Abstract
In their natural environment, bacteria often behave differently than they do under laboratory conditions. To gain insight into the physiology of bacteria in situ, dedicated approaches are required to monitor their adaptations and specific behaviors under environmental conditions. Optical microscopy is crucial for the observation of fundamental characteristics of bacteria, such as cell shape, size, and marker gene expression. Here, fluidic force microscopy (FluidFM) was exploited to isolate optically selected bacteria for subsequent identification and characterization. In this study, bacteriochlorophyll-producing bacteria, which can be visualized due to their characteristic fluorescence in the infrared range, were isolated from leaf washes. Bacterial communities from the phyllosphere were investigated because they harbor genes indicative of aerobic anoxygenic photosynthesis. Our data show that different species of Methylobacterium express their photosystem in planta, and they show a distinct pattern of bacteriochlorophyll production under laboratory conditions that is dependent on supplied carbon sources.
INTRODUCTION
The phyllosphere, or the above-ground parts of plants, is responsible for terrestrial photosynthesis and carbon dioxide fixation. It represents a large habitat that is colonized by a variety of different microorganisms, mostly bacteria that belong to a few predominant phyla (1). The phyllosphere provides a challenging environment for bacterial growth due to UV irradiation, drought stress, temperature fluctuations, and nutrient limitation. However, bacteria have evolved different strategies to overcome these challenges (1). For example, pigmentation reduces damage to DNA caused by irradiation. Nutrient limitation is countered by the expression of uptake systems for various carbohydrates in various bacteria and by the ability to use methanol as a carbon source, which requires dedicated one-carbon compound biochemistry, in more specialized methylotrophic bacteria (2, 3). Another strategy to counteract nutrient limitation might be to supplement energy production by using sunlight. Oxygenic and anoxygenic phototrophic bacteria are considered rare in the phyllosphere, in contrast to aquatic environments (4). However, it was recently shown that a substantial portion of phyllosphere bacteria harbor genes of aerobic anoxygenic phototrophs (AAnPs) (5). AAnPs cannot grow exclusively via photosynthesis but are dependent on organic carbon (6). They possess a single type of reaction center that contains bacteriochlorophyll a (BChl a), which exhibits an absorption in the infrared (IR) range (6). This fluorescence in the infrared (IR) range can be detected by epifluorescence microscopy, allowing the identification of anoxygenic phototrophs (7, 8). Known members of the AAnPs belong to the Proteobacteria and comprise a significant fraction of marine (9, 10) and freshwater (11, 12) microbial communities, where they play an important role in the cycling of organic and inorganic carbon (13–15). AAnPs have been described in terrestrial systems (16–19), but their role in terrestrial carbon cycling is not well studied. As mentioned above, leaf surfaces were investigated as a terrestrial habitat for AAnPs by mining metagenomic data for genes related to photosynthesis (5). It was shown that bacterial phyllosphere communities of different plants harbor specific genes, such as bchY, which encodes a subunit of the chlorophyllide oxidoreductase important for bacteriochlorophyll production, and pufLM, which encode the two core subunits of the photosynthesis reaction center. Microscopic analyses of bacteria washed from the phyllosphere further supported this finding by demonstrating the presence of bacteria exhibiting IR fluorescence (5). For a better understanding of the physiological process of aerobic anoxygenic phototrophy and its regulation, cultures of the organisms are required to obtain information that cannot be obtained by genomic information alone. The identification and characterization of the responsible organisms necessitates an approach by which one can optically identify the bacteriochlorophyll-expressing bacteria by their specific fluorescence in the IR range and subsequently isolate the targeted bacterium for clonal growth.
Several methods have been established to physically separate selected individual cells. A widely used method relies on fluorescence-activated cell sorting (FACS), which allows the high-throughput and rapid delivery of cells into tubes or microwell plates (20, 21). Although versatile, the method has not been shown to effectively sort cells with weak IR autofluorescence signals. Additionally, different cell shapes and sizes make it difficult to standardize sorting processes to obtain individual cells from complex mixtures. Other methods for isolating single bacterial cells under optical control include microfluidic devices that can be assisted by manipulation techniques such as optical tweezers (22; applied, e.g., in studies described in references 23, 24, and 25), electrode-based dielectrophoresis (DEP) (26), and optoelectronic tweezers (27); however, such approaches are not suitable for extracting cells directly from liquid and require additional steps for physical separation of target cells. They have, for instance, been successfully used in combination with PCR, and target cells are lysed within the compartments offered by the microfluidic set-up. A technique that allows direct extraction and transfer of target cells from a liquid sample is the use of micropipettes (28–31). Although tedious, micropipette systems have been used for physical separation of target cells prior to single-cell PCR (32–34).
In this study, we exploited the potential of the recently invented fluidic force microscope (FluidFM) (35) for single-cell isolation. This technology combines the force-controlled positioning of an atomic force microscope (AFM) with nanofluidics. The standardized microchanneled AFM cantilevers used for FluidFM have a defined opening and are fixed on a drilled probe holder, thus making it possible to use the cantilever as a force-controlled nanosyringe for gentle contact as well as for liquid manipulation (suction and dispensing). FluidFM is operated on top of an inverted optical microscope and enables the use of optical identification and epifluorescence (Fig. 1a). It was previously shown that chosen microorganisms adsorbed on a glass surface in liquid can be attached to the cantilever aperture by application of underpressure, moved, and released onto another defined position on the same surface by application of a short overpressure pulse (36). Here we demonstrate that this protocol can be extended to isolate bacteria from environmental samples. The bacteria can be lifted from liquid, transferred through air, and placed in the growth medium of choice for clonal growth and identification. After the feasibility of the method was demonstrated, bacteriochlorophyll-producing bacteria washed from leaf surfaces were isolated to identify bacteria that produce anoxygenic photosystems in situ.
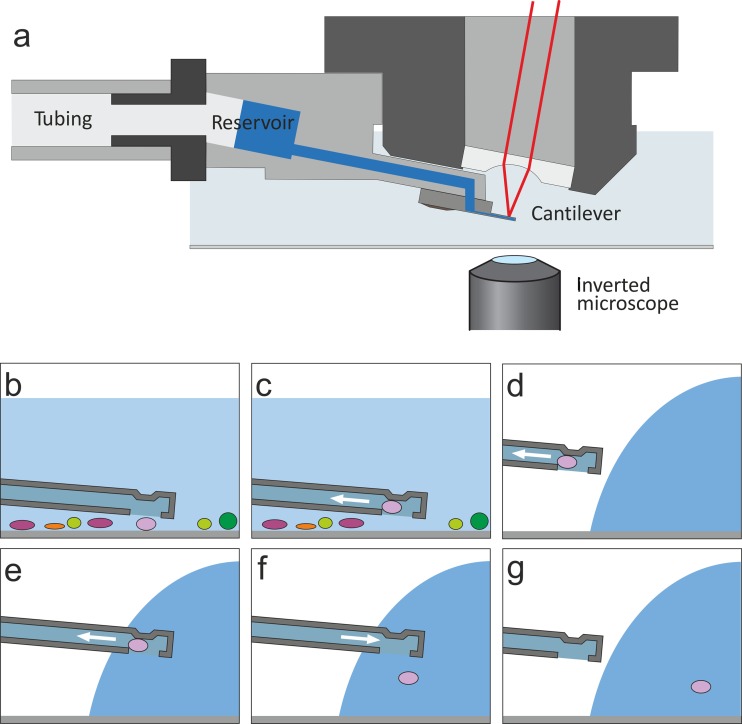
Fig 1.
(a) Schematic principle of the FluidFM technology displaying the hollow cantilever mounted on the AFM probe holder. The microchannel of the cantilever is connected to a pressure controller via a tubing system. (Adapted from reference 63 [copyright © 2013 Wiley-VCH Verlag GmbH & Co. KGaA, Weinheim, Germany].) (b to g) Schematic drawings of the isolation procedure showing the cantilever at different stages of the spatial manipulation procedure. (For photographs of the cantilever tip and procedure, see Fig. S1 in the supplemental material.)
MATERIALS AND METHODS
FluidFM technology and microchanneled cantilevers.
