Abstract
As global aquaculture fish production continues to expand, an improved understanding of how environmental factors interact in fish health and production is needed. Significant advances have been made toward economical alternatives to costly fishmeal-based diets, such as grain-based formulations, and toward defining the effect of rearing density on fish health and production. Little research, however, has examined the effects of fishmeal- and grain-based diets in combination with alterations in rearing density. Moreover, it is unknown whether interactions between rearing density and diet impact the composition of the fish intestinal microbiota, which might in turn impact fish health and production. We fed aquacultured adult rainbow trout (Oncorhynchus mykiss) fishmeal- or grain-based diets, reared them under high- or low-density conditions for 10 months in a single aquaculture facility, and evaluated individual fish growth, production, fin indices, and intestinal microbiota composition using 16S rRNA gene sequencing. We found that the intestinal microbiotas were dominated by a shared core microbiota consisting of 52 bacterial lineages observed across all individuals, diets, and rearing densities. Variations in diet and rearing density resulted in only minor changes in intestinal microbiota composition despite significant effects of these variables on fish growth, performance, fillet quality, and welfare. Significant interactions between diet and rearing density were observed only in evaluations of fin indices and the relative abundance of the bacterial genus Staphylococcus. These results demonstrate that aquacultured rainbow trout can achieve remarkable consistency in intestinal microbiota composition and suggest the possibility of developing novel aquaculture strategies without overtly altering intestinal microbiota composition.
INTRODUCTION
As aquaculture's contribution to global food fish consumption continues to increase (1), alternatives to fishmeal as the traditional protein source in aquaculture feeds need to be researched, refined, and adopted for sustainable industry growth (2, 3). Much research has focused on all-plant-protein diets and their impact on fish performance (4), palatability (5), and digestibility (6); water quality (7); intestinal inflammation (8); and the community of microorganisms residing in the intestine (microbiota) (9). Overall, significant advances have been made in alternative protein diet formulations in recent years, so that the growth performance of fish fed grain-based diets has been reported to be comparable to that of fish fed traditional fishmeal-based diets (7, 10). Limited research, however, has examined the effects of grain-based feeds in combination with alterations in fish-rearing density. Provided that a given aquaculture system's carrying capacity can support increases in fish biomass, larger harvests can, in theory, be attained by increasing the rearing density as fish are raised to market size. Inappropriately high rearing densities, however, can have negative effects on fish production and are commonly associated with decreased growth, decreased feed intake, reduced feed efficiency, and greater fin erosion (11). Whether these density-associated changes in performance and welfare are consistent when fish are fed either fishmeal- or grain-based diets remains unclear. Moreover, it remains unknown whether interactions between fish-rearing density and diet composition impact the composition of the intestinal microbiota. This gap in our knowledge is significant, because processes such as intestinal inflammation, dietary energy harvest, and behavior in other vertebrate species are due in part to alterations in intestinal microbiota composition (12–19).
A fundamental challenge in host-associated microbial ecology is determining the extent to which microbial lineages in a given host are shared among other hosts. Previous studies have shown that a subset of microbial lineages harbored by an individual host might also be found in many or all other individual hosts, a concept often referred to as a “core microbiota.” This term can be variably defined based on the taxonomic level or the degree of ubiquity and abundance among individual hosts under a given experimental condition, in a given environment, or in a given host species (20, 21). Although detection of a core microbiota is strongly affected by sample number, sampling depth, and many genetic and environmental factors, these factors can be addressed through careful experimental design. The relatively consistent environmental, dietary, and husbandry parameters inherent in aquaculture facilities provide attractive opportunities to explore the potential for a core microbiota in animal hosts. As new strategies for aquaculture enhancements are developed, it will be important to determine whether core microbiotas occur in aquaculture settings and whether such cores are affected by husbandry variation.
Our current information on the gut microbiota of rainbow trout (Oncorhynchus mykiss) is derived from analysis of culturable microorganisms (22–24) and culture-independent studies using fingerprinting and sequencing of 16S rRNA and other microbial genes (9, 23, 25–27). These studies have revealed that the rainbow trout gut microbiota is dominated by the bacterial phyla Proteobacteria and Firmicutes, the same phyla that dominate the intestines of many other fishes (16, 28, 29). In contrast to these methodologies, high-throughput pyrosequencing of 16S rRNA genes permits unbiased identification of rare, as well as abundant, bacterial members of the gut microbiota at low cost per sequence. The gut microbiota of aquacultured trout has previously been analyzed by pyrosequencing of the cpn60 gene (9, 30), but not the more commonly studied 16S rRNA gene. In this study, we tested whether long-term differences in rearing density and diet, alone and in combination, lead to alterations in animal performance, welfare, fillet quality, or gut microbiota using 16S rRNA gene pyrosequencing.
MATERIALS AND METHODS
Experimental treatments, fish performance data collection, and processing attributes.
All experiments involving rainbow trout were conducted in compliance with the requirements of the Animal Welfare Act (9CFR) and were approved by the Freshwater Institute's Institutional Animal Care and Use Committee.
