECT 7385b,d | Mussels | − | − | − | − | − | 4.24 | NI |
| CECT 7386Tb,d | Mussels | − | − | − | − | − | 5.00 | NI |
| A. trophiarum | ||||||||
| LMG 25535b | Pig feces | + | − | − | + | − | 5.08 | 4.10c |
| LMG 25534Tb | Pig feces | +a | − | − | +a | − | 4.08 | 3.04 |
| CECT 7650b | Chicken feces | + | − | − | + | − | 5.18 | 4.21c |
| A. defluvii | ||||||||
| SW28-7b | Sewage | + | + | − | − | − | 5.22 | 1.99 |
| CECT 7697Tb | Sewage | +a | +a | − | − | − | 4.82 | 1.68 |
| SW29-1 | Sewage | + | + | − | − | − | 5.58 | 3.08 |
| SW28-10 | Sewage | + | + | − | − | − | 5.04 | 2.49 |
| SW30-8 | Sewage | + | + | − | − | − | 4.85 | 1.57 |
| SW30-2b | Sewage | + | + | − | − | − | 5.23 | 2.17 |
| CH8-2b | Mussels | + | + | − | − | − | 5.87 | 1.92 |
| CC42b | Pig feces | + | + | − | − | − | 5.22 | 2.75 |
| A. molluscorum | ||||||||
| F91b,c | Mussels | + | − | − | − | − | 5.12 | NI |
| CECT 7696Tb,d | Mussels | +a | − | − | − | − | 4.94 | NI |
| F101-1b,d | Oysters | + | − | − | − | − | 4.26 | NI |
| A. ellisii | ||||||||
| F79-2b,d | Mussels | + | − | − | − | + | 5.64 | 1.87 |
| F79-6Tb,d | Mussels | +a | − | − | +a | +a | 5.25 | 2.80 |
| F79-7b,d | Mussels | + | − | − | + | + | 4.54 | 1.71 |
| A. bivalviorum | ||||||||
| F4Tb,d | Mussels | +a | − | − | +a | − | NA | NI |
| F118-2b,d | Mussels | + | − | − | − | − | NA | NI |
| F118-4b,d | Mussels | + | − | − | + | − | NA | NI |
| A. venerupis F67-11Tb,d | Clams | +a | − | − | − | − | 5.91 | NI |
| A. suis F41b,d | Pig meat | +a | − | − | − | − | 5.92 | NI |
| A. cloacae | ||||||||
| F26b,d | Mussels | + | − | − | + | − | 6.11 | 2.00 |
| SW28-13Tb,d | Sewage | +a | − | − | +a | − | 4.51 | 1.00 |
| Arcobacter sp. strain W63b,d | Seawater | +a | − | − | +a | − | NA | NI |
Confirmed by DNA sequencing.
The identity of these strains was confirmed by sequencing of the rpoB gene.
Invasion results equal to or higher than that for the positive control S. enterica (NCTC 3046).
Only available strains of these species.
Abbreviations: NA, no adhesion detected; NI, no invasion detected; ND, not determined; WWTP, wastewater treatment plant. The values for adhesion and invasion were proportionally calculated to an inoculum of 108 CFU ml−1 (8.0 log10 CFU ml−1) for each strain. Those results classified as high for adhesion (viable count, >5.0 log10 CFU ml−1) and invasion (>3.0 log10 CFU ml−1) are shown in bold.
The strains had been isolated from different sources: shellfish (n = 23), meat (n = 12), sewage (n = 11), and feces from pigs (n = 3), chickens (n = 3), and sheep (n = 1). Other miscellaneous environmental sources were seawater (n = 2), piggery effluent (n = 2), roots of Spartina alterniflora (n = 1), and also porcine abortion (n = 2). All strains were genetically identified using a multiplex PCR (m-PCR) (16) and the 16S rRNA restriction fragment length polymorphism (RFLP) methods specific for this genus (17, 18). The identity of 40 strains (Table 1) was confirmed by sequencing the rpoB gene as previously described (19). All strains of the same species showed unique profiles when genotyped by enterobacterial repetitive intergenic consensus (ERIC) PCR (20) (data not shown). The control strains for the adhesion and invasion assays, Salmonella enterica serovar Enteritidis (NCTC 3046) and Escherichia coli K-12 HB101 (Children's Hospital, Los Angeles, CA), were obtained from the Nottingham Trent University culture collection.
Preparation of bacterial suspensions.