FluidFM experiments were carried out using the FluidFM system, which consists of an AFM head, a hollow cantilever, a drilled probe holder, and a pressure controller (Cytosurge AG, Zurich, Switzerland, and Nanosurf AG, Liestal, Switzerland) as described by Potthoff et al. (37). The tipless hollow cantilevers (Cytosurge AG, Zurich, Switzerland) were made of silicon nitride (channel dimensions: length, 150 μm; height, 0.5 μm; width, 24 μm; diameter of circular aperture, 8 μm). The cantilever was subjected to plasma cleaning (plasma cleaner PDC-32G; Harrick Plasma, Ithaca, NY) for 2 min prior to use and were subsequently coated with 0.1 mg/ml of the grafter copolymer poly(l-lysine)-g-poly(ethylene glycol) [PLL(20)g(3.5)-PEG(2)] (Surface Solutions, Zurich, Switzerland) in 100 mM phosphate buffer for 1 h, as described previously (37, 38).
Isolation procedure.
White clover (Trifolium repens) plants grown outside were collected, and 20 trifoliates were sonicated for 5 min in 30 ml of 100 mM phosphate buffer pH 7.0 (8.66 g Na2HPO4, 6.08 g NaH2PO4 · 2H2O per liter). Part of the bacterial suspension thus obtained (0.5 ml) was then diluted with 7 ml of buffer in glass-bottom dishes (WillCo Wells, Amsterdam, Netherlands) that were subsequently placed on the stage of an inverted microscope (Zeiss AxioObserver D1). No actively swimming bacteria were observed after the bacteria had been allowed to sediment onto the glass surface for 1 h. The FluidFM equipped with a coated cantilever was placed onto the stage, and the probe was inserted into the liquid. Bacteria were optically chosen and sucked to the cantilever aperture, which was placed closely above a bacterium, by applying 50 kPa pressure. As soon as a bacterium was thus immobilized on the cantilever, the latter was removed from the liquid, and the probe holder and cantilever chip were dried with tissue paper. The WillCo dish containing the bacterial suspension was replaced with another dish containing a sterile cover glass of approximately 5 by 5 mm onto which a 4-μl droplet of buffer was deposited. The very end of the cantilever was then optically targeted into the side of the droplet a few hundred nanometers above the surface, and pressure was applied to release the bacterium from the probe. The bacterium was optically tracked throughout the process. The glass slide with the droplet containing the bacterium was then transferred into a sterile tube containing 2 ml of liquid medium. The entire protocol, from optical targeting to release, took about 5 to 8 min per bacterium. The same probe was used for isolation experiments during 6 to 8 h.
Bacterial culture conditions.
Isolated bacteria were grown in 2 ml liquid R2A in 15-ml tubes for up to 14 days at 28°C with 100 rpm. After this incubation time, a few microliters of the broth culture was streaked onto R2A agar (BD Biosciences). Agar plates were incubated at 28°C until the colonies were sufficiently large to perform colony PCR.
Methanol and succinate minimal media were prepared as described before (39). Propanediol medium was produced similarly to the methanol medium, but 5 ml/liter of propane-1,2-diol was added instead of methanol.
To detect fluorescence intensity after growth on different media, cells were grown in a day/night cycle of 8 h of light (200 μE/m2s) at 25°C and 16 h of dark at 22°C. Infrared fluorescence at the single-cell level was assessed after 4 h of light on the fourth day.
Identification by 16S rRNA and determination of photosynthesis genes.
The universal primers 27F (5′-AGAGTTTGATCCTGGCTCAG-3′) and 1492R (5′-GGTTACCTTGTTACGACTT-3′) were used to amplify 16S rRNA genes (40), and the previously described primers pufL67F (5′-TTCGACTTYTGGRTNGGNCC-3′) and pufM781R (5′-CCAKSGTCCAGCGCCAGAANA-3′) were used to amplify pufLM genes (41). The alternative puf gene primers pufLM_N97_for (5′-CTKTTCGACTTCTGGGTSG-3′) and pufLM_N97_rev (5′-CCCATSGTCCAGCGCCAGA-3′) were used to validate the absence of pufLM genes if no amplification could be achieved with the first primer pair (pufL67F/pufM781R) (42). To detect bchY genes, the universal primers bchY_fwd (5′-CCNCARACNATGTGYCCNGCNTTYGG-3′) and bchY_rev (5′-GGRTCNRCNGGRAANATYTCNCC-3′) were used (43). After gel isolation (NucleoSpin [gel and PCR cleanup]; Macherey-Nagel, Düren, Germany), PCR products were ligated into pGEM-T Easy vector (Promega, Fitchburg, WI). A clone was picked for plasmid purification (NucleoSpin [plasmid]; Macherey-Nagel) and was subjected to sequencing using standard primers T7 or SP6 (Microsynth AG, Switzerland). Isolates were then identified by an NCBI BLAST search.
Phylogenetic analyses were performed using the phylogeny.fr server (44). Briefly, multiple alignments were generated using MUSCLE (45) with default parameters and curated using Gblocks (46). Phylogeny was determined using the maximum-likelihood method PhyML (47), with the approximate likelihood ratio test for branch support estimation (48). Trees were drawn using TreeDyn (49). Trees were exported to Adobe Illustrator, and highly similar sequences (>99.5% sequence identity) were merged into a single node.
To generate the 16S rRNA gene tree, a sequence of more than 1,000 bp beginning in a conserved region after primer 27F (AAGTCCCG) and starting at approximately base pair 1050 (which corresponds to nucleotides A21 to G1036 for Methylobacterium extorquens AM1) was used. For the PufL tree, translated sequences from amino acid F30 to the stop codon, a length of 245 amino acids in M. extorquens AM1, was used. 16S rRNA reference sequences were downloaded from NCBI GenBank.
Microscopy and image analysis.
A Zeiss AxioObserver D1 microscope equipped with a Plan-Neofluar 40×/0.6 LD objective was used to optically track the FluidFM cantilever. The highest intensity setting of an EXFO X-Cite series 120Q illumination system was used for fluorescence imaging. For IR epifluorescence observations and determination, we used a custom filter setup: an excitation filter of 320 to 650 nm (BG39; Schott), a 650-nm dichroic mirror (650 dichroic longpass, extended reflection including the UV DCXRU; Chroma), and an emission filter of >850 nm (RG850; Schott) (50). Exposure times of 1,000 ms with a 2-fold gain were used for infrared fluorescence. Images were taken with an AxioCam MRm and the software Zeiss AxioVision 4.8.2. The fluorescence intensities of cells on different media were determined using ImageJ software. The maximal intensities of several representative single bacteria were determined by plotting the profile of a longitudinal line through the bacteria. The background was subtracted. Heat maps of the fluorescence of bacteria on different growth media were constructed based on fluorescence intensity values using the function heatmap.2 from the R package gplots. Standard settings were used.
RESULTS
Establishment of an isolation protocol for single bacteria using FluidFM.
To explore and establish FluidFM for single-bacterium isolation, different parameters were tested and optimized to ensure efficient isolation of a wide range of randomly chosen bacteria from environmental samples. Cantilevers with an aperture diameter of 8 μm were used; however, instead of cantilevers with a default channel height of 1 μm, as used in previous studies (36, 37, 51), cantilevers with a channel height of only 0.5 μm were used, because they ensured that the targeted bacteria remained confined at the aperture, where they could be easily spotted, and were not sucked into the channel of the probe. Plasma cleaning and coating the cantilever with PLL-g-PEG reduced the chance that a bacterium would irreversibly bind to the cantilever (38). For spatial manipulations (Fig. 1), the cantilever's aperture was positioned above the selected bacterium using the x-y movement of the microscopic stage, the cantilever was moved down, and the bacterium was sucked onto the aperture by applying underpressure using a pressure controller connected via a tubing system to the cantilever through a drilled probe holder (see Materials and Methods). Thereby, an underpressure of 50 kPa ensured that the bacteria remained attached at the cantilever aperture while leaving the sample liquid and were transferred through air. After lifting of the cell from the sample and spatial manipulation, the bacterium was released.
Different substrates for placing the selected bacteria were tested. Depositing the bacteria directly on thin agar layers clogged the cantilever after a few transferred bacteria, probably due to binding of the agar to the cantilever. Although placing them directly on glass was possible, it was more practical to release the bacteria into preformed droplets on glass slides (Fig. 1). Subsequently, the droplet containing the bacterium was transferred into liquid medium as described in Materials and Methods, and the cantilever could immediately be reused to pick up another cell.