A flowthrough fish culture system consisting of 12 circular 500-liter tanks was employed in this study, using water from a spring source with approximately constant 12.5°C temperature. Eyed rainbow trout eggs were procured from Troutlodge, Inc. (Sumner, WA); hatched alevins were then transferred to two of the 12 flowthrough tanks for introduction to feed. Fishmeal-based starter feed was used for all fish during this acclimation period. When the fish reached approximately 10 g, they were recombined in one tank and then randomly distributed in equal numbers to all 12 flowthrough tanks. The fish were subsequently fed either the experimental fishmeal- or grain-based feed (Table 1) for the remainder of the study and were reared in one of two density ranges: either 20 to 40 kg/m3 (low density) or 40 to 80 kg/m3 (high density). As the tanks approached the maximum density (40 or 80 kg/m3) for their specific treatment, fish were culled to reduce densities to low-end levels (20 or 40 kg/m3). The diet and density treatments were randomly allocated within the 12-tank system, so that each of the four diet/density treatment groups was replicated in three study tanks. Monthly length and weight assessments were made for each tank over the 10-month study to update their biomass increase and to guide density adjustments. All sampled fish were first anesthetized (75 mg/liter tricaine methanesulfonate [MS-222; Tricaine-S; Western Chemical Inc., Ferndale, WA]) prior to collection of performance data. Dead animals were removed and recorded daily to assess cumulative survival. Feed was administered by an in-house-designed computer-operated program to identical feeders for all 12 experimental tanks, with feeding events approximately once per hour. Daily feed levels were determined using standardized feed charts for rainbow trout; however, minor adjustments to daily feeding amounts were occasionally made based on visual observations of increased appetence or satiation. Overall thermal growth coefficients (TGC) and feed conversion ratios (FCR) were calculated for each tank at the end of the study period, based on the final performance data, and compared between treatments as follows: TGC = [(final mean weight1/3 − initial mean weight1/3)/(days during interval × mean temperature)] × 1,000 and FCR = feedcumulative/biomass gain, where weight is in grams, length is in millimeters, and temperature is in °C. At study's end (312 days posthatch), 5 randomly selected fish were removed from each tank, euthanized with an overdose (200 mg/liter) of MS-222, eviscerated, and processed to yield butterfly fillets. The butterfly fillet is produce when the head, viscera, and vertebral column and ribs have been removed. The dress yield (percent) (i.e., the head-on gutted yield) was calculated as follows: eviscerated weight/whole weight × 100. The pectoral girdle, belly flaps (approximately 1-cm strips along the ventral midline), and skin were removed from the butterfly fillets. The fillets were weighed, and the fillet yield (percent) was calculated as follows: fillet weight/whole weight × 100.
Table 1.
Nutritional compositions of the fishmeal- and grain-based experimental diets utilized
| Ingredient | Amt (g/kg) |
|
|---|---|---|
| Fishmeal diet | Grain-based diet | |
| Fish meala | 312.7 | |
| Blood mealb | 74.7 | |
| Soy protein concentratec | 289.1 | |
| Corn gluten meald | 251.7 | |
| Soybean meale | 192.4 | |
| Wheat gluten meale | 46.5 | |
| Wheat flourf | 284.0 | |
| Menhaden oilg | 112.0 | 167.4 |
| Vitamin premixh | 7.5 | 7.5 |
| Lysine | 11.1 | |
| Methionine | 2.8 | |
| Taurine | 5.0 | |
| Dicalcium phosphate | 36.5 | |
| Trace mineral premixi | 1.0 | 1.0 |
| Choline CL | 2.0 | 2.0 |
| Ascorbic acidj | 2.0 | 2.0 |
| Astazanthink | 0.2 | |
| Total protein (%) | 41.0 | 47.5 |
| Total fat (%) | 15.1 | 18.0 |
Omega Proteins; Menhaden Special Select; 628 g/kg protein.
IDF Inc.; 832 g/kg protein.
Solae; Pro-Fine VF; 693 g/kg crude protein.
Cargill; 602.0 g/kg protein.
ADM Inc.; 480 g/kg protein.
Manildra Milling; 120 g/kg protein.
Omega Proteins Inc.
United States Fish and Wildlife Service (USFWS) no. 30. Amounts contributed per kilogram of diet were as follows: vitamin A (as retinol palmitate), 10,000 IU; vitamin D3, 720 IU; vitamin E (as dl-α-tocopherol acetate), 530 IU; niacin, 330 mg; calcium pantothenate, 160 mg; riboflavin, 80 mg; thiamine mononitrate, 50 mg; pyridoxine hydrochloride, 45 mg; menadione sodium bisulfate, 25 mg; folacin, 13 mg; biotin, 1 mg; vitamin B12, 30 μg.
USFWS no. 3. Amounts contributed (mg/kg of diet) were as follows: zinc, 37; manganese, 10; iodine, 5; copper, 1.
Rovimix Stay-C; 35%; DSM Nutritional Products.
Carophyl Pink; DSM Nutritional Products.
16S rRNA gene sequencing and analyses.
