Abstract
The methanotrophs in rice field soil are crucial in regulating the emission of methane. Drainage substantially reduces methane emission from rice fields. However, it is poorly understood how drainage affects microbial methane oxidation. Therefore, we analyzed the dynamics of methane oxidation rates, composition (using terminal restriction fragment length polymorphism [T-RFLP]), and abundance (using quantitative PCR [qPCR]) of methanotroph pmoA genes (encoding a subunit of particulate methane monooxygenase) and their transcripts over the season and in response to alternate dry/wet cycles in planted paddy field microcosms. In situ methane oxidation accounted for less than 15% of total methane production but was enhanced by intermittent drainage. The dry/wet alternations resulted in distinct effects on the methanotrophic communities in different soil compartments (bulk soil, rhizosphere soil, surface soil). The methanotrophic communities of the different soil compartments also showed distinct seasonal dynamics. In bulk soil, potential methanotrophic activity and transcription of pmoA were relatively low but were significantly stimulated by drainage. In contrast, however, in the rhizosphere and surface soils, potential methanotrophic activity and pmoA transcription were relatively high but decreased after drainage events and resumed after reflooding. While type II methanotrophs dominated the communities in the bulk soil and rhizosphere soil compartments (and to a lesser extent also in the surface soil), it was the pmoA of type I methanotrophs that was mainly transcribed under flooded conditions. Drainage affected the composition of the methanotrophic community only minimally but strongly affected metabolically active methanotrophs. Our study revealed dramatic dynamics in the abundance, composition, and activity of the various type I and type II methanotrophs on both a seasonal and a spatial scale and showed strong effects of dry/wet alternation cycles, which enhanced the attenuation of methane flux into the atmosphere.
INTRODUCTION
Methanotrophs utilize methane as the sole carbon and energy source. Methane is a potent greenhouse gas in the atmosphere. The activity of methanotrophs is crucial for attenuation of methane emission from the biosphere into the atmosphere. They consume about 0.6 Gt methane annually, roughly equivalent to the total amount of methane emitted into the atmosphere (1). Although anaerobic oxidation of methane has been discovered in many anoxic sediments, it is the aerobic oxidation that is important for methane emission from rice field soil, oxidizing up to 90% of methane produced (2–5). Among the aerobic methanotrophs, proteobacterial methanotrophs play the dominant role, while verrucomicrobial methanotrophs are restricted to extreme environments (6). The aerobic oxidation of methane depends on methane monooxygenase (MMO) in the initial enzymatic reaction. This enzyme has two forms, a soluble type (sMMO) and a membrane-associated type (pMMO). All known bacterial methanotrophs except Methylocella and Methyloferula possess a pMMO (7, 8). The pmoA gene that encodes the subunit of membrane-bound MMO is highly conserved in proteobacterial methanotrophs and has been widely used as phylogenetic marker for ecological studies (9–14).
Proteobacterial methanotrophs can be divided into type I and type II, and type I can be further divided into types Ia and Ib based on the phylogeny of the pmoA gene (15, 16). The ecophysiology of the different types of methanotrophs remains largely unknown (12, 17). Many environmental factors, such as concentrations of methane and availability of N, can influence the composition and activity of methanotrophs (12, 17). An early study using agar diffusion columns showed that type I methanotrophs preferred lower methane and higher O2 concentrations than type II methanotrophs (18). Other studies using soils, however, revealed that both type I and type II methanotrophs dominated at high methane concentrations (19, 20). Recently, it was found that type II methanotroph Methylocystis sp. strain SC2 contains a novel isoenzyme, pMMO2, and can oxidize methane in low concentrations, even at the atmosphere level (21). Thus, the effects of methane concentrations on the composition and activity of the methanotrophic community are still unclear. Similarly, the effect of N availability on methanotrophs is also not yet completely clear (22).
A flooded rice field is a clearly structured ecosystem and contains three soil compartments: anoxic bulk soil, oxic surface soil, and oxic rhizosphere soil (12, 23). The capacity for methane oxidation exhibits niche differentiation, with a low capacity in bulk soil due to O2 limitation and a relatively high capacity in surface and rhizosphere soils. However, knowledge about the spatial distribution of methanotrophs on the soil compartment scale is still limited (12). It seems that type II methanotrophs are dominant in bulk soil whereas both type I and II methanotrophs exist in rhizosphere and surface soils (13, 24–28). Two separate studies of pmoA transcripts recently indicated that type I rather than type II methanotrophs were particularly active in the rhizosphere (29) and surface soils (30). The activity of methanotrophs (transcription of pmoA) in bulk soil has never been analyzed before, although bulk soil is the largest reservoir of methanotrophs in the rice field (31) and may become a habitat suitable for aerobic methanotrophs upon drainage (32). Thus, a systematic quantitative analysis of the presence and activity of methanotrophs in the different soil compartments is still lacking.
Methane oxidation in rice field microcosms exhibits seasonal variation, with higher rates at the early vegetative stages followed by a gradual decrease until negligible rates are reached at the late growth stages (27, 33). Such seasonal change has also been observed under field conditions with the same paddy soil (34). However, the population densities of total methanotrophs determined by the most-probable-number (MPN) technique seem to increase during the season and stay constant at the late growth stage (27). Relatively constant population densities of methanotrophs have also been found by quantification of pmoA copy numbers (29), but a systematic study of the activity and abundance of methanotrophs in the different soil compartments has not yet been done.
Intermittent drainage is the most promising strategy in rice agriculture management to mitigate methane emission (25, 35, 36). Drainage dramatically changes the physicochemical conditions of flooded paddy soil, for example, increasing diffusion of gases and availability of O2. Our previous study revealed that intermittent drainages resulted in both a decrease of methanogen and an increase of methanotroph population density, being partially responsible for the reduction of methane emission from rice field soil (25). Furthermore, the populations of total methanotrophs in the different soil compartments responded differently to intermittent drainage. However, it is still unclear how the active methanotrophs respond to drainage.
Therefore, we used rice field microcosms to study the abundance, activity, and composition of the aerobic methanotrophic bacterial community under conditions of two water regimens, i.e., continuous flooding and dry/wet alternation. We measured the methane oxidation rates in situ, the methane oxidation potential (MOP) in vitro, and the copy numbers of pmoA gene and pmoA transcripts and their compositions. We were interested to obtain a mechanistic understanding for how alternate dry/wet cycles regulate the activity and the population dynamics of methanotrophs on a seasonal as well as a spatial scale. We hypothesized that the different types of methanotrophs exhibit niche differentiation between bulk soil, surface soil, and rhizosphere soil and that this adaptation would influence methane oxidation activity.
MATERIALS AND METHODS
Rice microcosm and drainage treatment.