A colony of each strain was used to inoculate brain heart infusion (BHI; Difco, Becton, Dickinson and Company) broth, which was incubated under aerobic conditions for 48 h at 30°C for Arcobacter strains and overnight (15 ± 2 h) at 37°C for the control strains. After the incubation period, the cultures were diluted to an optical density (600 nm) of 0.08 (ca. 109 CFU ml−1 of bacterial cells) for Arcobacter strains and of 0.05 (ca. 108 CFU ml−1) for the control strains, as per previous studies (21). The cultures were centrifuged (5 min at 3,000 rpm, 4°C), and the resultant cell pellets were resuspended in the same volume of warm (37°C) Eagle's minimum essential medium (EMEM; M4655; Sigma) supplemented with 10% fetal bovine serum (FBS; F7524; Sigma) and 1% nonessential amino acids (NEAA; M7145; Sigma). The bacterial viable counts were determined on BHI agar supplemented with 5% sheep blood agar according to the Miles-Misra (22) method. The number of cells (CFU ml−1) of each bacterial suspension represented the mean from three enumerations.
Caco-2 adhesion and invasion assay.
The adhesion and invasion assays were as described previously (21, 23). Briefly, 0.5 ml of a suspension of 4 × 104 Caco-2 cells ml−1 in EMEM supplemented with penicillin (10,000 U) and streptomycin (10,000 μg ml−1) (P4333; Sigma) was added to each of the 24 wells of a microtiter plate which was then incubated for 48 h at 37°C under a 5% CO2 atmosphere (Sanyo CO2 incubator). When the cells had formed a confluent monolayer, the medium was removed, the wells were washed twice with phosphate-buffered saline (PBS; D8537; Sigma), and 0.5 ml of the bacterial suspension (ca. 109 CFU ml−1) was added. The plates were incubated for 2 h at 37°C to allow adhesion and invasion by the bacteria and were then washed twice with PBS to remove unbound bacteria. The cell monolayer was lysed with 1% Triton-X, and the total number of bacteria associated with the Caco-2 cells was counted as described above.
The number of adherent bacteria was calculated as the difference between the total number of bacteria associated with the Caco-2 cells and the number of intracellular bacteria. The latter was determined by inoculating another 24-well plate, which was washed twice with PBS and then supplemented with 0.5 ml of EMEM containing 125 mg ml−1 of gentamicin and incubated for 1 h at 37°C to kill extracellular bacteria. After incubation, the cells were washed twice with PBS and lysed with 1% Triton-X and the released bacteria were enumerated, as described above. All experiments were in triplicate. Results were expressed as the mean number of bacteria (log10 CFU ml−1) that adhered or invaded. The limit of detection for adhesion was 1.7 × 104 CFU ml−1 (4.23 log10 CFU ml−1), and the limit of detection for invasion was 1.7 × 102 CFU ml−1 (2.23 log10 CFU ml−1). Values above the detection limits were defined as adherent or invasive, respectively. In order to compare the adhesion and invasion results obtained for the different strains (Table 1) and species (Fig. 1) with those of the controls, the values for each strain or species were all proportionally calculated in relation to an initial inoculum of 1.0 × 108 CFU ml−1.
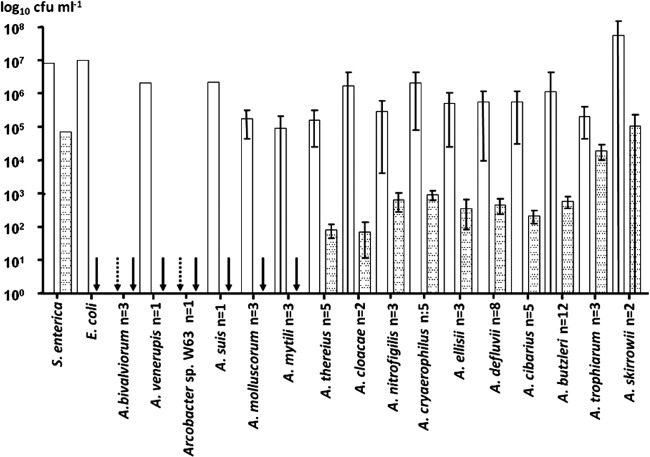
Fig 1.
Adhesion to (white bars) and invasion of (dotted bars) Caco-2 cells by 60 Arcobacter strains belonging to 15 species. Results of triplicate experiments are expressed as the mean (and standard deviation) and were proportionally calculated to an inoculum of 108 CFU ml−1. Control strains were Salmonella enterica, positive for both adhesion and invasion, and Escherichia coli, positive for adhesion and negative for invasion. Arrows indicate those cases in which adhesion (dotted) or invasion (black) was below the detection limit.