At 312 days posthatch (after 214 days under study treatment protocols), 3 fish/tank (2 or 3 tanks/treatment combination; 33 fish total) were randomly selected and euthanized with 200 mg/liter MS-222 (Western Chemical Inc., Ferndale, WA), and uniform 5-cm midintestine segments were carefully resected, flash frozen in liquid nitrogen, and stored at −80°C. Intestinal samples were shipped overnight on dry ice to the Core for Applied Genomics and Ecology, University of Nebraska (Lincoln, NE). Total genomic DNA was extracted from intestinal samples using Qiagen (Valencia, CA) Stool Kits. From the resulting DNA, the V1-V3 region of bacterial 16S rRNA genes was amplified using F8 and R518 primers tagged with the A and B Roche 454 Titanium sequencing adapters. The F8 primers were modified to contain an 8-base barcode unique to each sample (see Table S1 in the supplemental material). Pyrosequencing was performed by pooling all samples in a single region of a 2-region Titanium PicoTitre plate. Sequence data were filtered and analyzed with QIIME (31) using default parameters with the following exceptions: we removed sequences with ≥50 consecutive bases possessing an average quality score of <25 or with lengths of <150 or >1,000 bases. Sequences were then grouped by trout sample based on their barcodes; we used the QIIME denoiser algorithm (32) to denoise the sequences. The denoised sequences were binned by the UCLUST method into operational taxonomic units (OTUs) using a threshold of 97% or higher sequence identity. Representative sequences from each OTU were then aligned to the Greengenes core set (version gg_otus_4feb2011/taxonomies/greengenes_tax_rdp_train.txt) using PyNast (33). The representative sequences from each OTU were also taxonomically classified using the RDP Classifier program (34). Consensus lineages were assigned at each taxonomic level if ≥90% of the sequences in the OTU agreed with the classification. We also used the QIIME ChimeraSlayer algorithm to identify and exclude from subsequent analysis any OTUs with chimeric representative sequences. Additionally, OTUs assigned to the phylum Cyanobacteria were considered potential plant chloroplast contaminants and removed from the analysis. After the above filtering steps, a total of 185,216 high-quality bacterial 16S rRNA gene sequences remained for analysis. OTUs and their consensus lineages are tabulated in Table S4 in the supplemental material. To determine the relative abundance of each bacterial taxon, OTUs were binned according to their consensus lineages (see Table S3 in the supplemental material). To assess the degree of dissimilarity between the gut microbiotas of different samples, we conducted weighted and unweighted UniFrac analyses using 1,345 sequences from each sample. UniFrac distance matrices were graphically represented using principal-coordinates analysis (PCoA). Additionally, we calculated nonphylogenetic distances between samples by performing binary Jaccard analyses. To determine the bacterial diversity within individuals, we calculated Chao1, Shannon diversity index, and phylogenetic-distance values for each sample (see Table S2 in the supplemental material). LEfSe software (35) was used to identify discriminatory bacterial groups between conditions using sequences that had been taxonomically classified with RDP Classifier in QIIME. Taxa identified as discriminatory between two conditions were further subjected to two-way analysis of variance (ANOVA), followed by Bonferroni posttest using GraphPad Prism software. All analyses were performed using default parameters.
Fin quality assessments.
During the final sampling event, 25 fish from each tank were anesthetized and measured for fork length. Then, using digital microcalipers, the maximum length (i.e., the longest ray) of the following fins was measured to the nearest 0.1 mm: left and right pectoral, left and right pelvic, dorsal, ventral, and the top and bottom poles of the caudal fin. Fin indices (36) for all eight measured fins or fin components were then calculated by dividing their individual lengths by the fork length.
Fillet quality and contaminant analyses.
Fillet samples collected for processing attribute evaluation were sent to West Virginia University (Morgantown, WV) for the following assessments: cook yield, instrumental texture, proximate composition, and fatty acid profiles. Standard laboratory methods were used to determine the fillet cook yield and texture (37). Analyses of fillet moisture, fat, protein, and ash were performed according to AOAC-approved methods (38). Total lipids were extracted from muscle according to the method of Bligh and Dyer (39). Fatty acid analysis was performed on powdered muscle and minced visceral adipose tissue. Fatty acids were methylated using the method described by Fritshe and Johnston (40). Nonadecanoic acid (19:0) was used as an internal standard. Fatty acid methyl esters (FAMEs) were quantified using a Varian CP-3800 Gas Chromatograph (Varian Analytical Instruments, Walnut Creek, CA, USA) equipped with a flame ionization detector. FAMEs were identified based on comparison to retention times of standard FAMEs (Supelco quantitative standard FAME 37; Sigma-Aldrich, St. Louis, MO, USA). Peak area counts were computed by an integrator using the Star GC workstation version 6 software (Varian Inc.) and reported as percent fatty acid.
To determine pesticide and polychlorinated biphenyl (PCB) levels, at study's end, 3 fish were randomly selected from each of the 6 high-density tanks, euthanized with MS-222, and filleted. These 18 fillet samples were sent fresh on ice to Northeast Analytical Inc. (Schenectady, NY), where they were processed, homogenized, and analyzed. The Soxhlet extraction method (EPA Method 3540C) was employed for all fillet samples; analysis for organochlorine pesticides was performed by EPA Method 8081. Analysis for PCB congeners was performed by Comprehensive Quantitative Congener Specific PCB Method (Northeast Analytical Inc. Standard Operating Procedure NE133_02); a total of 209 PCB congeners were quantified. The pesticides quantified included aldrin, alpha-chlordane, alpha-benzene hexachloride (BHC), beta-BHC, chlordane, delta-BHC, dieldrin, endosulfan I, endosulfan II, endosulfan sulfate, endrin, endrin aldehyde, endrin ketone, gammachlordane, gamma-BHC, heptachlor, heptachlor epoxide, hexachlorobenzene, methoxychlor, p,p′-DDD (dichlorodiphenyldichloroethane), p,p′-DDE (dichlorodiphenyldichloroethylene), p,p′-DDT (dichlorodiphenyltrichloroethane), and toxaphene.