Rice microcosms were prepared following a procedure described previously (37). In brief, 1.4 kg of air-dried soil collected from an experimental rice field at China National Rice Research Institute (38) was weighed into 3.0-liter cylindrical plastic pots. Soil was flooded, and seedlings of an Indica rice variety (Oriza sativa [Jinzao 47]) were planted. A total of 48 microcosms were prepared, and a water layer of 2- to 3-cm depth above the soil surface was maintained till day 54. Thereafter, microcosms were divided into two groups: half of the pots were continuously flooded till the end of the experiment with soil water content of 70.7% to 73.3% (90 days) (referred to as FL treatment), and the other half were exposed to two periods of drainage from day 55 to day 62 and day 73 to day 80 (referred to as DR treatment). The water content of drained soil was 49.8% and 40.9% on day 62 and day 80, respectively. The DR soil was flooded again after each drainage event and the water content increased to a level similar to that of the FL soil. The experiments were carried out with three biological replicates.
Methane oxidation rate.
Difluoromethane (CH2F2) is considered to be a specific inhibitor of methane oxidation (39). In the present experiment, the methane oxidation rate was determined by analyzing the methane emission rate in the presence and absence of CH2F2. The difference was taken as the rate of methane oxidation expressed in mmol CH4 day−1 m−2 (40). Methane emission rates were determined using the closed static chamber approach (37). We added CH2F2 to the headspace of the static chamber at a concentration of 1% as described before (39, 40). For the last few measurements (from day 84 to day 89), the CH2F2 concentration was increased to 4% to overcome the possible limitation of diffusion of CH2F2 from the headspace to the rhizosphere.
Methane oxidation potential.
Destructive soil sampling of microcosms was conducted on days 54, 57, 62, 67, 72, 76, 80, and 90. Three subsamples were collected: (i) the surface soil was taken from the upper 5-mm soil layer; (ii) the bulk soil was taken from a zone that was free of rice roots; and (iii) the rhizosphere soil was stripped from the rice roots. Measurement of methane oxidation potential started within 1 h after soil sampling. About 2.0 g (wet weight) of fresh soil sample was mixed with 1.0 ml sterile demineralized water and placed into a 25-ml sterile glass tube. The air headspace was supplemented with about 50,000 ppm by volume (ppmv) CH4. The tubes were incubated for 100 h at 30°C and shaken at 120 rpm. Depletion of methane in the headspace was measured by the use of a gas chromatography-flame ionization detector (GC-FID) (41). Methane oxidation potentials were calculated from the slopes of regression lines of methane depletion with time and expressed as nmol CH4 h−1 per gram (dry weight) (gdw−1) of soil.
CH4 and NH4+ in soil porewater.
Rhizon samplers (Eijkelkamp, Giesbeek, Netherlands) were buried vertically into the soil of the microcosms close to rice roots, and the porewater samples were collected weekly (42). The concentration of dissolved methane in the porewater was analyzed according to a procedure described previously (14). The ammonium concentration was analyzed by using a Safire microplate reader (Tecan, Crailsheim, Germany) as described previously (43).
Nucleic acid extraction and cDNA synthesis.
An aliquot of the soil was frozen in liquid nitrogen immediately after sampling and stored at −80°C until molecular analysis. Total DNA and RNA of three replicated microcosm soil samples were extracted using a bead-beating method following a procedure described before (37). In brief, 0.5 g soil was extracted first with TPMS buffer (50 mM Tris-HCl [pH 7.0], 1.7% [wt/vol] polyvinylpyrrolidon K25, 20 mM MgCl2, 1% [wt/vol] sodium dodecyl sulfate) once and then twice with phenol-based lysis buffer (5 mM Tris-HCl [pH 7.0], 5 mM Na2EDTA, 1% [wt/vol] SDS, 6% [vol/vol] water-saturated phenol). The pooled supernatants were further extracted with water-saturated phenol, phenol-chloroform-isoamyl alcohol, and chloroform-isoamyl alcohol. The total nucleic acids were precipitated with 3 volume of cold ethanol, washed with cold 70% ethanol, and dissolved in TE buffer (10 mM Tris-HCl, 1 mM EDTA [pH 8.0]).
For RNA extraction, the coarse extracts were incubated at 37°C for 1 h with RNase-free DNase and a recombinant RNasin RNase inhibitor kit (Promega, Mannheim, Germany). Further purification of RNA was performed using an RNeasy Mini Kit (Qiagen, Hilden, Germany). The ImProm-II reverse transcription system (Promega) was used to synthesize the cDNA as described before (37). To verify the absence of DNA, a control PCR was performed with nuclease-free water instead of the reverse transcriptase. The details of the procedure have been described previously (37). DNA and cDNA samples were stored at −20°C until the further analysis.
T-RFLP analysis, cloning, and sequencing.
DNA and cDNA samples were used for PCR amplification of fragments of pmoA using the primer set A189 and mb661. For the analyses of terminal restriction fragment length polymorphism (T-RFLP), the forward primer was labeled with 6-carboxyfluorescein (FAM) (13). For cloning and sequencing, PCR amplification used the same primers without the FAM label. T-RFLP, cloning, and sequencing were performed according to protocols described previously (14). Randomly selected clones were sequenced, and the phylogenetic analysis was performed as described previously (14).
qPCR analysis.
Before real-time (quantitative) PCR (qPCR) analysis was performed, the possible inhibition by DNA impurity was reduced to a minimum by dilution. DNA and cDNA preparations were diluted by 25:1 and 5:1, respectively, for the gene copy and transcript quantification. The quantitative PCR of pmoA fragments was performed in an iCycler instrument (Bio-Rad, Munich, Germany) using SybrGreen Jumpstart Taq ReadyMix (Sigma, Munich, Germany) (10, 44). The A189 and mb661 primer set (same as for T-RFLP and cloning) was used. The thermal cycles and fluorescence signal acquisition followed the protocol described previously (10). The DNA standard was prepared from purified plasmid DNA of one pmoA clone, with the concentrations ranging from 1.0 × 101 to 1.0 × 108 copies μl−1 (14). The transcript standard was prepared from an in vitro transcript of one pmoA clone using Riboprobe in vitro transcription systems (Promega) according to the manufacturer's protocol (45). The concentrations of the transcript standard ranged from 1.0 × 101 to 1.0 × 106 transcripts μl−1. Each measurement was performed in triplicate.
Statistical analysis.
The ordination analysis of T-RFLP patterns of pmoA gene and transcript was done using CANOCO for Windows 4.5 software (Microcomputer Power, Ithaca, NY), and one-way analysis of variance (ANOVA) was performed to test significant differences between treatments using the SAS program (SAS Institute, Cary, NC). The details of the procedure have been described previously (25).