Data analysis.
The range of results between the detection limit and the mean obtained for the positive control (S. enterica) was divided into 3 categories defined as low, good, or high adhesion or invasion ability, as shown in Table 1.
The Mann-Whitney statistical test, corrected by using the Bonferroni multiple comparison test, was used to compare the results. For those strains where nonadhesion or noninvasion was detected, the respective detection limit value was assigned in the data set for statistical analysis. Significance was established at the P level of <0.05. The analyses were carried out using Prism version 5 (GraphPad) and SPSS version 20 (IBM) software.
Detection of virulence genes.
Bacterial DNA was extracted using the InstaGene DNA Purification Matrix (Bio-Rad Laboratories, Hercules, CA). The PCR methods used to detect the presence of ciaB, hecA, cj1349, cadF, and irgA genes used the primers and conditions previously described (12). PCR products were analyzed on a 2% agarose gel with Tris-borate-EDTA buffer at 80 V for 90 min using the 100-bp ladder (Fermentas) as a molecular mass marker. The gels were stained with SYBR Safe DNA gel stain (Invitrogen) and photographed using a UV transilluminator. A. butzleri LMG 10828T was used as the positive-control strain for all PCRs (12).
In order to confirm the identity of the amplicons, 28 PCR products from 5 genes of the different species were sequenced (Table 1). Sequences were obtained using amplification primers from Macrogen Corp. Europe (The Netherlands) and then compared with the A. butzleri RM4018 genome (GenBank NC_009850.1) using MEGA 5 software (24). Furthermore, a BLASTN comparison was carried out to confirm the presence of the studied genes in other deposited Arcobacter genomes.
Microscopic observation.
Strains representing those species that were adherent and invasive were selected for light and electron microscopy examination. The experiments were performed under the above-described conditions with the exception that Caco-2 cells were grown on coverslips placed into the 6 wells of the culture plates used. For light microscopy, cells were fixed with methanol and stained for 15 min with 10% Giemsa stain (Sigma-Aldrich) and then at least 10 fields per slide were visualized using an Olympus BX51 microscope. For transmission electron microscopy (TEM) and scanning electron microscopy (SEM), the cells were fixed by adding 2% glutaraldehyde solution in 0.1 M phosphate buffer (pH 7.4) for 2 h to the wells containing the coverslips. The cells were then rinsed with 0.1 M phosphate buffer (pH 7.4) and postfixed with 1% buffered osmium tetroxide for 1 h at 5°C in the dark. The fixed cells were washed in buffer and dehydrated by 15-min changes in a graded series of ethanol up to 100%. The samples for TEM and SEM were then separated. For TEM, loose cells were collected from the wells, transferred to Eppendorf tubes, and embedded in Spurr resin. Ultrathin sections for TEM were stained with uranyl acetate and lead citrate before examination using a JEOL 1011 microscope at 80 kV. The coverslip preparations were used for SEM and were subjected to serial mixtures of amylacetate-ethanol in a petri dish, in which the concentration of the first substance was gradually increased through six steps to 100%. The coverslips were critical point dried with CO2. After drying, specimens were mounted and coated with a thin layer of gold before examination using a JEOL JSM 6400 microscope at 15 kV.
Nucleotide sequence accession numbers.
Newly determined sequence data have been deposited in GenBank under accession numbers HF935040 to HF935067.