Histopathology evaluations.
At study's end, the 5 fish per tank randomly selected for processing attribute assessment also had standardized 3-cm sections of the posterior intestine removed and fixed in 10% neutral buffered formalin (3.7% formaldehyde). The fixed samples were sent to the Washington Animal Disease Diagnostic Laboratory (Pullman, WA) for histopathology evaluation. A 0- to 5-point grading scale was developed to quantify the extent and severity of intestinal inflammation, with 0 representing normal healthy tissue and 5 denoting severe inflammation with loss of mucosal integrity across most or all of the tissue evaluated. All animals displayed at least minimal inflammation, and severe inflammation (i.e., a score of 4) was observed in only one fish from the low-density, fishmeal diet group.
Statistical analysis.
Measurements of final fish performance, health, and yield were assessed for treatment effects using multivariable ANOVA, with diet, density, and diet-density interaction as independent variables. Contaminant data were analyzed with ANOVA for diet effects only. An alpha level of 0.05 was used to determine statistical significance. Relative abundances of bacterial taxa were considered significant by LEfSe (35) analysis if the Kruskal-Wallis test yielded an alpha value of <0.05, the pairwise Wilcoxon test yielded an alpha value of <0.05, and the logarithmic LDA effect score reached 2.0. LEfSe results were confirmed if two-way ANOVA yielded a P value of <0.05. Relative abundances of core OTUs were normalized by log10 transformation (41) prior to determining statistical significance using pairwise Student's t tests and a 5% false-discovery rate.
Nucleotide sequence accession number.
The 16S rRNA gene sequence data have been submitted to MG-RAST under accession number 4509015.3 (http://metagenomics.anl.gov/linkin.cgi?metagenome=4509015.3).
RESULTS
Rainbow trout intestines possess a large core microbiota that persists following long-term alteration in rearing density and diet.
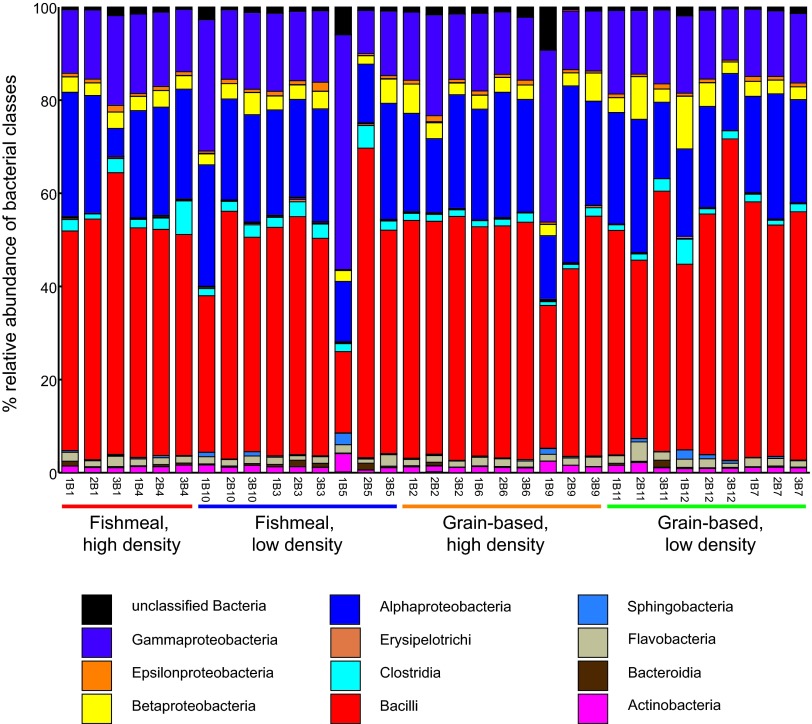
We sought to define the effects of diet composition and rearing density on the rainbow trout intestinal microbiota through 16S rRNA gene sequencing. Fish were raised together under identical conditions and fed a fishmeal-based diet until they averaged approximately 10 g, and then they were randomly distributed to flowthrough tanks and reared at high or low density and fed either fishmeal- or grain-based diets for 214 days. Genomic DNA was extracted from the midintestines of these animals, and their respective bacterial communities were evaluated using 454 pyrosequencing of the V1-V3 region of 16S rRNA genes (3 fish/tank; 2 or 3 tanks/condition; 5,612 ± 2,671 sequences/fish). We binned the resulting 185,216 16S rRNA gene sequences into 3,376 OTUs defined by 97% pairwise sequence identity and then classified the taxonomy of each OTU. We found that the relative diversities (see Table S2 in the supplemental material) and abundances of bacterial classes (Fig. 1) in the intestine were strikingly similar in most individuals across different diet and rearing density conditions. All bacterial communities were dominated by the classes Bacilli (48.6% ± 9.3% of sequences per sample), Alphaproteobacteria (21.8% ± 5.8%), Gammaproteobacteria (17.1% ± 7.6%), Betaproteobacteria (3.8% ± 2.0%), and Clostridia (2.2% ± 1.3%). This strong similarity among all samples at the class level raised the possibility that the rainbow trout intestines harbored a shared set of OTUs or a core gut microbiota.
