Abstract
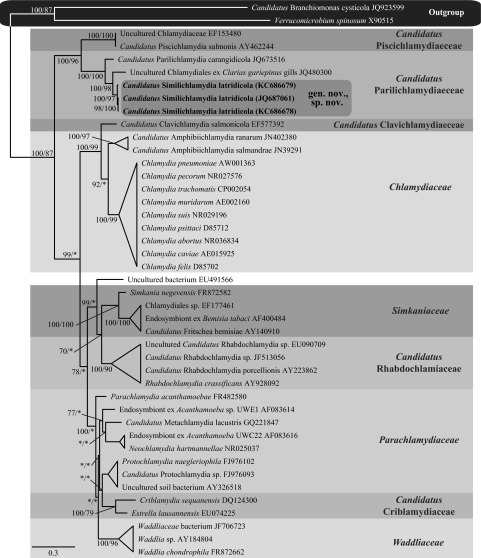
Histological analysis of gill samples taken from individuals of Latris lineata reared in aquaculture in Tasmania, Australia, and those sampled from the wild revealed the presence of epitheliocystis-like basophilic inclusions. Subsequent morphological, in situ hybridization, and molecular analyses were performed to confirm the presence of this disease and discovered a Chlamydia-like organism associated with this condition, and the criteria set by Fredericks and Relman's postulates were used to establish disease causation. Three distinct 16S rRNA genotypes were sequenced from 16 fish, and phylogenetic analyses of the nearly full-length 16S rRNA sequences generated for this bacterial agent indicated that they were nearly identical novel members of the order Chlamydiales. This new taxon formed a well-supported clade with “Candidatus Parilichlamydia carangidicola” from the yellowtail kingfish (Seriola lalandi). On the basis of sequence divergence over the 16S rRNA region relative to all other members of the order Chlamydiales, a new genus and species are proposed here for the Chlamydia-like bacterium from L. lineata, i.e., “Candidatus Similichlamydia latridicola” gen. nov., sp. nov.
INTRODUCTION
The striped trumpeter, Latris lineata (Forster 1801), is distributed in southern hemisphere waters from the Walters Shoals (43°50′E) and Amsterdam Island (77°33′E) in the Indian Ocean through the southern waters of Australia and then to Chatham Island (176°29′W) in the Pacific Ocean (http://www.fishbase.org/summary/FamilySummary.php?id=356). The overexploitation of this species throughout its range has led to a significant decrease in the wild population, with the total commercial catch decreasing in the last 20 years by almost 100 tonnes to 12.8 tonnes in 2009-2010 (1, 2). Because of the marked decline in wild stocks, the culture of L. lineata has been in development at the Tasmania Aquaculture and Fisheries Institute, Hobart, Tasmania, for 16 years. L. lineata is considered to be a suitable aquaculture candidate; however, issues associated with its complex and lengthy 9-month postlarval stage have been difficult to overcome (1). The life cycle of L. lineata has now been successfully closed, and established protocols exist for its reproduction and larval rearing (3). Despite this, a number of health issues were observed between 1994 and 2010 during the development of L. lineata for commercial aquaculture. Examples included abnormal swimming behavior, anorexia, swim bladder hyperinflation, skin lesions, and inflammation and swelling of gills in cultured juveniles due to infections from Kudoa neurophila and chondracanthid copepods (4, 5). In determining the causes of these health issues, epitheliocystis was also described in these fish (6). The latter disease is a condition of the skin and gills and is generally associated with infections by Chlamydia-like organisms (CLOs) (7–10). These CLOs are Gram-negative, intracellular bacteria that may cause cyst-like lesions in the gill lamellae (9, 11, 12). The lesions may lead to epithelial hyperplasia and inflammation of the infected tissues, increased mucus production, and respiratory distress, sometimes ending in death (12–15). Most reported losses in aquaculture attributed to epitheliocystis occur during the larval or juvenile culture stage (16).
Little is known about the epidemiology and pathogenesis of epitheliocystis agents. In an effort to understand this, researchers have turned from traditional microbiology methods to molecular techniques in an attempt to understand this condition. This has led to a move toward the fulfillment of Fredericks and Relman's molecular postulates instead of Koch's postulates (17). As a result, the primary method now used to describe and characterize unknown epitheliocystis agents taxonomically include phylogenetic analysis of DNA sequence data in combination with morphological descriptions. Following this trend, “Candidatus Parilichlamydia carangidicola” was recently recovered from the yellowtail kingfish in Australia by using molecular techniques and transmission electron microscopy as primary evidence (10).