Nucleotide sequence accession numbers.
The sequences determined in this study have been deposited in the GenBank database under accession numbers KF233886 to KF233940.
RESULTS
Seasonal variations of methane oxidation rate and potential.
Rates of methane oxidation increased with time, reached a peak on day 48 (corresponding to the tillering stage of rice plants) (Fig. 1A), and then decreased and became almost zero after day 60. A significant increase of the oxidation rate, however, was detected when the concentration of CH2F2 in the headspace was increased to 4% during the final growth stage (days 84 to 89). The first dry/wet cycle (days 55 to 72) did not affect the methane oxidation, but the second cycle (days 73 to 90) reduced the rate significantly.
Fig 1.
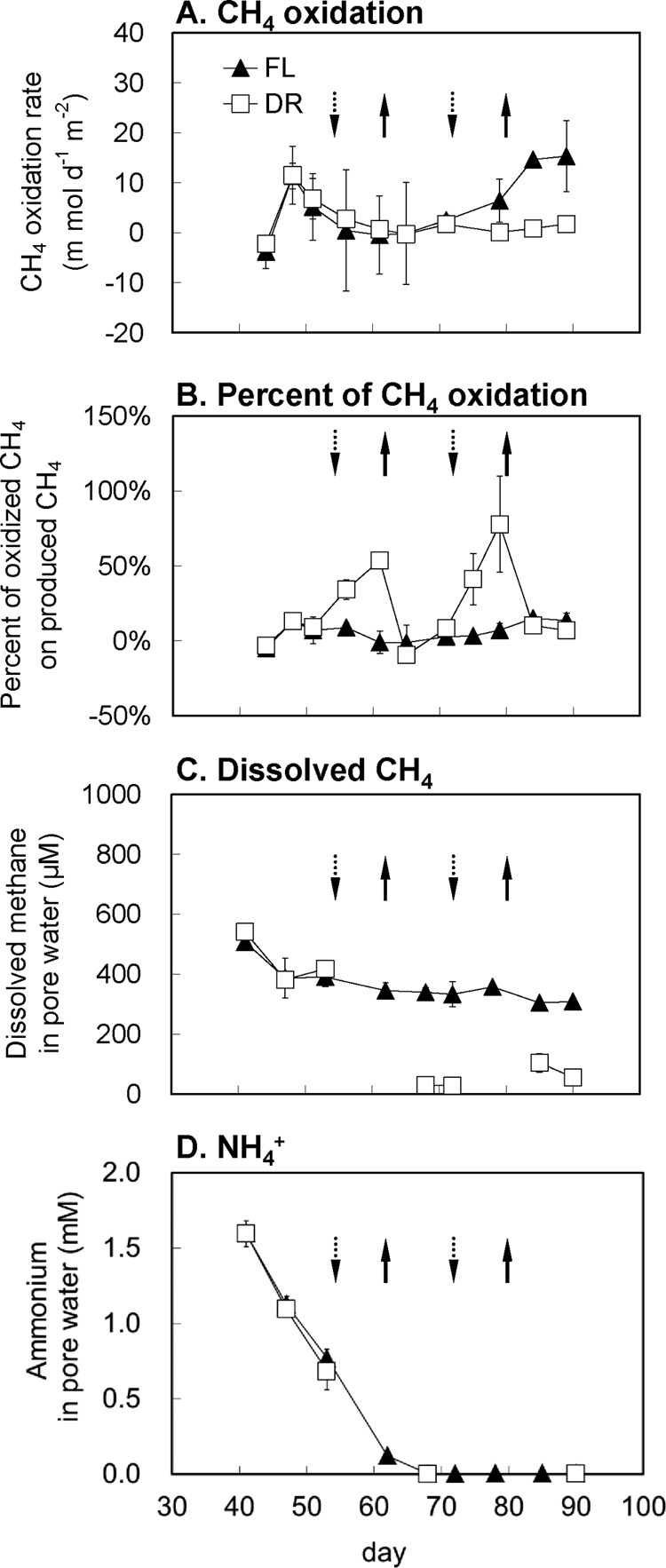
Methane oxidation rates in situ (A), percentage of oxidized methane accounted for produced methane (B), and the dissolved methane concentration (C) and NH4+ concentration (D) in pore water under conditions of treatments of continuous flooding (FL) and alternate dry/wet cycles (DR) (n = 3). Arrows pointing downward and upward indicate the beginning of draining (or redraining) and reflooding in the DR treatment, respectively.
We calculated the percentage of the anaerobically produced methane being oxidized. The percentage in the FL treatment never exceeded 15% within the entire experimental period, but the percentage in DR treatment was significantly higher at both drainage stages (Fig. 1B). For example, on both day 61 and day 79, the percentage was more than 50%, indicating that more than half of the methane produced was oxidized before being emitted into the atmosphere. These results indicated that drainages enhanced the relative contribution of methane oxidation to the total production.
The dissolved methane concentrations in the pore water of the FL treatment were about 500 μmol liter−1 and decreased by only about 40% during the season. However, drainage reduced the dissolved methane concentration to less than 50 μmol liter−1 (Fig. 1C). The methane concentrations then stayed low and never recovered, although the soil was flooded again after each drainage event. The ammonium concentrations in the pore water decreased to the detection limit after day 65 (Fig. 1D). There was no significant difference between the FL and DR treatments.
Methane oxidation potential (MOP) was determined by laboratory incubation. The highest MOP of FL treatment was detected in the rhizosphere soil, followed by that detected in surface soil (Fig. 2B and C). The bulk soil of FL treatment showed very low MOP throughout the experimental period (Fig. 2A). MOP in the rhizosphere and surface soils decreased markedly during the season in the later period.
Fig 2.