Fig 1.
16S rRNA gene sequences reveal similarities between intestinal microbiotas of rainbow trout raised under different diet and density conditions. Shown are relative abundances of bacterial classes in each sample. The labels under each column are sample names corresponding to individual fish. Numerical suffixes indicate the tank number, numerical prefixes identify biological replicates drawn from a given tank, and the letter B acts as a delimiter to separate the tank number from the fish number. The legend includes only taxa constituting 0.005% or more of at least one sample. For each treatment condition, samples were taken from 2 or 3 different tanks, and 3 biological samples were analyzed per tank.
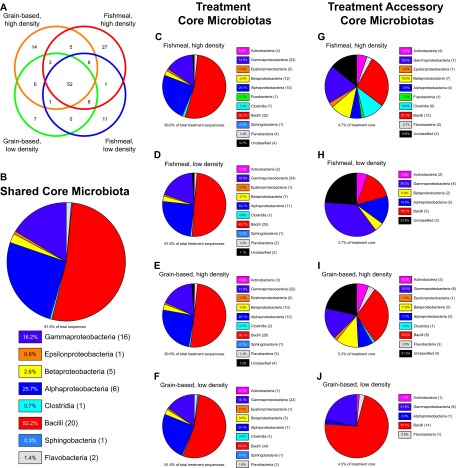
To determine the extent to which OTUs were shared across individuals and treatment groups, we first identified the OTUs present in every individual within a given treatment group (operationally defined here as a “treatment core”) and then evaluated the overlap between different treatment cores to identify those OTUs shared among all sequenced individuals (operationally defined here as the “shared core”) (Fig. 2A). Surprisingly, we found that the majority of OTUs within each treatment core was shared among all four conditions, yielding a shared core of 52 OTUs. This large shared core contained greater than half of the OTUs that appear in each treatment core (Fig. 2A) and constituted 81.6% of all sequences in this study (Fig. 2B). In agreement with the overall abundances of bacterial taxa (Fig. 1), we found that the shared core is composed primarily of the bacterial classes Bacilli, Alphaproteobacteria, and Gammaproteobacteria (Fig. 2B). As expected from the similarity in overall bacterial composition between samples (Fig. 1), treatment cores were not markedly different from each other or from the shared core (Fig. 2C to F; see Fig. S1 in the supplemental material). Additionally, sequences in OTUs within the shared core constituted 81.8% to 89.8% of all sequences in each treatment group (Fig. 2C to F). These results indicate that the tested variations in diet and rearing density did not exert large, long-term alterations on the gut microbiota of rainbow trout.
Fig 2.
Core microbiotas shared between rainbow trout raised under different diet and density conditions. (A) Numbers of OTUs (97% sequence identity) shared by all individuals within each specific treatment condition (treatment cores) and within all treatment conditions (shared core). The numbers indicate the OTUs shared by overlapping circles. Of the 3,376 OTUs identified in this study, 52 comprised the shared core, and an additional 87 OTUs were included in one or more treatment cores. (B) Composition of the shared core microbiota (the 52 OTUs present in all individuals) for all treatment conditions. The relative abundances of the bacterial classes present are shown in the chart legend; the numbers in parentheses following the legend labels denote the number of OTUs in the core microbiota belonging to the corresponding bacterial class. (C to F) Compositions of the treatment core microbiotas for each of the four treatment groups. The relative abundances of the bacterial classes present are shown in the chart legend; the numbers in parentheses following the legend labels denote the number of OTUs in the core microbiota belonging to the corresponding bacterial class. The text below the pie charts shows the contribution of the core microbiota to the entire microbiota of trout under each treatment condition. (G to J) Compositions of the accessory core microbiotas for each treatment condition (i.e., OTUs present in each individual under a given treatment condition but not in each individual under all treatment conditions). The relative abundances of the bacterial classes present are shown in the chart legend; the numbers in parentheses following the legend labels denote the number of OTUs in the core microbiota belonging to the corresponding bacterial class. The text below the pie charts shows the contribution of the accessory core microbiota to the treatment core microbiota of trout under each treatment condition. (See Tables S5 to S13 in the supplemental material.)
Variations in diet and rearing density cause minor changes to the rainbow trout gut bacterial community.