The objective of this study was to identify and characterize the agent causing epitheliocystis in L. lineata, both in cultured individuals and in fish obtained from the wild. Histological examination of epitheliocystis infections in the gill were confirmed by PCR of the 16S rRNA gene and in situ hybridization (ISH). Following this, Bayesian inference and maximum-likelihood phylogenetic analyses were performed by using 16S rRNA sequences to explore the relationships of the striped trumpeter epitheliocystis agent with other epitheliocystis agents in fish and with other members of the order Chlamydiales.
MATERIALS AND METHODS
Ethics statement.
Sampling of animals for this study was approved by the University of Tasmania Board of Animal Ethics, project number AEC0009926.
Sample collection.
L. lineata was reared in 20,000-liter recirculated and flowthrough tanks at the Tasmanian Aquaculture and Fisheries Institute, Hobart, Tasmania. Most fish were held in temperature- and light-controlled flowthrough recirculation tanks with 50% fresh seawater (sand and bag filtered [50 μm]) exchange three times a week. Some fish were in tanks on flowthrough seawater supply with only coarse particle filters. A total of 87 cultured fish were sampled at two time points, July 2010 (n = 8) and November 2010 (n = 79). The November 2010 samples were broodstock originally captured from southeastern and northeastern Tasmania and the F1 generation bred in captivity. All broodstock fish had been in captivity for at least 5 years and were not separated by origin. Fish were euthanized with 0.04% 2-phenoxyethanol, and then weight and length measurements were taken. In addition to the cultured striped trumpeter, wild fish (n = 6) were sampled from waters of southwestern Tasmania (43°32′48″S, 145°56′27″E). For all fish, the second gill arch on the sinistral side was sampled, with the first subsection fixed in seawater Davidson's fixative (cultured) or 10% neutral buffered formalin (wild) for histology and the second subsection frozen at −80°C (cultured) or placed in RNAlater (wild) for DNA extraction.
Histopathology.
Seawater Davidson's-fixed and formalin-fixed gills were routinely processed for histology. The gills were sectioned at 5 μm and stained with hematoxylin and eosin. The sections were examined by light microscopy to identify epitheliocystis inclusions and associated lesions (10).
DNA extraction, 16S rRNA amplification, and sequencing.
DNA was extracted from samples with the commercially available Epicentre MasterPure Complete DNA and RNA purification kit (Epicentre Biotechnologies, Madison, WI) according to the manufacturer's instructions and optimized as described previously (10). Extracted DNA from the July 2010 (n = 8), November 2010 (n = 5), and wild (n = 3) cohorts was screened by a conventional Chlamydiales-specific PCR assay targeting a 298-bp “signature sequence” region of the 16S rRNA gene with primers 16SIGF and 16SIGR as described previously (10, 18, 19). Expanded 800-bp (with primers 16SIGF and 806R) and nearly full-length (with primers 16SIGF and 16SB1) 16S rRNA sequences from selected representative samples were completed. PCR amplification, cycling conditions, purification, and sequencing for these assays were as previously described (10).
ISH.
Detection of the Chlamydia-like organism within the epitheliocystis cysts by ISH in 5-μm serial sections was conducted with Chlamydiales-specific antisense (digoxigenin [DIG]-ATG TG[T/C] TAC TAA CCC TTC CGC CAC TA-DIG) and sense (DIG-ATC CTA CGC TAC TAA GTC TCT CAT CA-DIG) DIG-labeled oligonucleotide probes as described previously (12, 20), with some modifications. Briefly, the sections were dewaxed, washed with phosphate-buffered saline (PBS) for 3 min, and digested for 30 min at 37°C in 5 mg/liter proteinase K (Sigma) in 0.1 mol/liter Tris-HCl (pH 7.6). The sections were washed twice with 0.2% glycine in PBS for 5 min, once with 0.01% Triton X-100 in PBS for 10 min, and twice with PBS for 5 min; dehydrated in 95 and 100% ethanol for 3 min each; and then dried. The hybridization mixture, containing 42% deionized formamide, 9.4% dextran sulfate, 5.8× saline-sodium citrate (SSC) buffer (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate), 4.7× Denhardt's solution, 94 mg/liter denatured salmon sperm DNA, and the probe(s) at a final concentration of 10.2 pmol/ml, was heated for 10 min at 100°C, cooled on ice, and added to the sections. The sections were incubated at 95°C for 5 min prior to overnight incubation at 55°C in a humid chamber. The sections were washed twice with 2× SSC for 15 min, once with 1× SSC for 5 min, once with 0.5× SSC for 5 min, and twice with Tris-HCl (pH 7.6) for 10 min. Blocking buffer (containing 0.1% Triton X-100, 2% normal sheep serum, and PBS) was added, and the sections were incubated for 30 min at room temperature. The hybridized probes were visualized by the addition of alkaline phosphatase-labeled anti-DIG Fab fragments, incubation in a humid chamber for 2 h, two washes with Tris-HCl (pH 9.5) for 10 min, chromogen (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside [BCIP]–Nitro Blue Tetrazolium) addition, and incubation in a humid chamber for 75 min. The sections were then counterstained with Nuclear Fast Red (Sigma) and mounted with Vectamount.