Bacterial contamination might arise from two potential sources during manipulation: first, during cantilever transfer to the target droplet, unwanted bacteria might stick to the outside of the cantilever surface and not be seen with the optical microscope (note that the cantilever itself was sterile due to the plasma cleaning procedure); second, contamination might occur from the air. To minimize contamination, only the very end of the cantilever was inserted into the target droplet (see Fig. S1 in the supplemental material), which minimized the risk of unwanted transfer of bacteria from the bacterial suspension, because only the optically observed part of the cantilever was in contact with the liquid of the target droplet. In addition, the transfer was performed within an incubator hood, minimizing airflow during manipulation; also, bacterial suspensions and target substrates were covered with sterile petri dishes whenever they were not being subjected to manipulation. To examine potential contamination, mock isolations were performed similarly to real transfer events, except that underpressure was applied without a bacterium being targeted and fixed to the cantilever opening. A total of 125 such mock transfers resulted in contamination in three cases. Although it is desirable to decrease this rate in the future, it was considered sufficiently low compared to the number of manipulation events carried out.
Isolation and identification of phyllosphere bacteria.
To demonstrate that FluidFM technology can be used to isolate bacteria, we first used clover leaf washes and randomly selected individual bacteria for subsequent isolation of liquid R2A medium (52). R2A medium was used to cultivate the bacteria, because a wide range of bacteria are able to multiply in this medium and it is commonly used to isolate various bacteria from environmental samples. Notably, from a total of more than 100 bacterial transfers to growth medium, about two-thirds of the isolates obtained from leaf samples grew to visible turbidity within 2 weeks. Subsequently, these bacteria were streaked on R2A agar, and all isolates that had demonstrated turbidity also grew on the solidified medium. Nutrient broth (NB) was also tested for cultivation of the isolated bacteria, and a recovery rate similar to that achieved with the R2A medium was obtained, further proving that a high recovery rate was possible with the described method. This high percentage of cultivable bacteria with individual isolation in separate compartments was in contrast to plating of dilution series on R2A medium, which resulted in a percentage of cultivable bacteria of about 10% compared to total cell counts using a Thoma chamber and was consistent with other studies describing a similarly low percentage of cultivable bacteria from the phyllosphere of 0.1 to 10% (53, 54).
Identification of isolates (69 in total) by 16S rRNA gene amplification and sequencing using the universal primers 27F and 1492R (55) showed that the bacterial species belong to several different phyla and classes (Table 1). Overall, the community composition resembled those determined previously by cultivation-independent approaches, such as metagenomic analysis (1, 2, 4). Approximately half of the isolates were Proteobacteria, mostly Alphaproteobacteria and several Beta- and Gammaproteobacteria. The dominating genera of the Proteobacteria were Methylobacterium (50%), Sphingomonas (30%), and Pseudomonas (15%). Another large fraction belonged to the Actinobacteria and included a wide range of species. Additionally, some representatives of Bacteroidetes and Firmicutes were found. The isolated methylobacteria were most closely related to Methylobacterium bullatum (13 isolates) and to Methylobacterium adhaesivum (3 isolates), with a sequence identity of more than 99.5%. Isolates from the genus Sphingomonas were also found to belong to different species, reflecting the phylogenetic diversity of the genus detected previously by cultivation-independent approaches (1, 2, 4).
Table 1.
Bacterial strains isolated from white clover leaf washesa
| Phylum (% of isolates) | Class (subclass) | Order (suborder) | Family | Genus | No. of isolates |
|---|---|---|---|---|---|
| Proteobacteria (50) | Alphaproteobacteria | Rhizobiales | Methylobacteriaceae | Methylobacterium | 16 |
| Sphingomonadales | Sphingomonadaceae | Sphingomonas | 10 | ||
| Betaproteobacteria | Burkholderiales | Comamonadaceae | Variovorax | 2 | |
| Oxalobacteraceae | Duganella | 1 | |||
| Gammaproteobacteria | Pseudomonadales | Pseudomonadaceae | Pseudomonas | 5 | |
| Xanthomonadales | Xanthomonadaceae | Xanthomonas | 1 | ||
| Firmicutes (3) | Bacilli | Bacillales | Bacillaceae | Bacillus | 1 |
| Paenibacillaceae | Paenibacillus | 1 | |||
| Bacteroidetes (7) | Sphingobacteria | Sphingobacteriales | Sphingobacteriaceae | Pedobacter | 2 |
| Flavobacteria | Flavobacteriales | Flavobacteriaceae | Flavobacterium | 2 | |
| Chryseobacterium | 1 | ||||
| Actinobacteria (40) | Actinobacteria (Actinobacteridae) | Actinomycetales (Micrococcineae) | Microbacteriaceae | Microbacterium | 4 |
| Plantibacter | 4 | ||||
| Frigoribacterium | 4 | ||||
| Curtobacterium | 4 | ||||
| Rathayibacter | 2 | ||||
| Labedella | 1 | ||||
| Leifsonia/Agreia | 1 | ||||
| Micrococcaceae | Kocuria | 2 | |||
| Micrococcus | 1 | ||||
| Rothia | 1 | ||||
| Actinomycetales (Corynebacterineae) | Nocardiaceae | Rhodococcus | 2 | ||
| Williamsia | 1 | ||||
| Corynebacteriaceae | Corynebacterium | 1 |
Isolates were assigned to their closest related genus by 16S rRNA sequencing.
Isolation and identification of single bacteriochlorophyll-expressing bacteria.
Next, the possibility of isolating a specific subset of bacteria from environmental samples using FluidFM was explored. To this end, bacteriochlorophyll-producing bacteria were identified by their specific fluorescence in the IR range (13, 56–58). Therefore, a custom-built filter set was used to detect fluorescence above 850 nm during excitation with light of less than 680 nm.
First, the fluorescence of bacteria known to express bacteriochlorophyll was assessed in the IR range. Among these were the facultative anaerobic anoxygenic phototrophic bacterium Rhodobacter sphaeroides DSM158 as well as the AAnP Methylobacterium extorquens PA1. As expected, these bacteria showed emission in the IR range, but other nonphototrophic bacteria, such as Escherichia coli DH5α, Bacillus subtilis BRB1, and Sphingomonas sp. FR1, did not show detectable IR fluorescence.
Bacterial suspensions were washed from clover leaves to investigate the presence of bacteriochlorophyll-producing bacteria. A small fraction (∼13%) exhibited IR fluorescence, while the rest appeared dark (Fig. 2), which is in line with previous microscopic observations (5). Some clover plants were kept in the dark for 24 h to determine if the bacteriochlorophyll expression decreased. Bacteria isolated from plants that were kept in the dark showed only a slight reduction in the occurrence and intensity of fluorescence (11.0% ± 2.5% of the bacteria showed IR fluorescence when plants were kept in the dark, compared to 13.2% ± 2.9% of those kept in the light; n > 500). Therefore, it was assumed that bacteria washed from leaves would not lose their fluorescence during the manipulation with FluidFM performed on the same day.
Fig 2.
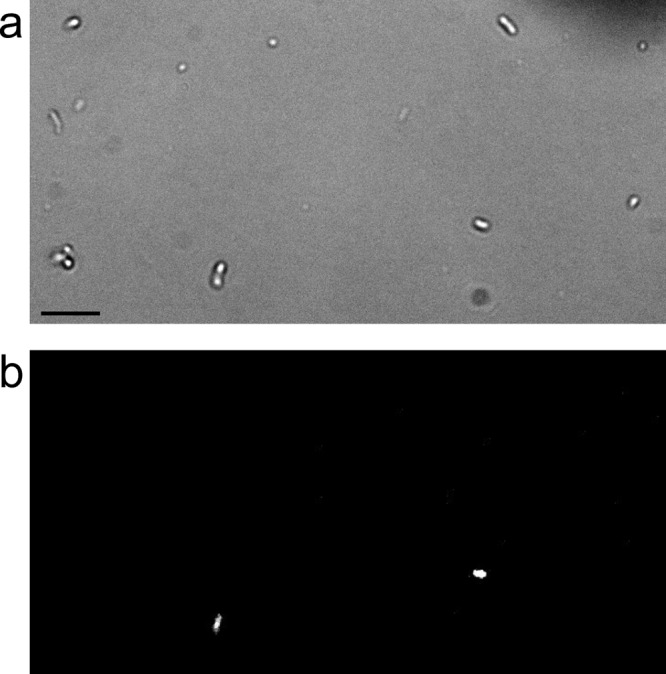
Bright-field (a) and infrared fluorescence (b) images of the same field of clover leaf washes, showing several bacteriochlorophyll-producing bacteria fluorescing while other bacteria exhibit no detectable fluorescence. Bar, 10 μm.