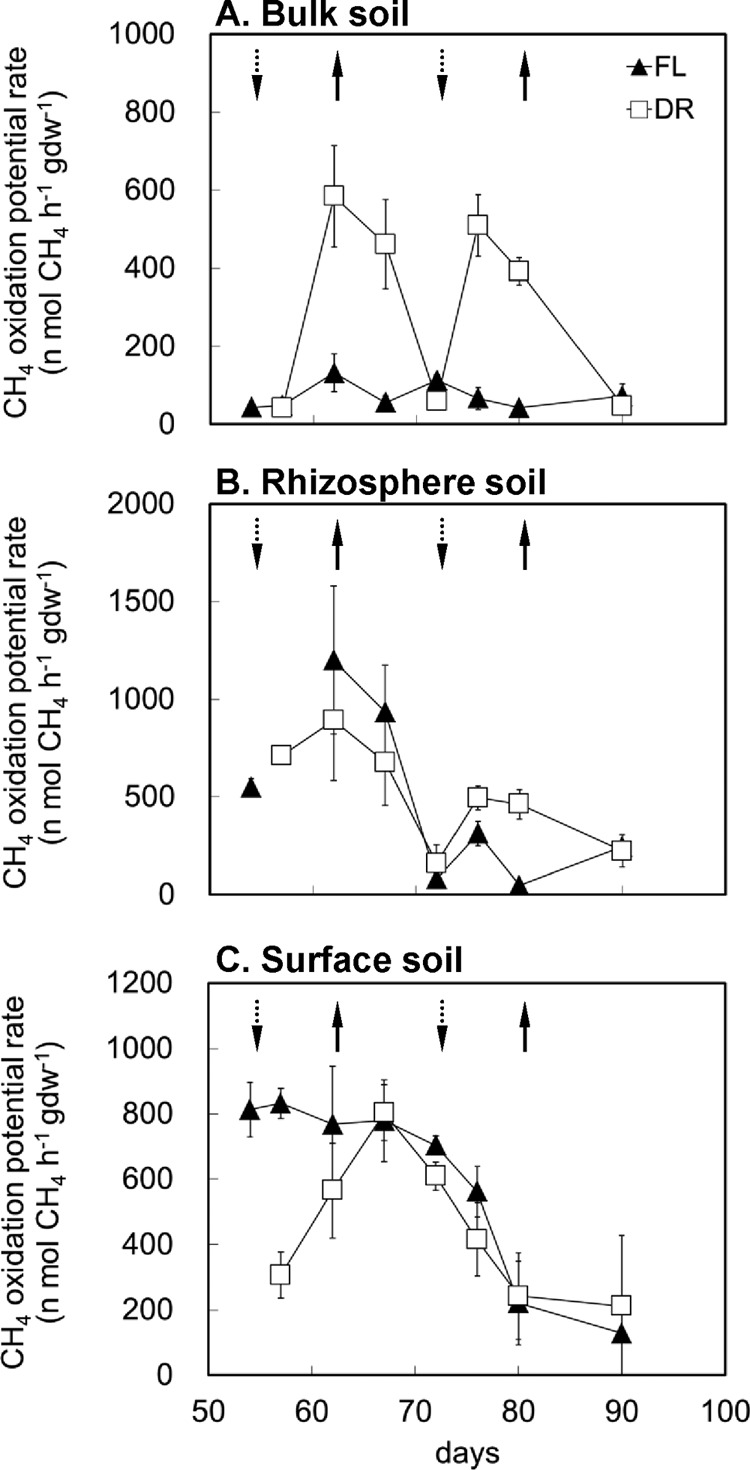
Rates of methane oxidation potential in bulk soil (A), rhizosphere soil (B), and surface soil (C) in the FL and DR treatments under aerobic incubation conditions (n = 3).
A remarkable stimulation of MOP was observed in the bulk soil after each drainage event (Fig. 2A). After the soil was reflooded, the MOP decreased again. These results indicate that drainage can substantially activate methane oxidation in the bulk soil. In contrast, the drainage events reduced MOP in the rhizosphere and surface soils during the early stage when MOP was high (Fig. 2B and C). In the later period, the drainage cycles apparently did not affect MOP in the surface soil but enhanced MOP in the rhizosphere soil.
Seasonal patterns of methanotrophic community.
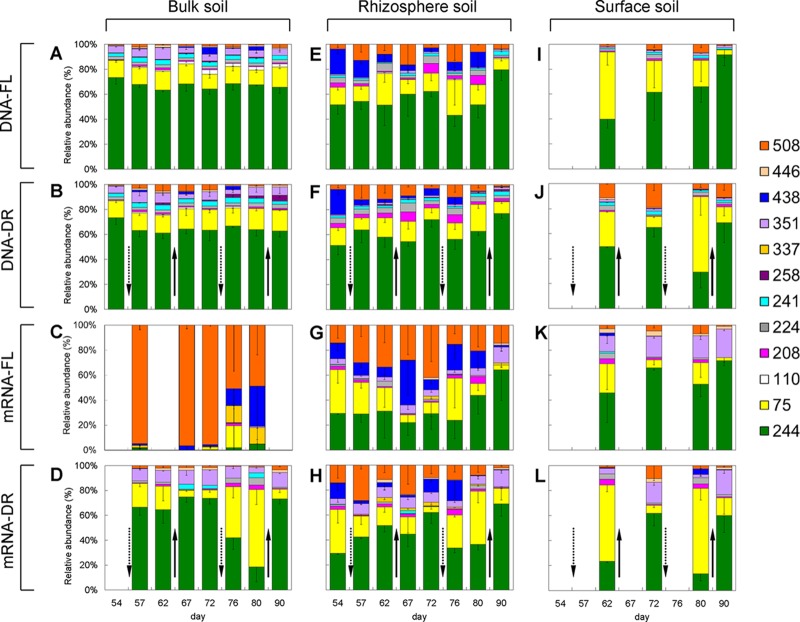
T-RFLP analyses of pmoA genes and transcripts were performed to investigate the population dynamics of methanotrophs in the rice soil (Fig. 3). Twelve major T-RFs were detected across samples. Five T-RFs (75, 244, 351, 438, and 508 bp) were prevalent, and seven (110, 208, 224, 241, 258, 337, and 446 bp) showed relatively low abundances. A clone library was constructed for the pmoA transcripts in the rhizosphere soil of FL treatment. Sequence analysis (see Table S1 and Fig. S1 in the supplemental material) indicated that the methanotrophic community in the soil comprised type Ib methanotroph genera Methylococcus and Methylocaldum (related to T-RF of 75 bp), type Ia genera Methylomonas (related to 438 bp) and Methylobacter (related to 508 bp), and type II genera Methylocystis and Methylosinus (related to T-RFs of 208 and 244 bp). This result was consistent with our previous studies (14, 25, 28). The assignments of T-RFs in the T-RFLP analyses are based on in silico analysis of clone sequences from this study and previous studies.
Fig 3.
Composition of the methanotrophic community in three soil compartments (bulk soil [A, B, C, and D], rhizosphere soil [E, F, G, and H], and surface soil [I, J, K, and L]) analyzed by T-RFLP targeting pmoA genes in FL treatment (A, E, and I) and DR treatment (B, F, and J) and pmoA transcripts in FL treatment (C, G, and K) and DR treatment (D, H, and L).