Molecular phylogenetic analysis.
The partial 16S rRNA regions sequenced here and data from additional Chlamydiales species and outgroup taxa obtained from GenBank were aligned as previously described (10).
The software jModelTest version 0.1.1 (21, 22) estimated TVM+I+G as the best nucleotide substitution model for this data set. Bayesian inference and maximum-likelihood analyses of the 16S rRNA data set were performed with MrBayes version 3.1.2 (23) and RAxML algorithm (24), respectively, run on the CIPRES portal (http://www.phylo.org/sub_sections/portal/) to explore relationships among these taxa under conditions set as previously described (10).
Quantification.
The prevalence (expressed as a percentage) and intensity (intensity = cysts per section/filaments per section) of epitheliocystis infection were calculated for each fish after visual inspection of hematoxylin-eosin-stained gill sections. The prevalence of CLOs detected by PCR was also calculated (expressed as a percentage). Statistical analyses were conducted with the IBM SPSS Statistics package, version 20.0.0.1 (2011). Levene's test was performed to ensure the assumption of homogeneity of variances. One-way analysis of variance was performed to test fish length and infection intensity against the sampling points.
Nucleotide sequence accession numbers.
The three 16S rRNA gene sequences of the L. lineata epitheliocystis agents determined in this study are available in the GenBank database under accession numbers JQ687061, KC686678, and KC686679.
RESULTS
Histopathology and prevalence of novel striped trumpeter Chlamydia-like organism.
The mean fish length and weight and the prevalence (percent) of the infectious agent at each sampling location are summarized in Table 1; the origins of the fish are also shown. There was no significant difference in fish length between the sampling points (F = 3.802, df = 1, P = 0.055). Epitheliocystis, as determined by histological examination, was present in fish sampled at all of the sampling points. The prevalence ranged from 50% in fish of wild origin (August 2011) to 100% in broodstock held in captivity (November 2010; Table 1). There was no significant difference in the intensity of epitheliocystis between the sampling points (F = 1.750, df = 2, P = 0.180). Infection intensity ranged from 0.02 to 3.37 cysts/filament in the November 2010 fish, from 0.07 to 2.43 cysts/filament in the July 2010 fish, and from 0.04 to 0.07 cysts/filament in the wild fish sampled in August 2011. Membrane-enclosed granulated basophilic cysts were present along the entire length of the filaments in affected fish from all three sampling points (Fig. 1B). In terms of the host response to these cysts, hyperplasia of epithelial cells could be observed in some, but not all, of the cultured fish from both sampling points while no responses were observed in infected wild fish (Fig. 1A and B).
Table 1.
Summary of the mean length and mean weight of the fish in this study, the prevalence and intensity of epitheliocystis striped trumpeter infections, the number of fish that were PCR positive, and their corresponding genotypes
| Parameter | July 2010, F1 generation | November 2010, broodstock | August 2011, wild |
|---|---|---|---|
| No. of samples | 8 | 79 | 6 |
| Mean length, mm (SE) | NDa | 539 (9.2)b | 605 (19.0)b |
| Mean wt, kg (SE) | ND | 2.80 (0.13) | ND |
| Prevalence, % (no. tested) | 100 (8/8) | 75.9 (60/79) | 50 (3/6) |
| Intensity (cysts/filaments) (SE) | 0.55 (0.28)b | 0.46 (0.16)b | 0.03 (0.01)b |
| % PCR positive (no. tested) | 100 (8) | 100 (5) | 100 (3) |
| Genotype | A/B | C/A | B |
| Origin | TAFI,c Hobart | TAFI, Hobartd | SWe Tasmaniaf |
ND, no data.