RESULTS
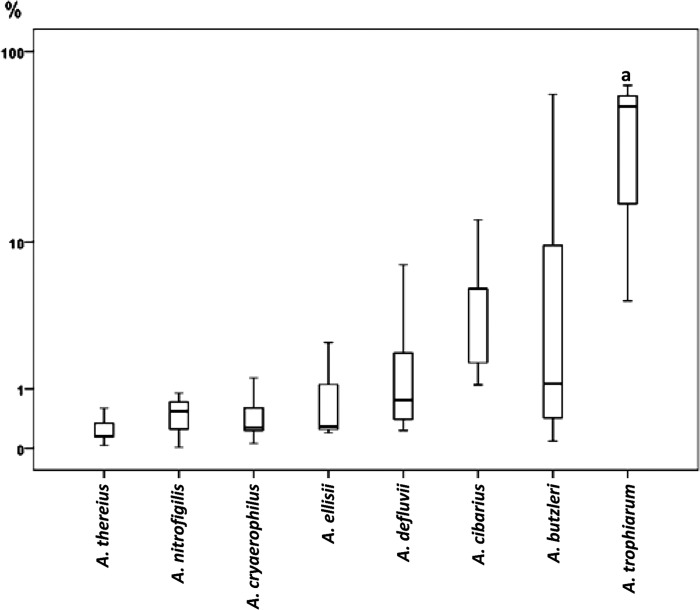
Nearly all (14/16) Arcobacter species adhered to Caco-2 cells, with the exceptions of A. bivalviorum and Arcobacter sp. strain W63, and a total of 10 Arcobacter species invaded cells (Fig. 1). The 8 most highly invasive strains belonged to the species A. trophiarum (3/3), A. skirrowii (1/2), A. cryaerophilus (1/5), A. butzleri (2/12), and A. defluvii (1/8). Most of these strains had been isolated from animal feces and sewage (Table 1). Two strains of A. trophiarum (LMG 25535 and CECT 7650) showed invasion capacities similar to that of the S. enterica positive control (Table 1). In fact, A. trophiarum was significantly (P < 0.05) more invasive than the other species (Fig. 2). On the other hand, only one strain of A. skirrowii (S7-1) showed a higher invasion capacity than did S. enterica (Table 1). Regarding the origin of strains, those recovered from fecal sources (animal feces and sewage) were significantly more invasive than those from other origins (P < 0.05) (Fig. 3).
Fig 2.
Box plots showing the invasion capacity of the Arcobacter species. Those species in which invasion was not detected and/or that included fewer than 3 strains, i.e., A. skirrowii, A. bivalviorum, A. molluscorum, A. mytili, A. venerupis, A. cloacae, A. suis, and Arcobacter sp. strain W63, are not represented. Results are expressed as percentage of invasion in relation to positive control. The length of the box shows the 50% interquartile range (25% to 75%) of the variable. The horizontal line in the box indicates the median, while vertical lines extending from the box show maximum and minimum values. “a” indicates a species more invasive (P < 0.05) than the other species.
Fig 3.
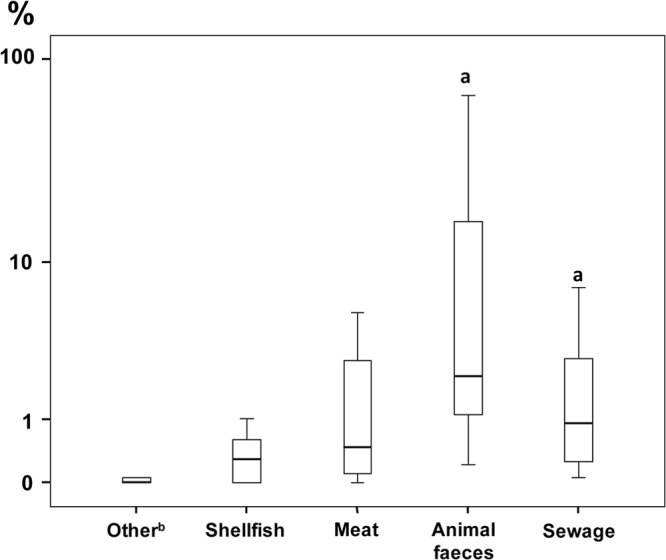
Box plots showing the invasion capacity of the Arcobacter strains grouped by origin. The 2 strains isolated from porcine abortion were not represented. Results are expressed as percentage of invasion in relation to positive control. The length of the box shows the 50% interquartile range (25% to 75%) of the variable. The horizontal line in the box indicates the median, while vertical lines extending from the box show maximum and minimum values. “a” indicates that strains from fecal sources (i.e., animal feces and sewage together) were significantly more invasive (P < 0.05) than were strains from other origins. “b” indicates that “Other” includes strains recovered from seawater, piggery effluent, and roots of Spartina alterniflora.
ciaB was the most prevalent virulence-associated gene detected (51/60, P < 0.05), followed by cj1349 (23/60) and cadF (15/60) (Table 1). Two strains of A. butzleri (F1 and F87) isolated from mussels and one strain of A. skirrowii (S7-1) from sewage were positive for four or five virulence genes. A. butzleri F1 and A. skirrowii S7-1 showed significant capacity to invade Caco-2 cells (Table 1). All A. defluvii, A. trophiarum, A. butzleri, A. skirrowii, and A. cryaerophilus strains possessed the ciaB gene, as did all strains considered highly invasive (Table 1). Some of these species possessed other genes, i.e., A. defluvii had the irgA gene (8/8); A. trophiarum had cj1349 (3/3); A. butzleri had cadF (12/12), cj1349 (11/12), irgA (2/12), and hecA (1/12); and A. skirrowii had cadF (1/2), cj1349 (1/2), and hecA (1/2). In contrast, all strains of A. thereius (n = 5) and A. mytili (n = 3) and one strain of A. cibarius were negative for all the tested genes (Table 1).