DNA- and mRNA-based T-RFLP fingerprints presumably represent the compositions of total and active methanotrophs, respectively. DNA T-RFLP revealed that the 244-bp T-RF (type II Methylocystis and Methylosinus) was dominant in the bulk soil, accounting for about 65% of total fluorescence frequency. T-RFLP profiles from the bulk soil did not change over the experimental period, suggesting stability of the methanotroph community with time (Fig. 3A). In the rhizosphere soil (Fig. 3E), the relative abundance of type I methanotroph T-RFs (438 and 508 bp) increased and that of type II T-RF (244 bp) decreased compared with the bulk soil. In addition, the temporal fluctuation of T-RFLP fingerprints increased in the rhizosphere soil. For the surface soil, T-RFLP analysis was performed only on days 62, 72, 80, and 90, corresponding to the end of each drainage or reflooding event. The T-RFLP profiles of the surface soil showed greater fluctuations than those of the bulk and rhizosphere soils (Fig. 3I). Drainage did not influence the T-RFLP profiles in the bulk and rhizosphere soils (Fig. 3B and F). However, drainage apparently increased the relative abundance of type I methanotrophs (T-RF of 75 bp) in the surface soil (Fig. 3J).
The patterns of mRNA T-RFLP differed markedly from those obtained with DNA T-RFLP. In the bulk soil with FL treatment (Fig. 3C), no PCR product could be obtained on days 54, 62, and 90 even when the number of amplification cycles was increased to 40. On the other days, the amplified products of pmoA transcripts mostly belonged to type I methanotrophs, in particular, Methylobacter (508-bp T-RF). This mRNA-based pattern differed greatly from the DNA-based pattern, possibly due to the weak representation of in vitro transcription with a low RNA quantity. Drainage facilitated PCR amplification of pmoA transcripts and generated T-RFLP patterns comparable to those of the DNA fingerprints (Fig. 3D). However, the relative abundance of the 75-bp T-RF (type I Methylococcus and Methylocaldum) substantially increased after each drainage event, especially after the second one, and decreased again after reflooding of the soil.
In the rhizosphere soil, the structure of the active methanotrophs was more dynamic and diverse than in the bulk soil (Fig. 3G). In particular, the relative abundance of T-RFs related to type I methanotrophs, especially those of Methylomonas (438 bp) and Methylobacter (508 bp), markedly increased. The relative abundance of Methylomonas (438 bp) and Methylobacter (508 bp) decreased in DR treatment relative to FL treatment (Fig. 3H).
In the surface soil, the 351-bp T-RF transcripts, which were probably indicative of the presence of type I Methylomicrobium (13, 33), became more abundant than the DNA fingerprints (Fig. 3K). Drainage resulted in a significant change of the active methanotroph community composition (Fig. 3L). The pmoA transcripts of Methylocystis and Methylosinus (244 bp) dominated under the flooding conditions and those of Methylococcus and Methylocaldum (75 bp) after the drainage events.
Correspondence analysis of all T-RFLP data revealed that the community structure of total methanotrophs did not respond to alternate dry/wet cycles (DNA level; see Fig. S2A in the supplemental material) but that the active methanotrophs changed greatly (mRNA level; see Fig. S2B in the supplemental material).
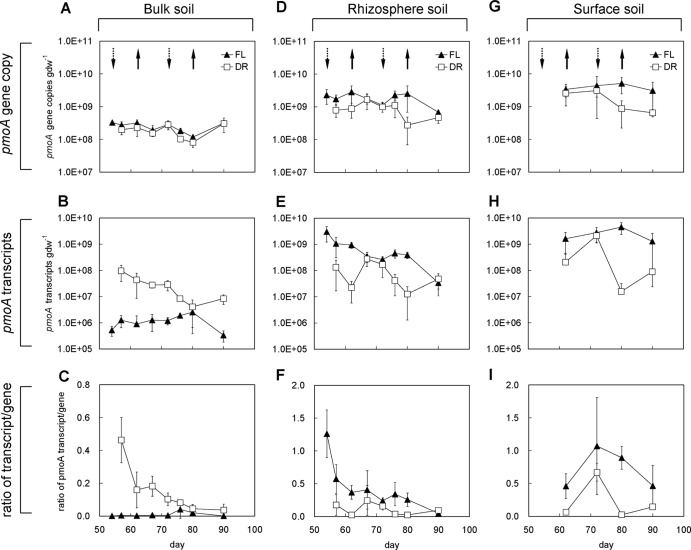
Quantification of pmoA genes and transcripts.
Quantitative PCR revealed that the surface soil (Fig. 4G) and the rhizosphere soil (Fig. 4D) contained pmoA copy numbers that were about 1 order of magnitude higher than in the bulk soil (Fig. 4A). The gene copy numbers in the three compartments with FL treatment did not change much over the experimental period. While drainage did not influence the gene copy numbers in the bulk soil, it reduced them in the rhizosphere and surface soils after the second drainage cycle by 1 order of magnitude.
Fig 4.
Dynamics of methanotrophic abundance shown by qPCR analyses of pmoA gene copies in bulk soil (A), rhizosphere soil (D), and surface soil (G); pmoA transcript copies in bulk soil (B), rhizosphere soil (E), and surface soil (H); and transcript-to-gene ratios in bulk soil (C), rhizosphere soil (F), and surface soil (I).
The pmoA transcript copy numbers in the FL treatments were different in the three soil compartments. In the bulk soil (Fig. 4B), they were about 2 to 3 orders of magnitude lower than in the rhizosphere soil (Fig. 4E) and surface soil (Fig. 4H). The transcript numbers remained more or less constant over time in the bulk and surface soils but decreased gradually in the rhizosphere soil.
Drainage treatment increased the pmoA transcript copy numbers in the bulk soil (Fig. 4B) but decreased them in the rhizosphere soil (Fig. 4E) and the surface soil (Fig. 4H). In the rhizosphere and surface soils, the dynamics of the pmoA transcript copy numbers paralleled the drainage and reflooding events, while in the bulk soil, pmoA transcript copy numbers were generally higher under DR than under FL conditions.
The transcript-to-gene ratio is a potential indicator of the relative expression levels of the pmoA gene. In the bulk soil, this ratio was on the average only 0.01 in FL treatment. In the DR treatment, it was significantly higher (Fig. 4C), indicating stimulation of methanotroph activity by drainage. In the flooded rhizosphere and surface soils, the ratio was still higher but decreased upon drainage (Fig. 4F and I).
DISCUSSION
This is a comprehensive and systematic report about the seasonal and spatial dynamics of methane oxidation, the population and activity of methanotrophic bacteria, and their responses to the dry/wet cycles in a planted paddy soil.
Seasonal dynamics.