Statistically significantly different.
TAFI, Tasmanian Aquaculture & Fisheries Institute.
Broodstock fish were held in captivity for >5 years and were originally from waters around Flinders Island (northeastern Tasmania) and Tasman Island (southeastern Tasmania).
SW, southwestern.
Fish were caught at 43°55′480S, 145°56′272E.
Fig 1.
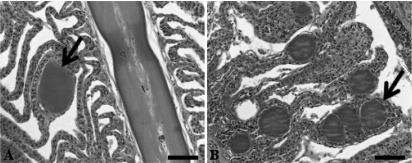
Epitheliocystis in striped trumpeter (L. lineata) gills stained with hematoxylin and eosin. Panels: A, single membrane-enclosed basophilic granular cyst (arrow) with no host response; B, multiple membrane-enclosed basophilic granular cysts (arrow) along individual lamellae with a hyperplasic-epithelium host response. Scale bars, 50 μm.
Molecular identification and phylogenetic analysis of the novel Chlamydia-like organism.
Preliminary screening with a Chlamydiales-specific 16S rRNA PCR assay revealed that 100% (n = 16) of the striped trumpeter samples screened PCR positive for chlamydial DNA (July 2010, n = 8; November 2010, n = 5; August 2011, n = 3). Pairwise alignments of sequences revealed three distinct genotypes with >99% nucleotide sequence similarity to each other. Six single-nucleotide polymorphisms (SNPs) were present in the three nearly full-length genotype sequences (1,396 bp). The SNPs were consistently found at the same positions within the gene whether using the 16SIGF/16SIGR, the 16SIGF/806R, or the 16SIGF/16SBI primer pair and were sequenced in multiple samples. In addition, these SNPs were positioned within the variable region of the signature sequence (bp 40 to 337 of the 16S rRNA gene) as outlined by Everett (18). Genotypes A and B were found multiple times and in fish from multiple origins. Genotype A was found in both July 2010 and November 2010 samples (Table 1). The fish sampled in July 2010 were also positive for genotype B, which was sequenced from the wild fish sampled in August 2011. Finally, genotype C was sequenced from fish sampled in November 2010 only (Table 1).
BLAST-n analysis of the consensus L. lineata CLO sequences against the NCBI database revealed these sequences to be novel, showing 93.7 to 94.0% sequence similarity to the next closest 16S rRNA sequence, “Ca. Parilichlamydia carangidicola,” a recently reported novel Chlamydia-like epitheliocystis agent in the yellowtail kingfish, Seriola lalandi (10). The next closest sequence (88% sequence similarity) identified belonged to “Ca. Piscichlamydia salmonis” from the Atlantic salmon (accession no. AY462244 [25] and EU326495 [26]) and the Arctic charr (GQ302987 [27]).
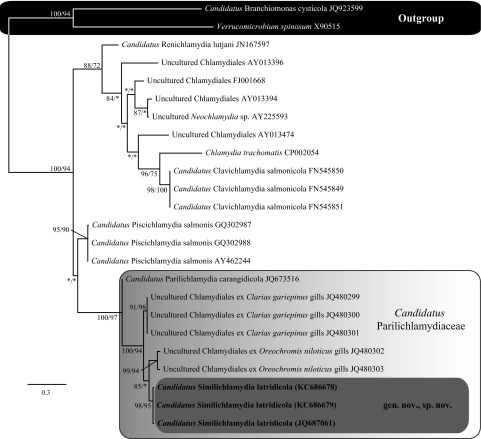
Alignment of the 16S rRNA data generated from the epitheliocystis agent isolated from the striped trumpeter (L. lineata) and the remainder of the Chlamydiales taxa and outgroups examined here yielded 1,127 characters of analysis. Bayesian inference and maximum-likelihood analyses resulted in phylograms with markedly similar topologies, with all of the currently recognized and candidate families within the order Chlamydiales forming relatively well-supported clades (Fig. 2). Alignment and analysis of the shorter signature sequence region of the 16S rRNA showed that the epitheliocystis agent reported here from L. lineata grouped together in a strongly supported clade that was sister to the sequences available for “Candidatus Parilichlamydia carangidicola,” multiple sequences available for “Candidatus Piscichlamydia salmonis,” and the sequence available for “Candidatus Renichlamydia lutjani” in GenBank, which have all been reported from teleosts (Fig. 3).