We next sought to determine whether variations in diet and rearing density evoked any consistent alterations in gut bacterial community composition. Although treatment cores were highly similar to each other and to the shared core (Fig. 2B to F), we did identify OTUs within each treatment core that were not observed in the shared core (operationally defined here as the “treatment accessory cores”) (Fig. 2G to J). OTUs within each treatment accessory core constituted a small fraction of the sequences within their respective treatment cores (3.7% to 5.3%), but comparison of treatment accessory cores revealed distinct differences between diet and rearing density treatments. Although the relative abundances of bacterial classes were similar in both high-density accessory cores, we observed a relative increase in Clostridia abundance and diversity in the accessory core of the fishmeal high-density treatment (Fig. 2G and I; see Fig. S1 in the supplemental material). In contrast, we observed pronounced differences between the two low-density accessory cores compared to each other and to the high-density accessory cores. For example, the grain-based low-density accessory core displayed marked increases in the abundance and diversity of the class Bacilli compared to other accessory cores (Fig. 2J; see Fig. S1 in the supplemental material). These results suggest that the tested variations in diet and rearing density are sufficient to induce specific alterations in the diversity and proportional abundance of relatively rare members of the gut microbiota.
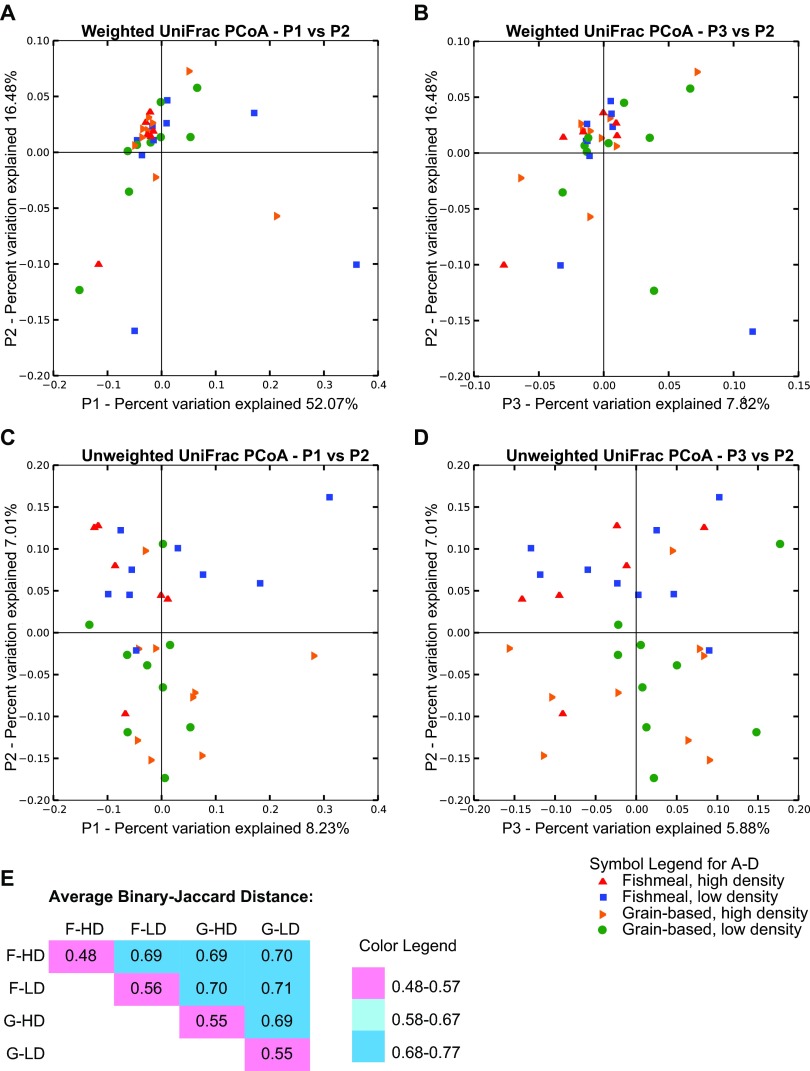
We next sought to determine whether the different diet-by-rearing-density treatments were sufficient to evoke alterations in the overall composition of gut bacterial communities. To do so, we compared diversities between samples from different treatment groups (i.e., beta diversity). PCoA of weighted UniFrac distances (an evaluation of community structure) showed that samples clustered together, regardless of diet or rearing density (Fig. 3A and B) and consistent with our observation of a large shared core microbiota. In contrast, PCoA of unweighted UniFrac distances (an evaluation of community membership that does not consider abundances) showed slight clustering of samples from the same treatment group (Fig. 3C and D). In accord with this, binary Jaccard analysis (a nonphylogenetic measure of community similarity) revealed that microbial communities from individual samples within the same treatment group were more similar to each other than to those from other treatment groups (Fig. 3E).
Fig 3.
Beta diversity estimates of the rainbow trout intestinal microbiota. (A to D) Use of UniFrac to measure phylogenetic distances between the gut microbiotas of individual trout from all treatment groups. (A and B) Weighted UniFrac PCoA plotted against the PC1 versus PC2 axes (A) and the PC2 versus PC3 axes (B). (C and D) Unweighted UniFrac PCoA plotted against the PC1 versus PC2 axes (C) and the PC2 versus PC3 axes (D). (E) Average binary Jaccard (nonphylogenetic) distances between the gut microbiotas of individuals in the same treatment group and between individuals from different treatment groups. F-HD, fishmeal, high density; F-LD, fishmeal, low density; G-HD, grain based, high density; G-LD, grain based, low density.