Methane oxidation rates in intact microcosms reached a peak at the tillering stage of rice plants and then decreased gradually. This tendency of decreasing rates has been observed previously (27, 40) and is here corroborated with the measurement of methane oxidation potentials showing a similar decreasing tendency in the rhizosphere and surface soils (Fig. 2B and C). The qPCR analysis also showed a substantial decrease in pmoA transcript number in the rhizosphere soil. The population size (i.e., total copy number of pmoA) remained fairly constant over the experimental period, and the composition of the methanotrophic community (i.e., T-RFLP patterns) also changed only slightly, as previously observed in a study of Italian rice field soil (33). Hence, although there were some seasonal dynamics of the methanotrophic populations, the seasonal change in activity was much more pronounced. The decrease of pmoA transcription activity during the experimental period was very possibly related to N limitation and the reduced capacity of O2 release by aging roots in the later stages (34, 40, 46). The concentrations of ammonium in pore water (Fig. 1D) and the extractable ammonium in the bulk, rhizosphere, and surface soils (data not shown) decreased to the detection limit after day 65. Although the effect of ammonium is still in debate, it is likely that ammonium addition stimulates the activity of methanotrophs in rice field soil (47, 48).
However, unexpectedly, a higher rate of methane oxidation was detected in the last days with 4% CH2F2. It was probable that 1% CH2F2 was insufficient to inhibit methane oxidation in the planted rice-soil system (49). Alternatively, 4% CH2F2 probably caused the inhibition of not only methane oxidation but also methane production. More tests might be necessary to define the proper use of CH2F2 for a best estimation of in situ methane oxidation in rice field soil.
Spatial dynamics.
To the best of our knowledge, the transcript numbers of pmoA genes among different soil compartments were quantified and compared for the first time in the present study, although the cell numbers of methanotrophs were counted by the MPN technique before (27, 31, 46). We found that the pmoA copy number was approximately 1 order of magnitude higher and the transcript number was 2 to 3 orders higher in the rhizosphere and surface soils than in the anoxic bulk soil. This spatial difference is highly consistent with the methane oxidation potential. The rhizosphere and surface soils are suitable niches for methanotrophs, since both O2 and methane are available there. However, the methanotrophic activity in the bulk soil is normally suppressed by the lack of O2 (12).
For the community structures, type II methanotrophs dominated the methanotrophic community in all soil samples, albeit with a decrease of relative abundance in the rhizosphere soil and a high fluctuation in the surface soil. The compositions of active methanotrophs differed markedly from the community structure. Type I methanotrophs produced significantly greater amounts of transcripts than type II, especially in the rhizosphere soil, where methane oxidation was active. Thus, type I methanotrophs are functionally more active than the type II group in the rice soil, in agreement with our previous study in the same soil (28) and with the studies in Italian rice field soils (30, 33).
Effects of dry/wet cycles.
A clear effect of drainage on methane oxidation potential was detected, and the levels of this effect differed among soil compartments. Drainage appeared to reduce methane oxidation potential in the rhizosphere and surface soils, especially in the early period (Fig. 2). This result appears to contradict our previous report showing the stimulation of rhizosphere methane oxidation by drainage (25). The discrepancy is probably due to the extent of soil dryness. In the earlier experiment, only a mild drainage was applied which kept soil water content above 65%, while the drainage was more extensive in the present experiment, with the soil water content decreasing to 41%. Most of methane was possibly emitted directly into the atmosphere after extensive drainage and became a limiting factor for methanotrophic activity in the rhizosphere and surface soils. Nevertheless, the methane oxidation potential in the bulk soil was significantly enhanced, in similarity to the observation in the rhizosphere soil in the previous study (25). The increase of O2 availability after drainage together with methane trapped in the bulk soil probably maintained the methanotrophic activity after drainage management.
In correspondence with the methane oxidation potential, drainage displayed distinct effects on methanotrophic population dynamics among the different soil compartments. Drainage resulted in a marked increase in the total transcript numbers as well as in the transcript-to-gene ratio in the bulk soil, while in the rhizosphere and surface soils the transcription dynamics showed a tight coupling with the dry/wet cycle, as declining and resuming levels paralleled drainage and reflooding events. The contrasting effects in soil compartments are probably related to the interplay of O2 and methane availability. While O2 was lacking in the anoxic bulk soil, drainage improved O2 availability. In the rhizosphere and surface soils, where both O2 and methane are not limiting under water-flooding conditions, drainage possibly caused depletion of methane due to direct emission and resulted in reduction of methanotrophic activity and growth. It has been hypothesized that rice field soil retains a seed bank of methanotrophs (represented by DNA), that different members are activated under different conditions (31, 50), and that drainage results in increase of type I methanotrophs (32). Our results indicate that the seed bank of methanotrophs is not influenced so much by the soil dry/wet cycle, while the active members respond much more strongly. Such a tight coupling between methanotrophic activity (transcripts) and a management factor (alternate flooding and drainage) has not been shown before to the best of our knowledge.
Type I and type II groups showed distinct transcriptional responses to the soil dry/wet cycle. Total transcripts increased after drainage in the bulk soil. The T-RFLP pattern of pmoA transcripts was similar to that of the gene fingerprints in the first drainage, but type I methanotrophs produced more transcripts in the second drainage cycle than type II methanotrophs. This result indicated that in the first drainage, type I and type II methanotrophs were both activated in the bulk soil but in the second drainage, type I methanotrophs were stimulated to a greater extent. In the rhizosphere and surface soils, the dynamics of total transcription was coupled to the drainage and reflooding cycle. Type I methanotrophs, especially the genera Methylococcus and Methylocaldum (i.e., 75-bp T-RF), appeared relatively resistant when total transcripts declined during drainage. On the other hand, reflooding of soil resulted in recovery of total transcripts, and it appeared that type II methanotrophs recovered faster than the type I group. The differential responses are probably related to the alternating O2 and methane conditions. Drainage increased O2 availability but possibly depleted methane. Type I methanotrophs are more resistant to this niche shift. Reflooding returned the soil to anoxic conditions, and methane concentration probably increased due to the recovery of methanogenesis (37). Type II methanotrophs are apparently better adapted to this niche shift than the type I group.
In summary, methane oxidation rate as well as oxidation potential showed a decreasing tendency over time. We demonstrated that a drainage-reflooding cycle in rice field soil resulted in dramatically different effects on the methanotrophs in the different soil compartments. Drainage substantially activated the activity of methanotrophs in the bulk soil but suppressed the activity and growth in the rhizosphere and surface soils. The response of pmoA transcription was stronger than that of the population dynamics. The decline and recovery of pmoA transcripts in the rhizosphere and surface soils paralleled the drainage and reflooding cycle, demonstrating the tight coupling of methanotrophic activity with environmental condition shifts. In the rhizosphere and surface soils, type I methanotrophs were more resistant when total transcripts declined during drainage, whereas the type II group showed a faster recovery after reflooding of soil. This differential response is possibly related to different adaptations to O2 and methane availability that change depending on drainage and reflooding management. In the anoxic bulk soil, which normally is not a suitable habitat for aerobic methanotrophs, drainage notably stimulated the activity of type I methanotrophs to a greater extent than that of the type II group and also stimulated potential methane oxidation. Although this stimulation was not reflected by methane oxidation under in situ conditions (which were apparently dominated by the activity in the rhizosphere), it resulted in strong enhancement of the percentage of anaerobically produced methane being oxidized.