Fig 2.
Relationships among the epitheliocystis agents isolated from the striped trumpeter, L. lineata, and the remainder of the Chlamydiales taxa and outgroups examined here based on Bayesian inference and maximum-likelihood analyses of the 16S rRNA data set. Posterior probability and bootstrap support values (respectively) are shown at the nodes, with values of <70% indicated by asterisks. Clades of representative genera have been collapsed.
Fig 3.
Relationships between the 298-bp (16S rRNA) signature sequence of the epitheliocystis agent detected in L. lineata gills and epitheliocystis agents detected in other fish species, Chlamydia trachomatis, and Piscirickettsia salmonis based on Bayesian inference and maximum-likelihood analyses. Posterior probability and bootstrap support values (respectively) are shown at the nodes, with values of <70% indicated by asterisks.
ISH.
The presence of Chlamydia-like sequences detected in epitheliocystis-positive striped trumpeter gill samples by PCR was confirmed by ISH with the Chlamydiales-specific probes. Epitheliocystis cysts reacted strongly to the antisense probe, with cysts staining a dark purple/black color. Epitheliocystis cysts that were incubated with the sense probe showed no reactivity (Fig. 4A and B).
Fig 4.
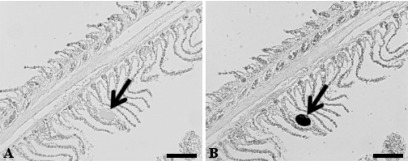
Detection of Chlamydiales bacteria in epitheliocystis-affected striped trumpeter (L. lineata) gills by ISH. Panels: A, reaction of epitheliocystis gill with the sense ISH probe (note the absence of a reaction [arrow] with the cyst); B, reaction of epitheliocystis cysts with the antisense ISH probe (note the dark coloration of the cyst [arrow], indicating a positive reaction). Scale bars, 100 μm.
DISCUSSION
Epitheliocystis is now known to affect >80 different species of fish (7, 10), and intriguingly, from all of these reports, it appears that the host responses considered to be severe or as having a hyperinfection are all from farmed fish (14–16, 27–32). Molecular diagnostics confirmed that this CLO, from both cultured and wild sources, is a member of the order Chlamydiales, based on the >80% sequence similarity to other species in the order (18). The nearly identical, nearly full-length 16S rRNA sequences of the CLO obtained over the three sampling periods and from both wild and cultured fish are strong evidence that it is the agent causing epitheliocystis in striped trumpeter fish. The detection of a Chlamydiales-specific 16S rRNA gene in striped trumpeter epitheliocystis gill inclusions by ISH with DIG-labeled probes provides further evidence that the cysts identified by light microscopy are the source of the amplified novel 16S rRNA gene sequence(s).
The prevalence of epitheliocystis and the response of the striped trumpeter to infection reported in this study are in line with those previously reported for this species (6). The response seen in this study, however, was not as severe as that in previous reports of hyperinfection in the largemouth bass, Micropterus salmoides, and the Atlantic salmon, Salmo salar (9, 28). Epithelial hyperplasia, epithelial lifting along the lamellae, and lamellar fusion in the striped trumpeter were observed, although the filling of the interlamellar spaces was not as severe as that in previous reports. This differs from the nearly complete filling of the interlamellar spaces reported in both the largemouth bass (28) and the Arctic charr (15, 27), which resulted in a severely compromised respiratory system.
Striped trumpeter fish were recently reported to be affected by epitheliocystis (6). While that was the first report of epitheliocystis in the family Latridae, the condition has been reported in other species in the superfamily Cirrhitoidea, including the rock cale, Crinodus lophodon, and the red morwong, Cheilodactylus fuscus (33). Like the striped trumpeter, both of these species originate from Australian waters. In these other species, benign cysts with little or no host response were observed in the gills, which matched that of the wild fish and some of the cultured striped trumpeter fish in this study (33). Unfortunately, no molecular data on the identity of the epitheliocystis agent from infections in these species are available for comparison, as the molecular techniques used here were not common practice at that time.