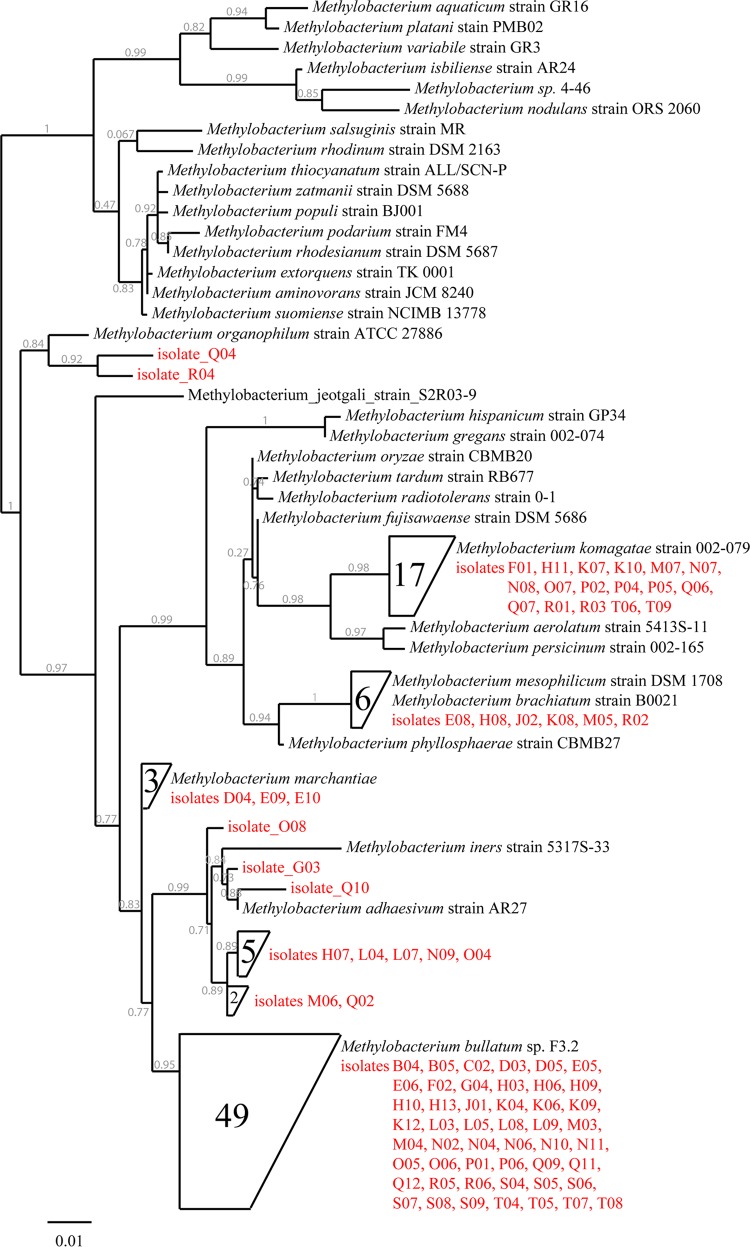
Subsequently, bacteria expressing bacteriochlorophyll were isolated and identified by 16S rRNA gene amplification and sequencing as described above. Consistent with the results from the random isolation experiments, 65% of the IR-fluorescent bacteria (92 out of 142) grew. Of these 92, 87 isolates were identified as belonging to the genus Methylobacterium (Fig. 3; also, see Fig. S2 in the supplemental material). Among these methylobacteria, 17 were nearly identical (>99.5%) to Methylobacterium komagatae, 6 to Methylobacterium mesophilicum, and 3 to Methylobacterium marchantiae. A group of 10 isolates formed a subgroup whose closest described relative was M. adhaesivum (98.5 to 99.5% sequence identity). The largest subgroup, consisting of 49 representatives, belonged to the recently described species M. bullatum (59) (>99.5% sequence identity), and similar 16S rRNA genes were previously identified from samples recovered from clover and other plants (e.g., Clover 1a clone 2_A08 [2] and Methylobacterium sp. strain 85 isolated from an Arabidopsis thaliana leaf [60]). While most Methylobacterium isolates were thus closely related to known species, isolate Q04 and R04 were more distantly related to described strains, with the closest relative being M. organophilum (isolate Q04, 97.7% and isolate R04, 97.9% sequence identity). While there are some more similar sequences (16 entries) of uncultured bacteria in the NCBI database for isolate R04 (up to 99.8% sequence identity), the closest entry for isolate Q04 (uncultured bacterium clone 1-3E; GenBank accession no. EU289432.1) demonstrates 97.9% sequence identity. The remaining 5 isolates from the pool of 92 did not exhibit IR fluorescence under different light regimens and in four different media. Attempts to amplify the pufLM and bchY genes by PCR with established primer pairs (41, 43) were unsuccessful, so it remains undetermined whether they possess divergent genes that could not be amplified with established primers (and do not express bacteriochlorophyll on the growth media tested) or represent false positives (from 142 bacterial transfers).
Fig 3.
Phylogenetic tree of partial 16S rRNA genes, including all 87 Methylobacterium isolates (red) compared to reference species from the NCBI GenBank database. Highly similar subgroups (>99.5% sequence identity) were combined in a single node. The scale bar represents the difference in percent sequence difference, and the likelihood of a branch is specified (gray numbers). A full version of the phylogenetic tree can be found in Fig. S2 in the supplemental material.
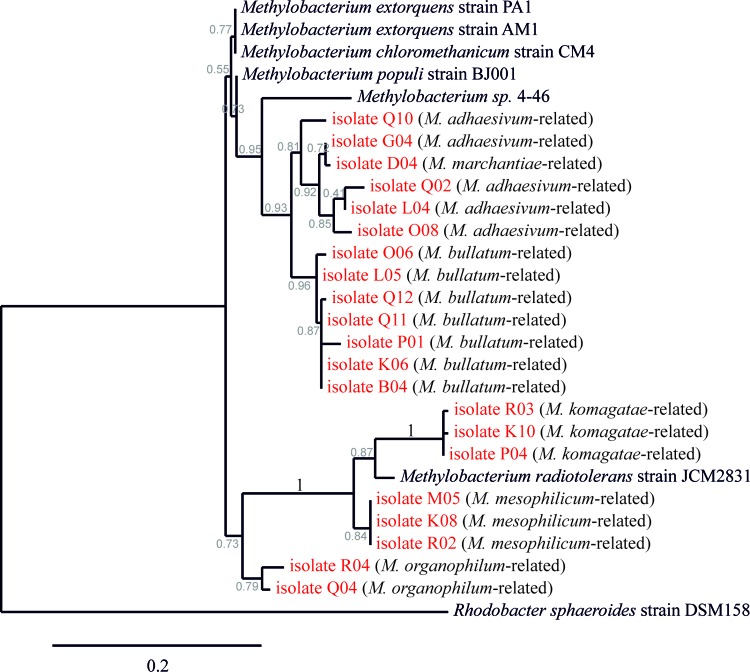
All of the Methylobacterium isolates were validated as AAnPs based on their IR fluorescence, and all of them were found to produce bacteriochlorophyll on agar plates under at least one condition (see below). In addition, validation was performed by amplification of the pufLM genes using a described primer set (41). The PCR products of representatives of each subgroup were sequenced and used to generate a phylogenetic tree of the PufL proteins (Fig. 4). All PufL sequences of the analyzed Methylobacterium isolates clustered together, with the PufL protein sequence of R. sphaeroides being more distantly related to the Methylobacterium cluster. The phylogenetic tree of the Methylobacterium PufL proteins appeared to be analogous to the tree generated from the 16S rRNA genes. Isolates that were closely related according to their 16S rRNA gene sequence also showed more similar PufL proteins, suggesting divergent evolution of the latter.
Fig 4.
Phylogenetic tree of partial PufL protein sequences of selected Methylobacterium isolates compared to reference strains from the NCBI GenBank database. The most closely related species according to the 16S rRNA analysis is given in parentheses. The scale bar represents the difference in percent sequence difference, and the likelihood of a branch is specified (gray numbers).
Characterization of isolates.