Supplementary Material
ACKNOWLEDGMENTS
This study was partially supported by the National Basic Research Program of China (2011CB100505), the Natural Science Foundation of China (41130527, 40830534, and 41101239), and the Max Planck Institute in a form of partner lab program.
Footnotes
Published ahead of print 14 June 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.00850-13.
REFERENCES
- 1. Thauer RK. 2010. Functionalization of methane in anaerobic microorganisms. Angew. Chem. Int. Ed. Engl. 49:6712–6713 [DOI] [PubMed] [Google Scholar]
- 2. Chanton JP, Whiting GJ, Blair NE, Lindau CW, Bollich PK. 1997. Methane emission from rice: stable isotopes, diurnal variations, and CO2 exchange. Global Biogeochem. Cycles 11:15–27 [Google Scholar]
- 3. Holzapfel-Pschorn A, Conrad R, Seiler W. 1985. Production, oxidation and emission of methane in rice paddies. FEMS Microbiol. Ecol. 31:343–351 [Google Scholar]
- 4. Conrad R, Rothfuss F. 1991. Methane oxidation in the soil surface-layer of a flooded rice field and the effect of ammonium. Biol. Fert. Soils 12:28–32 [Google Scholar]
- 5. Groot TT, van Bodegom PM, Harren FJM, Meijer HAJ. 2003. Quantification of methane oxidation in the rice rhizosphere using 13C-labelled methane. Biogeochemistry 64:355–372 [Google Scholar]
- 6. Op den Camp HJM, Islam T, Stott MB, Harhangi HR, Hynes A, Schouten S, Jetten MSM, Birkeland NK, Pol A, Dunfield PF. 2009. Environmental, genomic and taxonomic perspectives on methanotrophic Verrucomicrobia. Environ. Microbiol. Rep. 1:293–306 [DOI] [PubMed] [Google Scholar]
- 7. Dedysh SN, Liesack W, Khmelenina VN, Suzina NE, Trotsenko YA, Semrau JD, Bares AM, Panikov NS, Tiedje JM. 2000. Methylocella palustris gen. nov., sp. nov., a new methane-oxidizing acidophilic bacterium from peat bags, representing a novel subtype of serine-pathway methanotrophs. Int. J. Syst. Evol. Microbiol. 50:955–969 [DOI] [PubMed] [Google Scholar]
- 8. Vorobev AV, Baani M, Doronina NV, Brady AL, Liesack W, Dunfield PF, Dedysh SN. 2011. Methyloferula stellata gen. nov., sp. nov., an acidophilic, obligately methanotrophic bacterium that possesses only a soluble methane monooxygenase. Int. J. Syst. Evol. Microbiol. 61:2456–2463 [DOI] [PubMed] [Google Scholar]
- 9. Holmes AJ, Roslev P, McDonald IR, Iversen N, Henriksen K, Murrell JC. 1999. Characterization of methanotrophic bacterial populations in soils showing atmospheric methane uptake. Appl. Environ. Microbiol. 65:3312–3318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Kolb S, Knief C, Stubner S, Conrad R. 2003. Quantitative detection of methanotrophs in soil by novel pmoA-targeted real-time PCR assays. Appl. Environ. Microbiol. 69:2423–2429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. McDonald IR, Bodrossy L, Chen Y, Murrell JC. 2008. Molecular ecology techniques for the study of aerobic methanotrophs. Appl. Environ. Microbiol. 74:1305–1315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Conrad R. 2007. Microbial ecology of methanogens and methanotrophs. Adv. Agron. 96:1–63 [Google Scholar]
- 13. Horz HP, Yimga MT, Liesack W. 2001. Detection of methanotroph diversity on roots of submerged rice plants by molecular retrieval of pmoA, mmoX, mxaF, and 16S rRNA and ribosomal DNA, including pmoA-based terminal restriction fragment length polymorphism profiling. Appl. Environ. Microbiol. 67:4177–4185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Ma K, Qiu QF, Lu YH. 2010. Microbial mechanism for rice variety control on methane emission from rice field soil. Glob. Change Biol. 16:3085–3095 [Google Scholar]
- 15. Hanson RS, Hanson TE. 1996. Methanotrophic bacteria. Microbiol. Rev. 60:439–471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Lüke C, Frenzel P. 2011. Potential of pmoA amplicon pyrosequencing for methanotroph diversity studies. Appl. Environ. Microbiol. 77:6305–6309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Semrau JD, DiSpirito AA, Yoon S. 2010. Methanotrophs and copper. FEMS Microbiol. Rev. 34:496–531 [DOI] [PubMed] [Google Scholar]
- 18. Amaral JA, Archambault C, Richards SR, Knowles R. 1995. Denitrification associated with groups I and II methanotrophs in a gradient enrichment system. FEMS Microbiol. Ecol. 18:289–298 [Google Scholar]
- 19. Bodrossy L, Stralis-Pavese N, Murrell JC, Radajewski S, Weilharter A, Sessitsch A. 2003. Development and validation of a diagnostic microbial microarray for methanotrophs. Environ. Microbiol. 5:566–582 [DOI] [PubMed] [Google Scholar]
- 20. Henckel T, Roslev P, Conrad R. 2000. Effects of O2 and CH4 on presence and activity of the indigenous methanotrophic community in rice field soil. Environ. Microbiol. 2:666–679 [DOI] [PubMed] [Google Scholar]
- 21. Baani M, Liesack W. 2008. Two isozymes of particulate methane monooxygenase with different methane oxidation kinetics are found in Methylocystis sp. strain SCZ. Proc. Natl. Acad. Sci. U. S. A. 105:10203–10208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Bodelier PLE. 2011. Interactions between nitrogenous fertilizers and methane cycling in wetland and upland soils. Curr. Opin. Environ. Sustain. 3:379–388 [Google Scholar]
- 23. Liesack W, Schnell S, Revsbech NP. 2000. Microbiology of flooded rice paddies. FEMS Microbiol. Rev. 24:625–645 [DOI] [PubMed] [Google Scholar]
- 24. Wu LQ, Ma K, Lu YH. 2009. Rice roots select for type I methanotrophs in rice field soil. Syst. Appl. Microbiol. 32:421–428 [DOI] [PubMed] [Google Scholar]
- 25. Ma K, Lu YH. 2011. Regulation of microbial methane production and oxidation by intermittent drainage in rice field soil. FEMS Microbiol. Ecol. 75:446–456 [DOI] [PubMed] [Google Scholar]