Bioinformatics analysis of the putative genes in sequenced Arcobacter strains agreed with the laboratory studies. A. butzleri strain ED-1 (a recently released genome [GenBank NC_017187.1]) possessed the 5 tested genes, as did A. butzleri F1 (Table 1). Arcobacter sp. strain L (GenBank NC_017192.1), which groups with A. defluvii on the basis of its 16S rRNA gene sequence (29), possessed the ciaB and irgA genes, which are also present in all A. defluvii strains. A. nitrofigilis strain DSM 7299T (GenBank NC_014166.1) possessed only the ciaB gene, as was also determined experimentally (Table 1).
Both light and electron microscopy demonstrated the presence of extracellular bacteria closely associated with the membrane of Caco-2 cells and intracellular bacterial cells (see Fig. S1 and S2 in the supplemental material). In general, all Arcobacter species showed a homogeneous distribution of bacterial cells on the Caco-2 surface without any specific pattern of adhesion. Strains of A. trophiarum appeared to form clusters inside the Caco-2 cells (see Fig. S1).
DISCUSSION
This is the first study of Arcobacter virulence potential which has included representatives of all accepted Arcobacter species (except A. marinus and A. halophilus) and a potential new Arcobacter species (strain W63). It has shown that most species (14/16) adhered to Caco-2 cells while 10/16 were invasive. All strains of A. cryaerophilus, A. butzleri, and A. skirrowii adhered to the human intestinal Caco-2 cells, and most invaded the cell line: 5/5, 11/12, and 1/2, respectively. Previous studies with these 3 species showed that overall 55/99 adhered and 9/44 invaded Caco-2, CHO, HeLa, HEp-2, INT407, IPI-2I, or Vero cell lines (reference 3 and references therein), but only 3 studies were performed on Caco-2 cells (13, 21, 25). Ho et al. (21) tested 4 strains of A. cryaerophilus, 2 of A. skirrowii, and 1 of A. butzleri, mainly isolated from newborn piglets or sow amniotic fluid, and also the type strain of A. cibarius (LMG 21996T) isolated from chicken carcasses. Although the 8 strains adhered to Caco-2 cells, only two strains of A. cryaerophilus were able to invade. In relation to A. cibarius, in our study the type strain (CECT 7203T) and strain NC81 showed adhesion but no invasion capacity. Although this was in agreement with previous results (21), the remaining three A. cibarius strains showed an invasion capacity. Houf and Stephan (25) determined the ability of only 7 A. cryaerophilus strains (isolated from feces of healthy human carriers) to attach to Caco-2 cells, of which only 2 adhered. The higher adhesion and invasion capacity (5/5) observed in our study could be due to the different origins of strains, as previously proposed (9, 13). A recent study compared the adhesion and invasion capacities of 3 isolates of A. butzleri from chicken meat and 3 of human origin for Caco-2 and HT-29 cells. All the isolates showed adhesion and invasion of Caco-2 cells, while only 4 showed adhesion to HT-29 cells and 3 invaded the cell line (13). Two isolates from chicken and one from human showed the highest adhesion to and invasion of Caco-2 and HT-29 cells, while the other two human isolates were less adhesive to and less invasive of Caco-2 cells. Coincidentally, the latter isolates were noninvasive of HT-29 cells, and therefore, it was concluded that the results were strain dependent.
In our study, all strains of the recently described species A. trophiarum, A. defluvii, A. ellisii, and A. cloacae were able to invade Caco-2 cells. Furthermore, the strains of A. trophiarum (all from feces of pig and chicken) were significantly more invasive than the others (P < 0.05) (Fig. 2). It is notable that 100% of strains of A. trophiarum (3/3) and A. thereius (5/5) adhered, whereas 100% and 80%, respectively, invaded. This is despite both species being previously described as unable to grow at 37°C under laboratory conditions (26, 27) yet having been isolated from warm-blooded animals: pig feces (26) and porcine abortion and cloacal content of ducks (27). Interestingly, the strains of these species remained viable or even grew when incubated at 37°C for 2 h in EMEM while in BHI they showed a slight decrease in viable counts after incubation (data not shown). Our results could indicate that EMEM and the Caco-2 cells simulate the natural intestinal habitat better than does BHI.