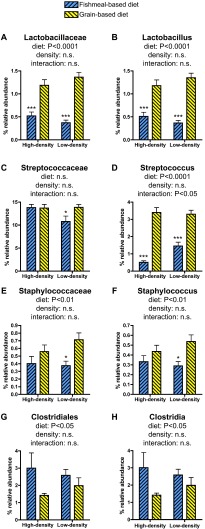
We next determined whether the similarity of gut bacterial communities within each treatment group is associated with differential abundances of specific bacterial taxa using LEfSe software (35), followed by 2-way ANOVA. This analysis identified several taxa within the phylum Firmicutes that were significantly discriminatory for diet type. The relative abundance of the family Lactobacillaceae and its included genus Lactobacillus were significantly enriched in fish fed a grain-based diet under both density conditions (Fig. 4A and B). Although there was no significant effect of diet on the relative abundance of the family Streptococcaceae, the included genus Streptococcus was enriched in fish fed a grain-based diet and was the only taxon to display a significant interaction between diet and density conditions (Fig. 4C and D). The relative abundances of the family Staphylococcaceae and its included genus Staphylococcus were significantly enriched in fish fed a grain-based diet, with the major effect being observed under low-density conditions (Fig. 4E and F). In contrast, the relative abundances of the family Clostridiales and its included genus Clostridia were significantly affected by diet, with a trend toward increased relative abundance in fishmeal-fed animals (Fig. 4G and H). Together, these results indicate that the tested diet and rearing-density combinations caused consistent alterations in a limited number of bacterial community members and that differences between treatments were sufficient to create treatment-specific bacterial community profiles in these animals.
Fig 4.
Bacterial taxa identified as discriminatory between experimental conditions. Bacterial taxa identified by LEfSe as discriminatory between experimental conditions were subjected to 2-way ANOVA and Bonferroni posttests. The taxa that were confirmed as significant by 2-way ANOVA are shown (with the exception of panel C). The data are plotted as mean percent relative abundance ± standard error of the mean (SEM), with the ANOVA P value summary for diet, density, and interaction between diet and density shown above each graph. The asterisks indicate significant differences between low-density and high-density samples under the same diet conditions identified by Bonferroni posttest (*, P < 0.05; ***, P > 0.001). (See Tables S14 to S19 in the supplemental material.)
Long-term alteration in diet and rearing density do not impact intestinal histopathology.
Because grain-based diets have previously been associated with intestinal inflammation in fish (42, 43), we next sought to determine if alterations in diet and rearing density were sufficient to alter intestinal histopathology. All animals displayed at least a minimal level of intestinal inflammation, but intestinal inflammation was not affected by treatment (P > 0.05) (see Table S20 in the supplemental material). These data suggest that the tested diets and rearing densities were not sufficient to significantly alter the severity of intestinal inflammation.
Rainbow trout performance, survival, and fin condition are significantly affected by diet and rearing density.
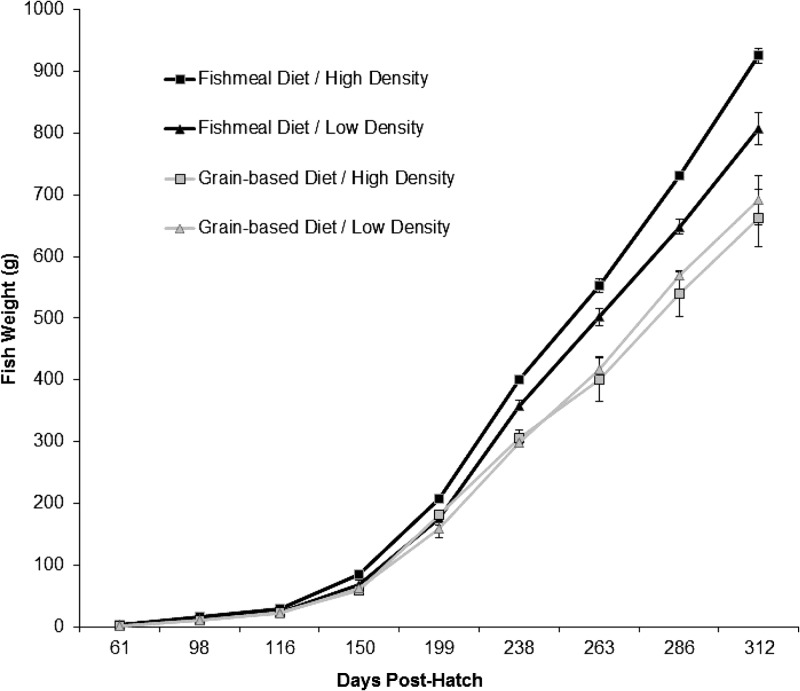
Additionally, we determined if variations in diet and rearing conditions impacted fish performance and health. By study's end, statistically significant differences in fish weights were detected, with higher weights being observed in fishmeal diet groups relative to grain-based-diet groups. Rearing density, however, did not significantly affect the final fish weight (Table 2 and Fig. 5). Despite lower final weights, however, fish fed grain-based diets were better able to utilize dietary energy for growth, as indicated by the significantly greater feed efficiency (i.e., lower feed conversion rates) in these groups (Table 2). Again, no significant association between density and feed conversion was determined. Survival was generally high (>96%) among all treatment groups; however, significantly higher survival was observed in grain-based-diet treatment groups (Table 2).