- 26. Lüke C, Krause S, Cavigiolo S, Greppi D, Lupotto E, Frenzel P. 2010. Biogeography of wetland rice methanotrophs. Environ. Microbiol. 12:862–872 [DOI] [PubMed] [Google Scholar]
- 27. Eller G, Frenzel P. 2001. Changes in activity and community structure of methane-oxidizing bacteria over the growth period of rice. Appl. Environ. Microbiol. 67:2395–2403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Qiu QF, Noll M, Abraham WR, Lu YH, Conrad R. 2008. Applying stable isotope probing of phospholipid fatty acids and rRNA in a Chinese rice field to study activity and composition of the methanotrophic bacterial communities in situ. ISME J. 2:602–614 [DOI] [PubMed] [Google Scholar]
- 29. Shrestha M, Shrestha PM, Frenzel P, Conrad R. 2010. Effect of nitrogen fertilization on methane oxidation, abundance, community structure, and gene expression of methanotrophs in the rice rhizosphere. ISME J. 4:1545–1556 [DOI] [PubMed] [Google Scholar]
- 30. Reim A, Lueke C, Krause S, Pratscher J, Frenzel P. 2012. One millimetre makes the difference: high-resolution analysis of methane-oxidizing bacteria and their specific activity at the oxic-anoxic interface in a flooded paddy soil. ISME J. 6:2128–2139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Eller G, Krüger M, Frenzel P. 2005. Comparing field and microcosm experiments: a case study on methano- and methylo-trophic bacteria in paddy soil. FEMS Microbiol. Ecol. 51:279–291 [DOI] [PubMed] [Google Scholar]
- 32. Henckel T, Jackel U, Conrad R. 2001. Vertical distribution of the methanotrophic community after drainage of rice field soil. FEMS Microbiol. Ecol. 34:279–291 [DOI] [PubMed] [Google Scholar]
- 33. Shrestha M, Abraham WR, Shrestha PM, Noll M, Conrad R. 2008. Activity and composition of methanotrophic bacterial communities in planted rice soil studied by flux measurements, analyses of pmoA gene and stable isotope probing of phospholipid fatty acids. Environ. Microbiol. 10:400–412 [DOI] [PubMed] [Google Scholar]
- 34. Krüger M, Eller G, Conrad R, Frenzel P. 2002. Seasonal variation in pathways of CH4 production and in CH4 oxidation in rice fields determined by stable carbon isotopes and specific inhibitors. Glob. Change Biol. 8:265–280 [Google Scholar]
- 35. Sass RL, Fisher FM, Wang YB, Turner FT, Jund MF. 1992. Methane emission from rice fields: the effect of floodwater management. Global Biogeochem. Cycles 6:249–262 [Google Scholar]
- 36. Yagi K, Tsuruta H, Kanda K, Minami K. 1996. Effect of water management on methane emission from a Japanese rice paddy field: automated methane monitoring. Global Biogeochem. Cycles 10:255–267 [Google Scholar]
- 37. Ma K, Conrad R, Lu YH. 2012. Responses of methanogen mcrA genes and their transcripts to an alternate dry/wet cycle of paddy field soil. Appl. Environ. Microbiol. 78:445–454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Lu WF, Chen W, Duan BW, Guo WM, Lu YH, Lantin RS, Wassmann R, Neue HU. 2000. Methane emissions and mitigation options in irrigated rice fields in southeast China. Nutr. Cycl. Agroecosyst. 58:65–73 [Google Scholar]
- 39. Miller LG, Sasson C, Oremland RS. 1998. Difluoromethane, a new and improved inhibitor of methanotrophy. Appl. Environ. Microbiol. 64:4357–4362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Krüger M, Frenzel P, Conrad R. 2001. Microbial processes influencing methane emission from rice fields. Glob. Change Biol. 7:49–63 [Google Scholar]
- 41. Bodelier PLE, Hahn AP, Arth IR, Frenzel P. 2000. Effects of ammonium-based fertilisation on microbial processes involved in methane emission from soils planted with rice. Biogeochemistry 51:225–257 [Google Scholar]
- 42. Lu YH, Wassmann R, Neue HU, Huang CY. 2000. Dynamics of dissolved organic carbon and methane emissions in a flooded rice soil. Soil Sci. Soc. Am. J. 64:2011–2017 [Google Scholar]
- 43. Murase J, Noll M, Frenzel P. 2006. Impact of protists on the activity and structure of the bacterial community in a rice field soil. Appl. Environ. Microbiol. 72:5436–5444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Kolb S, Knief C, Dunfield PF, Conrad R. 2005. Abundance and activity of uncultured methanotrophic bacteria involved in the consumption of atmospheric methane in two forest soils. Environ. Microbiol. 7:1150–1161 [DOI] [PubMed] [Google Scholar]
- 45. Dumont MG, Pommerenke B, Casper P, Conrad R. 2011. DNA-, rRNA- and mRNA-based stable isotope probing of aerobic methanotrophs in lake sediment. Environ. Microbiol. 13:1153–1167 [DOI] [PubMed] [Google Scholar]
- 46. Gilbert B, Frenzel P. 1995. Methanotrophic bacteria in the rhizosphere of rice microcosms and their effect on porewater methane concentration and methane emission. Biol. Fertil. Soils 20:93–100 [Google Scholar]
- 47. Bodelier PLE, Roslev P, Henckel T, Frenzel P. 2000. Stimulation by ammonium-based fertilizers of methane oxidation in soil around rice roots. Nature 403:421–424 [DOI] [PubMed] [Google Scholar]
- 48. Mohanty SR, Bodelier PLE, Floris V, Conrad R. 2006. Differential effects of nitrogenous fertilizers on methane-consuming microbes in rice field and forest soils. Appl. Environ. Microbiol. 72:1346–1354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Vicca S, Flessa H, Loftfield N, Janssens IA. 2009. The inhibitory effect of difluoromethane on CH4 oxidation in reconstructed peat columns and side-effects on CO2 and N2O emissions at two water levels. Soil Biol. Biochem. 41:1117–1123 [Google Scholar]
- 50. Krause S, Lüke C, Frenzel P. 2012. Methane source strength and energy flow shape methanotrophic communities in oxygen-methane counter-gradients. Environ. Microbiol. Rep. 4:203–208 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.