Questions remain about the origin of this infection and the potential impact that this novel epitheliocystis agent may have on the health and productivity of the striped trumpeter. In the cultured environment, the water source and treatment and the tank environmental conditions (temperature, salinity, dissolved oxygen, and nitrates) are controlled and monitored daily by staff. However, since infections were found in broodstock from both southwestern and northwestern Tasmanian waters and in fish sampled from the wild, it is reasonable to conclude that epitheliocystis occurs naturally in the environment and was introduced into the culture systems either with the fish or with the seawater supply. Although the bacterial sequences obtained from the different sources were of three distinct genotypes, they are extremely closely related. The development of in vitro methods for culturing these bacteria is needed to help answer questions on how variable these organisms naturally are, how these organisms are transmitted within a population, and what environmental factors, if any, may lead to hyperinfection.
Because of the current inability to culture Chlamydia-like bacteria in vitro, an alternative to Koch's postulates must be used. The molecular postulates of Fredericks and Relman were therefore used in this study (17). The nearly identical sequences identified here from both wild and cultured sources were present in all cases of disease observed in histology. The nature of the CLOs detected here is consistent with the known biological characteristics of Chlamydia; that is, they are intracellular bacteria requiring a host cell to replicate. The order specificity of the sequences was detected within the epitheliocystis cysts through ISH; and all of these results are repeatable. These results are in line with the molecular postulates of disease causation (17) and provide strong evidence that the epitheliocystis agent of the striped trumpeter is of Chlamydiales origin. On the basis of its novel 16S rRNA signature sequence, the sequence divergence from other Chlamydiales species and the observed phylogenetic relationships of this bacterium to other taxa within the order according to their classification (18), we propose the name “Candidatus Similichlamydia latridicola” (gen. nov., sp. nov.) for the Chlamydia-like epitheliocystis agent infecting the striped trumpeter.
Taxonomy.
“Candidatus Similichlamydia latridicola” gen. nov., sp. nov., recovered from the striped trumpeter (L. lineata). Similichlamydia gen. nov.; Si.mi.li.chla.my′di.a. L. adj. similis, resembling; N.L. fem. n. Chlamydia, a bacterial genus name; N.L. fem. n. Similichlamydia, resembling Chlamydia. latridicola sp. nov.; la.tri.di.′co.la. N.L. n. Latris -idis, a zoological genus name; L. suff. -cola (from L. n. incola), inhabitant, dweller; N.L. n. latridicola, Latris dweller, isolated from the striped trumpeter (Latris lineata).
Obligate intracellular bacteria infecting fish gills. Membrane-bound inclusions present as granular and tightly packed, staining basophilic under hematoxylin and eosin. Inclusions are found along the gill filament at the base, middle, and tip of the lamellae and incite a host response of cellular hyperplasia. Inclusions react with ISH 16S rRNA probes and stain purple/black. The new genus and species 16S rRNA sequence is 6.0 to 6.3% different from the 16S rRNA of “Candidatus Parilichlamydiaceae,” placing it within this family, but not as a member of the genus “Candidatus Parilichlamydia,” according to the classification scheme of Everett (18).
ACKNOWLEDGMENTS
We thank Stephen Battaglene for giving us access to cultured striped trumpeter samples, providing information on the culture conditions, and commenting on the manuscript. We acknowledge the following people for their assistance with this work: Philip Crosbie, Anna Overweter, and Melanie Leef for sampling cultured striped trumpeter; Troy Gaston for sampling wild striped trumpeter; Karine Cadoret for assistance with histology and ISH; Andrew Bridle for assistance with ISH troubleshooting; James Marsh for preliminary PCR analysis; Eileen Roulis and Martina Jelocnik for assisting with additional PCR analyses; and Jean Euzeby for assistance with the Latin formation of new bacterial names.