The abilities of bacterial pathogens to adhere to and invade mammalian cell lines are studied because these abilities are necessary for successful colonization and infection of the host (25). The present study indicates that many Arcobacter spp., including the recently discovered ones, have the potential ability to colonize and enter human cells.
The putative virulence genes showed an order of prevalence for A. butzleri (n = 12), A. cryaerophilus (n = 5), and A. skirrowii (n = 2), i.e., 85.0% ciaB, 38.3% cj1349, 25.0% cadF, 16.7% irgA, and 3.3% hecA, similar to that previously reported (12, 13). A. butzleri showed the highest prevalence of virulence genes (100% ciaB, 91.7% cj1349, 91.7% cadF, 16.7% irgA, and 8.3% hecA [n = 12]). Similar results have been obtained for the latter species in previous studies, because the ciaB, cj1349, and cadF genes were detected in 100% of isolates while irgA was detected in 17.3% to 30% of the strains and hecA was detected in 13.5% to 25.8% of strains (12, 13). Furthermore, in our study, the only strain (F1) that possessed all 5 genes belonged to A. butzleri.
It is plausible that there was a detection bias toward A. butzleri strains, as the primers were designed from the A. butzleri RM4018 genome (GenBank NC_009850.1). Nevertheless, there was correlation between the absence of virulence genes and the lack of invasion of Caco-2 cells, given that the 7 strains of A. thereius (3 strains), A. mytili (3 strains), and A. cibarius which were negative for all tested genes were either low or noninvasive (Table 1). In contrast, A. skirrowii S7 showed the highest adhesion and invasion values and possessed the four virulence-related genes (ciaB, hecA, cj1349, and cadF). This strain, and 2 strains of A. trophiarum, showed invasion values similar to or higher than (P < 0.05) those of S. enterica (used as the positive control), and their virulence genotype included at least the ciaB and cj1349 genes. The occurrence of putative virulence genes in the different Arcobacter species did correlate with those in the published whole-genome sequences. Karadas et al. (13) observed no correlation between virulence gene patterns and adhesion to or invasion of Caco-2 and HT-29 cell lines. They also observed that the putative functional domains of ciaB, cadF, and cj1349 genes did not change depending on the adhesion or invasion capacity. Those results were in part explained by the low number of isolates compared (n = 6), and it was indicated that further strains needed to be tested (13).
With respect to possible associations between strain origin and virulence, it was notable that strains from fecal sources were the most invasive (P < 0.05), followed by those from shellfish and meat (Fig. 3). Furthermore, the strains from fecal sources (n = 18) carried a higher proportion of virulence genes (100% with ciaB, 38.9% with irgA, 33.3% with cj1349, 11.1% with cadF, and 5.6% with hecA) than did those from food (n = 35; 88.6% with ciaB, 45.7% with cj1349, 37.1% with cadF, 8.6% with irgA, and 2.9% with hecA) and the rest of the strains (n = 7; 42.9% with ciaB and 14.3% with cj1349 and without irgA, hecA, and cadF). The irgA gene was more prevalent in strains from sewage (54.5%) than in others (8.2%, P < 0.05), and the cadF gene was more prevalent in those from food (37.1%) than in others (8.0%, P < 0.05). It is plausible that such traits are species related, since 8 of the 10 strains positive for the irgA gene were strains of A. defluvii. Of these, 75.0% (6/8) were from sewage. Eleven of the 15 food strains positive for the cadF gene belonged to A. butzleri, of which 90.9% (10/11) were from food.
The A. nitrofigilis type strain CECT 7204T, isolated from roots of Spartina alterniflora (28), did not adhere to or invade Caco-2 cells; however, it possessed the ciaB gene as shown by PCR, and this was also confirmed when analyzing its genome. In contrast, the two other A. nitrofigilis strains (F74 and F2176) isolated from mussels, which are considered a potential source of Arcobacter infection (3), showed adhesion and invasion abilities and were also ciaB+. In this respect, the role of ciaB and the other genes associated with Arcobacter virulence needs to be further studied.
This is the first study that demonstrates the presence of putative virulence genes associated with both adhesion and invasion and complementary in vitro tissue culture analysis for nearly all the Arcobacter species, using strains isolated from various sources. On the basis of these results, most Arcobacter species were confirmed as potential human pathogens, with some strains of A. butzleri, A. cryaerophilus, A. skirrowii, and the recently described A. trophiarum and A. defluvii potentially being more virulent. Further studies are warranted to further characterize these virulence traits and confirm their role in infection.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported in part by the project AGL2011-30461-C02-02 of the Ministerio de Ciencia e Innovación (MICINN), Spain, and by funding from the European Union Seventh Framework Programme (FP7/2007-2013 under grant agreement 311846). A.L. thanks Universitat Rovira i Virgili for a doctoral grant and CONICYT, Chile, for financial support through Becas Chile.