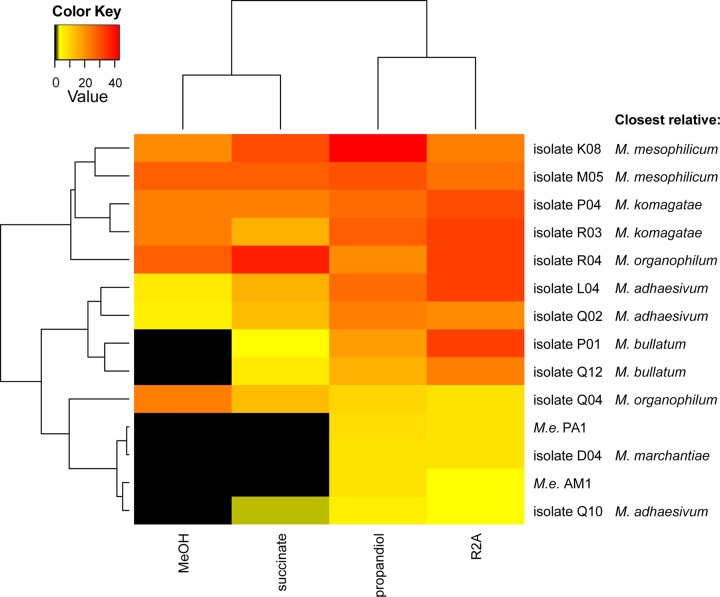
Bacteriochlorophyll production in M. extorquens AM1 (formally Pseudomonas AM1) has been shown to be dependent on culture conditions, including growth substrate and light (61, 62). In order to test whether the Methylobacterium strains isolated from the phyllosphere also show a dependency on the growth medium for bacteriochlorophyll expression, representative Methylobacterium strains from each of the identified subgroups were grown on different solidified media and tested for IR fluorescence after growth during diurnal cycles at the single-cell level. Isolates belonging to similar subgroups behaved alike under similar conditions (Fig. 5). The closely related M. bullatum-like, M. marchantiae-like, and M. adhaesivum-like strains showed pronounced differences in IR fluorescence on the different media tested. IR fluorescence was very low or not detectable on minimal medium supplemented with methanol or succinate and much higher on R2A or minimal medium supplemented with propanediol, as measured by single-cell fluorescence. This effect was most apparent for the M. bullatum-like isolates P01 and Q12. Although these isolates strongly fluoresced on R2A, no fluorescence was detectable on methanol. In contrast, isolate Q04 showed stronger fluorescence on minimal medium supplemented with methanol than on R2A.
Fig 5.
Heat map cluster analysis of the relative fluorescence intensity of Methylobacterium isolates grown on different solidified media after 3 days with a day/night cycle. The closest related species according to the 16S rRNA analysis is indicated. M.e., M. extorquens.
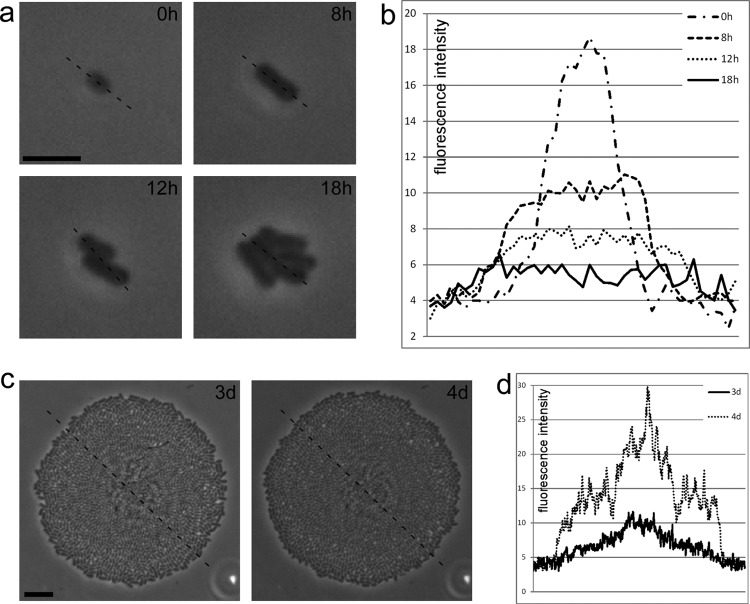
The differences in bacteriochlorophyll expression in the Methylobacterium isolates suggested that the tested methylobacteria regulate bacteriochlorophyll production in a manner that is dependent on the growth conditions. We observed that upon a switch to fresh medium of the same kind, bacteriochlorophyll was regulated in some of the tested isolates. Isolate Q12 was chosen to monitor this induction-or-repression phenomenon of bacteriochlorophyll production. To observe bacteriochlorophyll formation, individual bacteria were grown on R2A agar, and colony development from a single bacterium was monitored by time-lapse microscopy. As the bacterium divided, fluorescence weakened, and no fluorescence was detectable after a few cell divisions (Fig. 6a and b). This suggested that the bacteriochlorophyll was diluted and that no new reaction centers were formed during exponential growth. Notably, after 3 days, when the colony reached a diameter of about 100 μm, it became fluorescent again. Fluorescence further increased after 4 days (Fig. 6c and d), when the density of the colony reached its maximum.
Fig 6.
Growth of a single bacterium from isolate Q12 on 1:10-diluted R2A agar. A plot of fluorescence intensity is shown for each time point (b and d), as indicated by the dashed line in the corresponding bright-field images (a and c). As the cell divides from a single cell (a), the infrared fluorescence slowly fades and is not detectable after 1 day (b). When the colony reaches its maximum size (c), the infrared fluorescence reappears after 3 days and becomes even stronger after 4 days (d). Bars, 5 μm (a) and 10 μm (c).
DISCUSSION
Microscopy is a crucial instrument in microbiology for inspecting phenotypic differences. However, it is often underexploited in the postgenomic era, and further improvements in the method are required to link the physiology of bacteria with their identity at the single cell level. The results presented here demonstrate that FluidFM is a promising tool to facilitate single-cell isolation by picking optically selected bacteria. FluidFM cantilevers are more precise than micropipettes with regard to tip size and the volume of pipetted liquid. Moreover, the bacterium stays at the aperture of an FluidFM tip and is therefore optically tractable. In contrast to FACS, optical microscopic control using micromanipulation is possible, and weak fluorescence (such as that of BChl a) is sufficient to identify cells of interest for single-cell isolation. While we used FluidFM to isolate bacteria from leaf washes, it will be interesting to explore whether the technology can be used to pick up and place individual bacteria directly from the environment, such as the leaf surface. We expect such an approach to be challenging and to require the use of pyramidal tips (63), but with a modified tip geometry.
In this work we isolated bacteria from leaf washes. Notably, approximately two-thirds of the bacteria were cultivable, which is a higher rate than that reported for other habitats, such as soil, where the rate was 0.1 to 1% (64). A cultivation success rate of two-thirds is, however, also higher than that obtained with a parallel determination of CFU, about 10% after dilution from the same bacterial leaf wash suspensions, and that reported in other studies regarding culturability of phyllosphere bacteria with 0.1 to 10% (e.g., references 53 and 54). The isolation of individual bacteria in this work may have helped to avoid antibiosis effects that normally inhibit the success of isolation approaches from this environment during plate isolation. It will thus be interesting to determine whether strains from other plant hosts can also be isolated with a comparably high recovery rate and to perform systematic dilution-to-extinction cultivations with phyllosphere bacteria. The high isolation rate from the samples analyzed in this study resulted in a high congruence with respect to the identification of bacterial genera and species and their relative population sizes based on previous cultivation-independent analyses (1, 2, 4). Overall, the isolation performed in this study indicates that most bacteria from the phyllosphere are readily cultivable and that representative strains can be obtained for targeted analysis, an aspect that will be important for studying the physiology of the bacteria, understanding their coexistence, and building representative synthetic bacterial communities for model systems.