Footnotes
Published ahead of print 14 June 2013
REFERENCES
- 1. Battaglene SC, Cobcroft JM. 2007. Advances in the culture of striped trumpeter larvae: a review. Aquaculture 268:195–208 [Google Scholar]
- 2. Hartmann K, Lyle JM. 2011. Tasmanian scalefish fishery—2009/10. Institute for Marine and Antarctic Studies, University of Tasmania, Sandy Bay, Tasmania [Google Scholar]
- 3. Battaglene SC, Cobcroft JM. 2010. Enhanced hatchery production of striped trumpeter, Latris lineata, in Tasmania through system design, microbial control and early weaning. Aquafin Cooperative Research Centre, Hobart, Tasmania [Google Scholar]
- 4. Cobcroft JM, Battaglene SC. 2013. Ultraviolet irradiation is an effective alternative to ozonation as a seawater treatment to prevent Kudoa neurophila infection of striped trumpeter. J. Fish Dis. 369:57–65 [DOI] [PubMed] [Google Scholar]
- 5. Andrews M, Battaglene SC, Cobcroft JM, Adams MB, Noga EJ, Nowak BF. 2010. Host response to the chondracanthid copepod Chondracanthus goldsmidi, a gill parasite of the striped trumpeter, Latris lineata (Forster), in Tasmania. J. Fish Dis. 33:211–220 [DOI] [PubMed] [Google Scholar]
- 6. Lai CC, Crosbie PBB, Battaglene SC, Nowak BF. 2013. Effects of epitheliocystis on serum lysozyme activity and osmoregulation in cultured juvenile striped trumpeter Latris lineata (Forster). Aquaculture 388-. 391:99–104 [Google Scholar]
- 7. Nowak BF, LaPatra SE. 2006. Review: epitheliocystis in fish. J. Fish Dis. 29:573–588 [DOI] [PubMed] [Google Scholar]
- 8. Karlsen M, Nylund A, Watanabe K, Helvik JV, Nylund S, Plarre H. 2008. Characterisation of “Candidatus Clavochlamydia salmonicola”: an intracellular bacterium infecting salmonid fish. Environ. Microbiol. 10:208–218 [DOI] [PubMed] [Google Scholar]
- 9. Mitchell SO, Steinum T, Rodger H, Holland C, Falk K, Colquhoun DJ. 2010. Epitheliocystis in Atlantic salmon, Salmo salar L., farmed in fresh water in Ireland is associated with “Candidatus Clavochlamydia salmonicola” infection. J. Fish Dis. 33:665–673 [DOI] [PubMed] [Google Scholar]
- 10. Stride MC, Polkinghorne A, Miller TL, Groff JM, LaPatra SE, Nowak BF. 2013. Molecular characterization of “Candidatus Parilichlamydia carangidicola,” a novel Chlamydia-like epitheliocystis agent in yellowtail kingfish, Seriola lalandi (Valenciennes), and the proposal of a new family, “Candidatus Parilichlamydiaceae” fam. nov. (order Chlamydiales). Appl. Environ. Microbiol. 79:1590–1597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Draghi A., II 2006. Molecular characterization of chlamydia-like bacteria associated with epitheliocystis in farmed salmonids, Atlantic salmon (Salmo salar) and Arctic char (Salvelinus alpinus), and environmental chlamydiae from aquatic environments. Ph.D. thesis University of Connecticut, Storrs, CT [Google Scholar]
- 12. Meijer A, Roholl PJM, Ossewaarde JM, Jones B, Nowak BF. 2006. Molecular evidence for association of Chlamydiales bacteria with epitheliocystis in leafy seadragon (Phycodurus eques), silver perch (Bidyanus bidyanus), and barramundi (Lates calcarifer). Appl. Environ. Microbiol. 72:284–290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Lewis EJ, McLaughlin SM, Bodammer JE, Sawyer TK. 1992. Epitheliocystis in ten new host species of marine fish. J. Fish Dis. 15:267–271 [Google Scholar]
- 14. Crespo S, Zarza C, Padros F, Marin de Mateo M. 1999. Epitheliocystis agents in sea bream Sparus aurata: morphological evidence for two distinct Chlamydia-like developmental cycles. Dis. Aquat. Organ. 37:61–72 [DOI] [PubMed] [Google Scholar]
- 15. Draghi A, Bebak IIJ, Popov VL, Noble AC, Geary SJ, West A, Byrne P, Frasca S., Jr 2007. Characterization of a Neochlamydia-like bacterium associated with epitheliocystis in cultured Arctic charr Salvelinus alpinus. Dis. Aquat. Organ. 76:27–38 [DOI] [PubMed] [Google Scholar]
- 16. Syasina I, Park IS, Kim JM. 2004. Epitheliocystis disease in red sea bream Pagrus major and induced hybrid, red sea bream Pagrus major female black sea bream Acanthopagrus schlegeli male, cultured in Korea. Bull. Eur. Assoc. Fish Pathol. 24:260–267 [Google Scholar]