The authors are solely responsible for the content of this publication. It does not represent the opinion of the European Commission. The European Commission is not responsible for any use that might be made of data appearing therein.
We thank Kurt Houf (Ghent University, Belgium) and Nalini Chinivasagam (Animal Research Institute, Queensland, Australia) for kindly providing Arcobacter strains and Pilar Hernandez from IISPV for support with the statistics. A.L. is thankful to the School of Science and Technology, Nottingham Trent University, for their tissue culture facilities and to Susan Joseph for her kind support.
None of the authors report any potential conflicts of interest.
Footnotes
Published ahead of print 14 June 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.01073-13.
REFERENCES
- 1. Vandamme P, Falsen E, Rossau R, Hoste B, Segers P, Tytgat R, De Ley J. 1991. Revision of Campylobacter, Helicobacter, and Wolinella taxonomy: emendation of generic descriptions and proposal of Arcobacter gen. nov. Int. J. Syst. Bacteriol. 41:88–103 [DOI] [PubMed] [Google Scholar]
- 2. Vandamme P, Dewhirst FE, Paster BJ, On SLW. 2005. Genus II. Arcobacter Vandamme, Falsen, Rossau, Segers, Tytgat and De Ley 1991a, 99VP, p 1161–1165 In Brenner DJ, Kreig NP, Staley JT, Garrity GM. (ed), Bergey's manual of systematic bacteriology, 2nd ed Springer, New York, NY [Google Scholar]
- 3. Collado L, Figueras MJ. 2011. Taxonomy, epidemiology and clinical relevance of the genus Arcobacter. Clin. Microbiol. Rev. 24:174–192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Figueras MJ, Levican A, Collado L, Inza MI, Yustes C. 2011. Arcobacter ellisii sp. nov., isolated from mussels. Syst. Appl. Microbiol. 34:414–418 [DOI] [PubMed] [Google Scholar]
- 5. Levican A, Collado L, Aguilar C, Yustes C, Diéguez AL, Romalde JL, Figueras MJ. 2012. Arcobacter bivalviorum sp. nov. and Arcobacter venerupis sp. nov., new species isolated from shellfish. Syst. Appl. Microbiol. 35:133–138 [DOI] [PubMed] [Google Scholar]
- 6. Levican A, Collado L, Figueras MJ. 2013. Arcobacter cloacae sp. nov. and Arcobacter suis sp. nov., two new species isolated from food and sewage. Syst. Appl. Microbiol. 36:22–27 [DOI] [PubMed] [Google Scholar]
- 7. Vandenberg O, Dediste A, Houf K, Ibekwem S, Souayah H, Cadranel S, Douat N, Zissis G, Butzler JP, Vandamme P. 2004. Arcobacter species in humans. Emerg. Infect. Dis. 10:1863–1867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Ho HT, Lipman LJ, Gaastra W. 2006. Arcobacter, what is known and unknown about a potential foodborne zoonotic agent! Vet. Microbiol. 115:1–13 [DOI] [PubMed] [Google Scholar]
- 9. Wesley IV, Miller WG. 2010. Arcobacter: an opportunistic human food-borne pathogen?, p 185–211 In Scheld WM, Grayson ML, Hughes JM. (ed), Emerging infections 9. ASM Press, Washington, DC [Google Scholar]
- 10. Miller WG, Parker CT, Rubenfield M, Mendz GL, Wösten MM, Ussery DW, Stolz JF, Binnewies TT, Hallin PF, Wang G, Malek JA, Rogosin A, Stanker LH, Mandrell RE. 2007. The complete genome sequence and analysis of the epsilonproteobacterium Arcobacter butzleri. PLoS One 2:e1358. 10.1371/journal.pone.0001358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Dasti JI, Tareen AM, Lugert R, Zautner AE, Gross U. 2010. Campylobacter jejuni: a brief overview on pathogenicity-associated factors and disease-mediating mechanisms. Int. J. Med. Microbiol. 300:205–211 [DOI] [PubMed] [Google Scholar]