Our attempts to specifically isolate AAnPs from the phyllosphere resulted in a collection of Methylobacterium spp. The isolation of Methylobacterium spp. was not unexpected, since these previously described AAnPs (61, 62) are known to occur in high numbers in the phyllosphere (1, 2, 4, 65–68) and harbor genes for a photosynthetic reaction center as well as genes for bacteriochlorophyll biosynthesis based on completely genome-sequenced strains of this genus (with the exception of the nonpigmented nodulating root symbiont Methylobacterium nodulans) (69, 70). However, the exclusive isolation of methylobacteria was not anticipated, because the diversity observed at the genomic level by metagenome studies suggested a higher diversity of pufM genes than was expected to be derived solely from Methylobacterium spp. (5). However, the phylogenic source of the pufM genes should be considered with caution due to potential horizontal gene transfer, as has been described previously (5, 71). A possible explanation for the exclusive isolation of Methylobacterium species could be that they were the only AAnPs that were active and that produced reaction centers above the limit of detection under the analyzed conditions. Notably, a previous study estimated that approximately 50% of the bacteria on clover harbored the genes for anoxygenic photosynthesis, but only 7% showed detectable fluorescence in the IR range (5). Given that 13% of the bacteria in the present study exhibited IR fluorescence and that 23% methylobacteria were isolated upon random selection and isolation (Table 1), these numbers do not contradict the possibility that all fluorescent bacteria observed belonged to the genus Methylobacterium. They rather suggest that not all methylobacteria living in the phyllosphere produce bacteriochlorophyll at detectable levels. Our observation that the methylobacteria isolated from in planta samples varied greatly with respect to bacteriochlorophyll production levels supports this idea. Of the 16 Methylobacterium isolates obtained by random selection, 13 were related to M. bullatum and 3 were related to M. adhaesivum, indicating that these species are dominant. However, isolation of AAnPs also frequently identified bacteria related to M. komagatae and M. mesophilicum. A possible explanation is that these species produced high levels of bacteriochlorophyll in planta, which is in line with the detection of IR fluorescence on all tested media (Fig. 5), and that not all isolates related to M. bullatum and M. adhaesivum produce sufficient amounts of bacteriochlorophyll to be detectable in planta. Interestingly, these isolates showed a higher dependency on bacteriochlorophyll production dependent on the growth medium. While Sato et al. had already found that the formation of bacteriochlorophyll of strain M. extorquens AM1 is dependent on the growth substrate and day/night cycle (61, 62), the variability and strain dependence in the bacteriochlorophyll production by different strains observed here suggests that different Methylobacterium strains have distinct regulatory cues to regulate bacteriochlorophyll expression as photoheterotrophs. Regulation of the photosynthetic trait has not yet been studied in Methylobacterium spp. at the molecular level and it will be of particular interest to unravel the regulatory mechanisms underlying the control of the bacteriochlorophyll production in dependence of the nature and amount of alternative carbon substrates and the role of a predicted bacteriophytochrome encoded in the genome of the model strain M. extorquens PA1 (66, 69) and other Methylobacterium spp.
Dedicated work will also be required to understand the diversity of Methylobacterium strains regarding their in situ physiology. Gathering spatial information will be important, since different species might occupy distinct microniches in the phyllosphere. These sites might offer various exposures to light, for instance on the upper and lower sides of leaves, and different levels of nutrient availability, which is known to be heterogeneous (72, 73). In consequence, the demand for supplementing energetic needs by photosynthesis might differ depending on available alternative carbon substrates at different sites. In this context, it will also be of interest to characterize the contribution of energy produced from light under different environmental conditions, given the autotrophic potential recently predicted by the genome-scale model of M. extorquens (74). Moreover, the underlying regulatory mechanisms triggering the apparent arrest in bacteriochlorophyll production described for strain Q12 and its production after a higher density of cells has been reached (Fig. 6) is of interest. The phenotypic switch observed here upon transfer to fresh medium might depend on repression of transcription of genes related to photosynthesis when carbon sources become available after starvation and/or quorum-sensing systems, which have been shown to exist in Methylobacterium spp. (75–77).
In conclusion, the single-cell isolation approach using FluidFM defined here revealed the presence of methylobacteria as bacteriochlorophyll-expressing AAnPs in the clover phyllosphere and a new facet of interspecies diversity within the genus Methylobacterium. The study also demonstrated the power of using FluidFM for single-cell isolation.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by ETH Zurich and by the Swiss Innovation Promotion Agency KTI-CTI (11722.1 PFNM-NM).
We thank Natacha Bodenhausen for help with the heatmap analysis, Mitja Remus-Emsermann for help with the phylogenetic tree analysis, and Eva Potthoff and Orane Guillaume-Gentil for fruitful discussions.
Footnotes
Published ahead of print 14 June 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.01087-13.
REFERENCES
- 1. Vorholt JA. 2012. Microbial life in the phyllosphere. Nat. Rev. Microbiol. 10:828–840 [DOI] [PubMed] [Google Scholar]
- 2. Delmotte N, Knief C, Chaffron S, Innerebner G, Roschitzki B, Schlapbach R, von Mering C, Vorholt JA. 2009. Community proteogenomics reveals insights into the physiology of phyllosphere bacteria. Proc. Natl. Acad. Sci. U. S. A. 106:16428–16433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Sy A, Timmers AC, Knief C, Vorholt JA. 2005. Methylotrophic metabolism is advantageous for Methylobacterium extorquens during colonization of Medicago truncatula under competitive conditions. Appl. Environ. Microbiol. 71:7245–7252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Knief C, Delmotte N, Chaffron S, Stark M, Innerebner G, Wassmann R, von Mering C, Vorholt JA. 2012. Metaproteogenomic analysis of microbial communities in the phyllosphere and rhizosphere of rice. ISME J. 6:1378–1390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Atamna-Ismaeel N, Finkel OM, Glaser F, von Mering C, Vorholt JA, Koblizek M, Belkin S, Beja O. 2012. Bacterial anoxygenic photosynthesis on plant leaf surfaces. Environ. Microbiol. Rep. 4:209–216 [DOI] [PubMed] [Google Scholar]
- 6. Yurkov VV, Beatty JT. 1998. Aerobic anoxygenic phototrophic bacteria. Microbiol. Mol. Biol. Rev. 62:695–724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Albrecht-Buehler G. 1997. Autofluorescence of live purple bacteria in the near infrared. Exp. Cell Res. 236:43–50 [DOI] [PubMed] [Google Scholar]
- 8. Pierson BK, Howard HM. 1972. Detection of bacteriochlorophyll-containing micro-organisms by infrared fluorescence photomicrography. J. Gen. Microbiol. 73:359–363 [Google Scholar]
- 9. Ritchie AE, Johnson ZI. 2012. Abundance and genetic diversity of aerobic anoxygenic phototrophic bacteria of coastal regions of the pacific ocean. Appl. Environ. Microbiol. 78:2858–2866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Beja O, Suzuki MT, Heidelberg JF, Nelson WC, Preston CM, Hamada T, Eisen JA, Fraser CM, DeLong EF. 2002. Unsuspected diversity among marine aerobic anoxygenic phototrophs. Nature 415:630–633 [DOI] [PubMed] [Google Scholar]