- 17. Fredericks DN, Relman DA. 1996. Sequence-based identification of microbial pathogens: a recondiseration of Koch's postulates. Clin. Microbiol. Rev. 9:18–33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Everett KDE, Bush RM, Anderson AA. 1999. Emended description of the order Chlamydiales, proposal of Parachlamydiceae fam. nov. and Simkaniaceae fam. nov., each containing one monotypic genus, revised taxonomy of the family Chlamydiceae, including a new genus and five new species, and standards for the identification of organisms. Int. J. Syst. Bacteriol. 49:415–440 [DOI] [PubMed] [Google Scholar]
- 19. Polkinghorne A, Schmidt-Posthaus H, Meijer A, Lehner A, Vaughan L. 2010. Novel Chlamydiales associated with epitheliocystis in a leopard shark Triakis semifasciata. Dis. Aquat. Organ. 91:75–81 [DOI] [PubMed] [Google Scholar]
- 20. Meijer A, van der Vliet JA, Roholl PJM, Gielis-Proper SK, de Vries A, Ossewaarde JM. 1999. Chlamydia pneumoniae in abdominal aortic aneurysms: abundance of membrane components in the absence of heat shock protein 60 and DNA. Arterioscler. Thromb. Vasc. Biol. 19:2680–2686 [DOI] [PubMed] [Google Scholar]
- 21. Posada D. 2008. jModelTest: phylogenetic model averaging. Mol. Biol. Evol. 25:1253–1256 [DOI] [PubMed] [Google Scholar]
- 22. Guindon S, Gascuel O. 2003. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst. Biol. 52:696–704 [DOI] [PubMed] [Google Scholar]
- 23. Ronquist F, Huelsenbeck JP. 2003. MRBAYES 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19:1572–1574 [DOI] [PubMed] [Google Scholar]
- 24. Stamatakis A, Hoover P, Rougemont J. 2008. A rapid bootstrap algorithm for the RAxML web servers. Syst. Biol. 57:758–771 [DOI] [PubMed] [Google Scholar]
- 25. Draghi A, Popov VL, Kahl MM, Stanton JB, Brown CC, Tsongalis GJ, West A, Frasca S. 2004. Characterization of “Candidatus Piscichlamydia salmonis” (order Chlamydiales), a Chlamydia-like bacterium associated with epitheliocystis in farmed Atlantic salmon (Salmo salar). J. Clin. Microbiol. 42:5286–5297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Nylund A, Watanabe K, Nylund S, Karlsen M, Saether PA, Arnesen CE, Karlsbakk E. 2008. Morphogenesis of salmonid gill poxvirus associated with proliferative gill disease in farmed Atlantic salmon (Salmo salar) in Norway. Arch. Virol. 153:1299–1309 [DOI] [PubMed] [Google Scholar]
- 27. Draghi A, Bebak J, Daniels S, Tulman ER, Geary SJ, West A, Popov VL, Frasca S., Jr 2010. Identification of “Candidatus Piscichlamydia salmonis” in Arctic charr Salvelinus alpinus during a survey of charr production facilities in North America. Dis. Aquat. Organ. 89:39–49 [DOI] [PubMed] [Google Scholar]
- 28. Goodwin AE, Park E, Nowak BF. 2005. Short communication: successful treatment of largemouth bass, Micropterus salmoides (L.), with epitheliocystis hyperinfection. J. Fish Dis. 28:623–625 [DOI] [PubMed] [Google Scholar]
- 29. Szakolczai J, Vetesi F, Pitz 1999. Epitheliocystis disease in cultured pacu (Piaractus mesopotamicus) in Brazil. Acta Vet. Hung. 47:311–318 [DOI] [PubMed] [Google Scholar]
- 30. Bradley TM, Newcomer CE, Maxwell KO. 1988. Epitheliocystis associated with massive mortalities of cultured lake trout Salvelinus namaycush. Dis. Aquat. Organ. 4:9–17 [Google Scholar]
- 31. Company R, Sitja-Bobadilla A, Pujalte MJ, Garay E, Alvarez-Pellitero P, Perez-Sanchez J. 1999. Bacterial and parasitic pathogens in cultured common dentex, Dentex dentex L. J. Fish Dis. 22:299–309 [Google Scholar]
- 32. Katharios P, Papadaki M, Papandroulakis N, Divanach P. 2008. Severe mortality in mesocosm-reared sharpsnout sea bream Diplodus puntazzo larvae due to epitheliocystis infection. Dis. Aquat. Organ. 82:55–60 [DOI] [PubMed] [Google Scholar]
- 33. Nowak BF. 1996. Health of red morwong, Cheilodactylus fuscus, and rock cale, Crinodus lophodon, from Sydney cliff-face sewage outfalls. Mar. Pollut. Bull. 33:281–292 [Google Scholar]