- 12. Douidah L, de Zutter L, Baré J, De Vos P, Vandamme P, Vandenberg O, Van den Abeele AM, Houf K. 2012. Occurrence of putative virulence genes in Arcobacter species isolated from humans and animals. J. Clin. Microbiol. 50:735–741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Karadas G, Sharbati S, Hänel I, Messelhäußer U, Glocker E, Alter T, Gölz G. 7 May 2013. Presence of virulence genes, adhesion and invasion of Arcobacter butzleri. J. Appl. Microbiol. [Epub ahead of print.] 10.1111/jam.12245 [DOI] [PubMed] [Google Scholar]
- 14. Donachie SP, Bowman JP, On SL, Alam M. 2005. Arcobacter halophilus sp. nov., the first obligate halophile in the genus Arcobacter. Int. J. Syst. Evol. Microbiol. 55:1271–1277 [DOI] [PubMed] [Google Scholar]
- 15. Kim HM, Hwang CY, Cho BC. 2010. Arcobacter marinus sp. nov. Int. J. Syst. Evol. Microbiol. 60:531–536 [DOI] [PubMed] [Google Scholar]
- 16. Houf K, Tutenel A, De Zutter L, Van Hoof J, Vandamme P. 2000. Development of a multiplex PCR assay for the simultaneous detection and identification of Arcobacter butzleri, Arcobacter cryaerophilus and Arcobacter skirrowii. FEMS Microbiol. Lett. 193:89–94 [DOI] [PubMed] [Google Scholar]
- 17. Figueras MJ, Collado L, Guarro J. 2008. A new 16S rDNA-RFLP method for the discrimination of the accepted species of Arcobacter. Diagn. Microbiol. Infect. Dis. 62:11–15 [DOI] [PubMed] [Google Scholar]
- 18. Figueras MJ, Levican A, Collado L. 2012. Updated 16S rRNA-RFLP method for the identification of all currently characterized Arcobacter spp. BMC Microbiol. 12:292. 10.1186/1471-2180-12-292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Collado L, Cleenwerck I, Van Trappen S, De Vos P, Figueras MJ. 2009. Arcobacter mytili sp. nov., an indoxyl acetate-hydrolysis-negative bacterium isolated from mussels. Int. J. Syst. Evol. Microbiol. 59:1391–1396 [DOI] [PubMed] [Google Scholar]
- 20. Houf K, De Zutter L, Van Hoof J, Vandamme P. 2002. Assessment of the genetic diversity among arcobacters isolated from poultry products by using two PCR-based typing methods. Appl. Environ. Microbiol. 68:2172–2178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Ho HT, Lipman LJ, Hendriks HG, Tooten PC, Ultee T, Gaastra W. 2007. Interaction of Arcobacter spp. with human and porcine intestinal epithelial cells. FEMS Immunol. Med. Microbiol. 50:51–58 [DOI] [PubMed] [Google Scholar]
- 22. Miles AA, Misra SS, Irwin JO. 1938. The estimation of the bactericidal power of the blood. J. Hyg. (Lond.) 38:732–749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Townsend S, Hurrel E, Forsythe S. 2008. Virulence studies of Enterobacter sakazakii isolates associated with a neonatal intensive care unit outbreak. BMC Microbiol. 8:64. 10.1186/1471-2180-8-64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. 2011. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 28:2731–2739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Houf K, Stephan R. 2007. Isolation and characterization of the emerging foodborne pathogen Arcobacter from human stool. J. Microbiol. Methods 68:408–413 [DOI] [PubMed] [Google Scholar]
- 26. De Smet S, Vandamme P, De Zutter L, On S, Douidah L, Houf K. 2011. Arcobacter trophiarum sp. nov. isolated from fattening pigs. Int. J. Syst. Evol. Microbiol. 63:356–361 [DOI] [PubMed] [Google Scholar]
- 27. Houf K, On S, Coenye T, Debruyne L, De Smet S, Vandamme P. 2009. Arcobacter thereius sp. nov., isolated from pigs and ducks. Int. J. Syst. Evol. Microbiol. 59:2599–2604 [DOI] [PubMed] [Google Scholar]
- 28. McClung CR, Patriquin DG, Davis RE. 1983. Campylobacter nitrofigilis sp. nov., a nitrogen fixing bacterium associated with roots of Spartina alterniflora Loisel. Int. J. Syst. Bacteriol. 33:605–612 [Google Scholar]
- 29. Collado L, Levican A, Perez J, Figueras MJ. 2011. Arcobacter defluvii sp. nov., isolated from sewage. Int. J. Syst. Evol. Microbiol. 61:1895–1901 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.