- 11. Waidner LA, Kirchman DL. 2007. Aerobic anoxygenic phototrophic bacteria attached to particles in turbid waters of the Delaware and Chesapeake estuaries. Appl. Environ. Microbiol. 73:3936–3944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Masin M, Nedoma J, Pechar L, Koblizek M. 2008. Distribution of aerobic anoxygenic phototrophs in temperate freshwater systems. Environ. Microbiol. 10:1988–1996 [DOI] [PubMed] [Google Scholar]
- 13. Kolber ZS, Plumley FG, Lang AS, Beatty JT, Blankenship RE, VanDover CL, Vetriani C, Koblizek M, Rathgeber C, Falkowski PG. 2001. Contribution of aerobic photoheterotrophic bacteria to the carbon cycle in the ocean. Science 292:2492–2495 [DOI] [PubMed] [Google Scholar]
- 14. Koblizek M, Masin M, Ras J, Poulton AJ, Prasil O. 2007. Rapid growth rates of aerobic anoxygenic phototrophs in the ocean. Environ. Microbiol. 9:2401–2406 [DOI] [PubMed] [Google Scholar]
- 15. Koblizek M. 2011. Role of photoheterotrophic bacteria in the marine carbon cycle, p 49–51 In Jiao N, Azam F, Sanders S. (ed), Microbial carbon pump in the ocean. AAAS, Washington, DC [Google Scholar]
- 16. Kim MK, Schubert K, Im WT, Kim KH, Lee ST, Overmann J. 2007. Sphingomonas kaistensis sp. nov., a novel alphaproteobacterium containing pufLM genes. Int. J. Syst. Evol. Microbiol. 57:1527–1534 [DOI] [PubMed] [Google Scholar]
- 17. Giraud E, Hannibal L, Fardoux J, Vermeglio A, Dreyfus B. 2000. Effect of Bradyrhizobium photosynthesis on stem nodulation of Aeschynomene sensitiva. Proc. Natl. Acad. Sci. U. S. A. 97:14795–14800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Csotonyi JT, Swiderski J, Stackebrandt E, Yurkov V. 2010. A new extreme environment for aerobic anoxygenic phototrophs: biological soil crusts. Adv. Exp. Med. Biol. 675:3–14 [DOI] [PubMed] [Google Scholar]
- 19. Fleischman D, Kramer D. 1998. Photosynthetic rhizobia. Biochim. Biophys. Acta 1364:17–36 [DOI] [PubMed] [Google Scholar]
- 20. Muller S, Nebe-von-Caron G. 2010. Functional single-cell analyses: flow cytometry and cell sorting of microbial populations and communities. FEMS Microbiol. Rev. 34:554–587 [DOI] [PubMed] [Google Scholar]
- 21. Martinez-Garcia M, Swan BK, Poulton NJ, Gomez ML, Masland D, Sieracki ME, Stepanauskas R. 2012. High-throughput single-cell sequencing identifies photoheterotrophs and chemoautotrophs in freshwater bacterioplankton. ISME J. 6:113–123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Ashkin A, Dziedzic JM. 1987. Optical trapping and manipulation of viruses and bacteria. Science 235:1517–15120 [DOI] [PubMed] [Google Scholar]
- 23. Leung K, Zahn H, Leaver T, Konwar KM, Hanson NW, Page AP, Lo CC, Chain PS, Hallam SJ, Hansen CL. 2012. A programmable droplet-based microfluidic device applied to multiparameter analysis of single microbes and microbial communities. Proc. Natl. Acad. Sci. U. S. A. 109:7665–7670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Marcy Y, Ouverney C, Bik EM, Losekann T, Ivanova N, Martin HG, Szeto E, Platt D, Hugenholtz P, Relman DA, Quake SR. 2007. Dissecting biological “dark matter” with single-cell genetic analysis of rare and uncultivated TM7 microbes from the human mouth. Proc. Natl. Acad. Sci. U. S. A. 104:11889–11894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Pamp SJ, Harrington ED, Quake SR, Relman DA, Blainey PC. 2012. Single-cell sequencing provides clues about the host interactions of segmented filamentous bacteria (SFB). Genome Res. 22:1107–1119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Pohl HA, Hawk I. 1966. Separation of living and dead cells by dielectrophoresis. Science 152:647–649 [DOI] [PubMed] [Google Scholar]
- 27. Chiou PY, Ohta AT, Wu MC. 2005. Massively parallel manipulation of single cells and microparticles using optical images. Nature 436:370–372 [DOI] [PubMed] [Google Scholar]
- 28. Chowdhury TK. 1969. Fabrication of extremely fine glass micropipette electrodes. J. Sci. Instrum. 2:1087–1090 [DOI] [PubMed] [Google Scholar]
- 29. Hochmuth RM. 2000. Micropipette aspiration of living cells. J. Biomech. 33:15–22 [DOI] [PubMed] [Google Scholar]
- 30. Frohlich J, Konig H. 1999. Rapid isolation of single microbial cells from mixed natural and laboratory populations with the aid of a micromanipulator. Syst. Appl. Microbiol. 22:249–257 [DOI] [PubMed] [Google Scholar]
- 31. Ishoy T, Kvist T, Westermann P, Ahring BK. 2006. An improved method for single cell isolation of prokaryotes from meso-, thermo- and hyperthermophilic environments using micromanipulation. Appl. Microbiol. Biotechnol. 69:510–514 [DOI] [PubMed] [Google Scholar]
- 32. Woyke T, Tighe D, Mavromatis K, Clum A, Copeland A, Schackwitz W, Lapidus A, Wu D, McCutcheon JP, McDonald BR, Moran NA, Bristow J, Cheng JF. 2010. One bacterial cell, one complete genome. PLoS One 5:e10314. 10.1371/journal.pone.0010314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Hongoh Y, Sharma VK, Prakash T, Noda S, Taylor TD, Kudo T, Sakaki Y, Toyoda A, Hattori M, Ohkuma M. 2008. Complete genome of the uncultured termite group 1 bacteria in a single host protist cell. Proc. Natl. Acad. Sci. U. S. A. 105:5555–5560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Stepanauskas R. 2012. Single cell genomics: an individual look at microbes. Curr. Opin. Microbiol. 15:613–620 [DOI] [PubMed] [Google Scholar]
- 35. Meister A, Gabi M, Behr P, Studer P, Voros J, Niedermann P, Bitterli J, Polesel-Maris J, Liley M, Heinzelmann H, Zambelli T. 2009. FluidFM: combining atomic force microscopy and nanofluidics in a universal liquid delivery system for single cell applications and beyond. Nano Lett. 9:2501–2507 [DOI] [PubMed] [Google Scholar]
- 36. Dörig P, Stiefel P, Behr P, Sarajlic E, Bijl D, Gabi M, Vörös J, Vorholt JA, Zambelli T. 2010. Force-controlled spatial manipulation of viable mammalian cells and micro-organisms by means of FluidFM technology. Appl. Phys. Lett. 97:023701 [Google Scholar]
- 37. Potthoff E, Guillaume-Gentil O, Ossola D, Polesel-Maris J, Leibundgut-Landmann S, Zambelli T, Vorholt JA. 2012. Rapid and serial quantification of adhesion forces of yeast and Mammalian cells. PLoS One 7:e52712. 10.1371/journal.pone.0052712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Huang NP, Michel R, Voros J, Textor M, Rolf Hofer Rossi A, Elbert DL, Hubbell JA, Spencer ND. 2001. Poly(l-lysine)-g-poly(ethylene glycol) layers on metal oxide surfaces: surface-analytical characterization and resistance to serum and fibrinogen adsorption. Langmuir 17:489–498 [Google Scholar]
- 39. Peyraud R, Kiefer P, Christen P, Massou S, Portais JC, Vorholt JA. 2009. Demonstration of the ethylmalonyl-CoA pathway by using 13C metabolomics. Proc. Natl. Acad. Sci. U. S. A. 106:4846–4851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Turner S, Pryer KM, Miao VP, Palmer JD. 1999. Investigating deep phylogenetic relationships among cyanobacteria and plastids by small subunit rRNA sequence analysis. J. Eukaryot. Microbiol. 46:327–338 [DOI] [PubMed] [Google Scholar]
- 41. Tank M, Thiel V, Imhoff JF. 2009. Phylogenetic relationship of phototrophic purple sulfur bacteria according to pufL and pufM genes. Int. Microbiol. 12:175–185 [PubMed] [Google Scholar]
- 42. Nagashima KV, Hiraishi A, Shimada K, Matsuura K. 1997. Horizontal transfer of genes coding for the photosynthetic reaction centers of purple bacteria. J. Mol. Evol. 45:131–136 [DOI] [PubMed] [Google Scholar]
- 43. Yutin N, Suzuki MT, Rosenberg M, Rotem D, Madigan MT, Suling J, Imhoff JF, Beja O. 2009. BchY-based degenerate primers target all types of anoxygenic photosynthetic bacteria in a single PCR. Appl. Environ. Microbiol. 75:7556–7559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Dereeper A, Guignon V, Blanc G, Audic S, Buffet S, Chevenet F, Dufayard JF, Guindon S, Lefort V, Lescot M, Claverie JM, Gascuel O. 2008. Phylogeny.fr: robust phylogenetic analysis for the non-specialist. Nucleic Acids Res. 36:W465–W469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Edgar RC. 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32:1792–1797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Talavera G, Castresana J. 2007. Improvement of phylogenies after removing divergent and ambiguously aligned blocks from protein sequence alignments. Syst. Biol. 56:564–577 [DOI] [PubMed] [Google Scholar]
- 47. Guindon S, Gascuel O. 2003. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst. Biol. 52:696–704 [DOI] [PubMed] [Google Scholar]
- 48. Anisimova M, Gascuel O. 2006. Approximate likelihood-ratio test for branches: a fast, accurate, and powerful alternative. Syst. Biol. 55:539–552 [DOI] [PubMed] [Google Scholar]
- 49. Chevenet F, Brun C, Banuls AL, Jacq B, Christen R. 2006. TreeDyn: towards dynamic graphics and annotations for analyses of trees. BMC Bioinformatics 7:439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Koblizek M, Ston-Egiert J, Sagan S, Kolber ZS. 2005. Diel changes in bacteriochlorophyll a concentration suggest rapid bacterioplankton cycling in the Baltic Sea. FEMS Microbiol. Ecol. 51:353–361 [DOI] [PubMed] [Google Scholar]
- 51. Stiefel P, Schmidt FI, Dörig P, Behr P, Zambelli T, Vorholt JA, Mercer J. 2012. Cooperative vaccinia infection demonstrated at the single-cell level using FluidFM. Nano Lett. 12:4219–4227 [DOI] [PubMed] [Google Scholar]
- 52. van der Linde K, Lim BT, Rondeel JM, Antonissen LP, de Jong GM. 1999. Improved bacteriological surveillance of haemodialysis fluids: a comparison between tryptic soy agar and Reasoner's 2A media. Nephrol. Dial. Transplant. 14:2433–2437 [DOI] [PubMed] [Google Scholar]
- 53. Yashiro E, Spear RN, McManus PS. 2011. Culture-dependent and culture-independent assessment of bacteria in the apple phyllosphere. J. Appl. Microbiol. 110:1284–1296 [DOI] [PubMed] [Google Scholar]
- 54. Rastogi G, Tech JJ, Coaker GL, Leveau JH. 2010. A PCR-based toolbox for the culture-independent quantification of total bacterial abundances in plant environments. J. Microbiol. Methods 83:127–132 [DOI] [PubMed] [Google Scholar]
- 55. Lane DJ. 1991. 16S/23S rRNA sequencing, p 115–175 In Stackebrandt E, Goodfellow M. (ed), Nucleic acid techniques in bacterial systematics. John Wiley and Sons, New York, NY [Google Scholar]
- 56. Boomer SM, Pierson BK, Austinhirst R, Castenholz RW. 2000. Characterization of novel bacteriochlorophyll-a-containing red filaments from alkaline hot springs in Yellowstone National Park. Arch. Microbiol. 174:152–161 [DOI] [PubMed] [Google Scholar]
- 57. Jiao N, Zhang Y, Chen Y. 2006. Time series observation based infrared epifluorescence microscopic (TIREM) approach for accurate enumeration of bacteriochlorophyll-containing microbes in marine environments. J. Microbiol. Methods 65:442–452 [DOI] [PubMed] [Google Scholar]
- 58. Masin M, Zdun A, Ston-Egiert J, Nausch M, Labrenz M, Moulisová V, Michal Koblíek M. 2006. Seasonal changes and diversity of aerobic anoxygenic phototrophs in the Baltic Sea. Aquat. Microb. Ecol. 45:247–254 [Google Scholar]
- 59. Hoppe T, Peters K, Schmidt F. 2011. Methylobacterium bullatum sp. nov., a methylotrophic bacterium isolated from Funaria hygrometrica. Syst. Appl. Microbiol. 34:482–486 [DOI] [PubMed] [Google Scholar]
- 60. Knief C, Dengler V, Bodelier PL, Vorholt JA. 2012. Characterization of Methylobacterium strains isolated from the phyllosphere and description of Methylobacterium longum sp. nov. Antonie Van Leeuwenhoek 101:169–183 [DOI] [PubMed] [Google Scholar]
- 61. Sato K. 1978. Bacteriochlorophyll formation by facultative methylotrophs, Protaminobacter ruber and Pseudomonas AM 1. FEBS Lett. 85:207–210 [DOI] [PubMed] [Google Scholar]
- 62. Sato K, Hagiwara K, Shimizu S. 1985. Effect of cultural conditions on tetrapyrrole formation, especially bacteriochlorophyll formation in a facultative methylotroph, Protaminobacter ruber. Agric. Biol. Chem. 49:1–5 [Google Scholar]
- 63. Guillaume-Gentil O, Potthoff E, Ossola D, Dorig P, Zambelli T, Vorholt JA. 2013. Force-controlled fluidic injection into single cell nuclei. Small 9:1904–1907 [DOI] [PubMed] [Google Scholar]
- 64. Handelsman J. 2004. Metagenomics: application of genomics to uncultured microorganisms. Microbiol. Mol. Biol. Rev. 68:669–685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Knief C, Frances L, Cantet F, Vorholt JA. 2008. Cultivation-independent characterization of Methylobacterium populations in the plant phyllosphere by automated ribosomal intergenic spacer analysis. Appl. Environ. Microbiol. 74:2218–2228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Knief C, Frances L, Vorholt JA. 2010. Competitiveness of diverse Methylobacterium strains in the phyllosphere of Arabidopsis thaliana and identification of representative models, including M. extorquens PA1. Microb. Ecol. 60:440–452 [DOI] [PubMed] [Google Scholar]
- 67. Knief C, Ramette A, Frances L, Alonso-Blanco C, Vorholt JA. 2010. Site and plant species are important determinants of the Methylobacterium community composition in the plant phyllosphere. ISME J. 4:719–728 [DOI] [PubMed] [Google Scholar]
- 68. Corpe WA, Rheem S. 1989. Ecology of the methylotrophic bacteria on living leaf surfaces. FEMS Microbiol. Lett. 62:243–249 [Google Scholar]
- 69. Marx CJ, Bringel F, Chistoserdova L, Moulin L, Farhan Ul Haque M, Fleischman DE, Gruffaz C, Jourand P, Knief C, Lee MC, Muller EE, Nadalig T, Peyraud R, Roselli S, Russ L, Goodwin LA, Ivanova N, Kyrpides N, Lajus A, Land ML, Medigue C, Mikhailova N, Nolan M, Woyke T, Stolyar S, Vorholt JA, Vuilleumier S. 2012. Complete genome sequences of six strains of the genus Methylobacterium. J. Bacteriol. 194:4746–4748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Vuilleumier S, Chistoserdova L, Lee MC, Bringel F, Lajus A, Zhou Y, Gourion B, Barbe V, Chang J, Cruveiller S, Dossat C, Gillett W, Gruffaz C, Haugen E, Hourcade E, Levy R, Mangenot S, Muller E, Nadalig T, Pagni M, Penny C, Peyraud R, Robinson DG, Roche D, Rouy Z, Saenampechek C, Salvignol G, Vallenet D, Wu Z, Marx CJ, Vorholt JA, Olson MV, Kaul R, Weissenbach J, Medigue C, Lidstrom ME. 2009. Methylobacterium genome sequences: a reference blueprint to investigate microbial metabolism of C1 compounds from natural and industrial sources. PLoS One 4:e5584. 10.1371/journal.pone.0005584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Igarashi N, Harada J, Nagashima S, Matsuura K, Shimada K, Nagashima KV. 2001. Horizontal transfer of the photosynthesis gene cluster and operon rearrangement in purple bacteria. J. Mol. Evol. 52:333–341 [DOI] [PubMed] [Google Scholar]
- 72. Leveau JH, Lindow SE. 2001. Appetite of an epiphyte: quantitative monitoring of bacterial sugar consumption in the phyllosphere. Proc. Natl. Acad. Sci. U. S. A. 98:3446–3453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Remus-Emsermann MNP, Leveau JHJ. 2010. Linking environmental heterogeneity and reproductive success at single-cell resolution. ISME J. 4:215–222 [DOI] [PubMed] [Google Scholar]
- 74. Peyraud R, Schneider K, Kiefer P, Massou S, Vorholt JA, Portais JC. 2011. Genome-scale reconstruction and system level investigation of the metabolic network of Methylobacterium extorquens AM1. BMC Syst. Biol. 5:189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Nieto Penalver CG, Cantet F, Morin D, Haras D, Vorholt JA. 2006. A plasmid-borne truncated luxI homolog controls quorum-sensing systems and extracellular carbohydrate production in Methylobacterium extorquens AM1. J. Bacteriol. 188:7321–7324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Poonguzhali S, Madhaiyan M, Sa T. 2007. Production of acyl-homoserine lactone quorum-sensing signals is widespread in gram-negative Methylobacterium. J. Microbiol. Biotechnol. 17:226–233 [PubMed] [Google Scholar]
- 77. Nieto Penalver CG, Morin D, Cantet F, Saurel O, Milon A, Vorholt JA. 2006. Methylobacterium extorquens AM1 produces a novel type of acyl-homoserine lactone with a double unsaturated side chain under methylotrophic growth conditions. FEBS Lett. 580:561–567 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.