Table 2.
Fish performance and processing and fillet quality attributes for each diet/density treatment group at study's end
| Parameter | Value (mean ± standard error) |
|||
|---|---|---|---|---|
| Fishmeal diet |
Grain-based diet |
|||
| High density | Low density | High density | Low density | |
| Fish performance | ||||
| Final wt (g)a | 925 ± 12 | 807 ± 26 | 663 ± 40 | 691 ± 46 |
| Survival (%)a | 96.4 ± 0.5 | 97.1 ± 0.7 | 97.9 ± 0.2 | 97.6 ± 0.5 |
| FCR (overall)a | 1.18 ± 0.02 | 1.15 ± 0.03 | 1.02 ± 0.06 | 1.10 ± 0.01 |
| TGC (overall) | 2.46 ± 0.04 | 2.45 ± 0.04 | 2.42 ± 0.07 | 2.33 ± 0.08 |
| Processing attributes | ||||
| Dress yield (%)a | 86.1 ± 0.4 | 85.4 ± 0.5 | 87.7 ± 0.4 | 88.9 ± 0.4 |
| Fillet index (%) | 49.8 ± 0.5 | 49.7 ± 0.6 | 50.3 ± 0.3 | 50.8 ± 0.5 |
| Fillet attributes | ||||
| Cook yield (%) | 84.3 ± 0.6 | 84.8 ± 0.5 | 85.0 ± 0.4 | 84.4 ± 0.5 |
| Texture (Kramer g/g wt)b | 340 ± 20 | 334 ± 18 | 316 ± 11 | 344 ± 11 |
| Proximate analysis | ||||
| Moisture (%) | 70.6 ± 0.3 | 70.4 ± 0.2 | 70.4 ± 0.2 | 70.8 ± 0.3 |
| Fat (%) | 8.8 ± 0.4 | 8.7 ± 0.3 | 9.1 0±.3 | 8.8 ± 0.4 |
| Protein (%)a | 20.2 ± 0.2 | 20.4 ± 0.1 | 20.6 ± 0.1 | 20.7 ± 0.1 |
| Ash (%) | 1.2 ± 0.0 | 1.3 ± 0.0 | 1.3 ± 0.0 | 1.3 ± 0.0 |
| Fatty acids (mg/g tissue) | ||||
| Omega-3 | ||||
| ALA (C18:3n3) | 0.75 ± 0.23 | 0.84 ± 0.19 | 0.86 ± 0.27 | 1.18 ± 0.04 |
| EPA (C20:5n3)a | 3.41 ± 0.13 | 3.05 ± 0.17 | 2.10 ± 0.09 | 2.19 ± 0.07 |
| DHA (C22:6n3)a | 11.3 ± 0.65 | 10.3 ± 0.44 | 8.92 ± 0.23 | 10.0 ± 0.77 |
| Total omega-3a | 15.5 ± 0.68 | 14.1 ± 0.34 | 11.9 ± 0.31 | 13.8 ± 0.57 |
| Omega-6 | ||||
| DGLA (C20:3n6) | 1.13 ± 0.17 | 1.01 ± 0.26 | 1.00 ± 0.24 | 1.13 ± 0.16 |
| Eicosadienoic acid (C20:2n6)a | 9.65 ± 0.24 | 9.55 ± 0.20 | 13.2 ± 0.24 | 13.7 ± 0.14 |
| Total omega-6a | 10.8 ± 0.13 | 10.6 ± 0.25 | 14.2 ± 0.16 | 14.8 ± 0.23 |
Parameter showed a statistically significant difference between diet treatment groups using ANOVA (P < 0.05); no statistical differences were determined between density treatment groups, and no statistical interactions between treatments were detected.
Kramer shear force is a measure of sample firmness. Samples are sheared by blades and force-deformation curves, from which the Kramer values are derived, are obtained.
Fig 5.
Average rainbow trout weights for the duration of the experiment. The data points represent means of 20 to 60 trout sampled at each monthly growth performance assessment up to 312 days posthatch. The error bars represent standard errors.
Because fin erosion is an established indicator of fish welfare under culture conditions (44, 45), we measured fin indices (i.e., the length of the longest ray of each rayed fin relative to the fork length) as a means of evaluating fish welfare. Although no major fin erosion was noted qualitatively on any of the sampled fish, fin indices were significantly higher in grain-based-diet treatment groups for all measured fins (Table 3), indicating healthier fins overall in these groups. Statistical interaction between diet and density was observed when modeling these main effects and their associations with indices for the pectoral (left and right), dorsal, and pelvic (left and right) fins. In these cases, the overall trend was an increase in the fin index when fish were fed grain-based diets but a decrease in the fin index associated with the increased-density treatment. Together, these data show that the tested alterations in diet and rearing density were sufficient to independently and interactively modify rainbow trout health and performance.
Table 3.
Indices of rainbow trout fins in each treatment group
| Fin | Index (mean ± standard error) |
|||
|---|---|---|---|---|
| Fishmeal diet |
Grain-based diet |
|||
| High density | Low density | High density | Low density | |
| Pectoral (left)a,b | 0.104 ± 0.002 | 0.108 ± 0.001 | 0.114 ± 0.001 | 0.117 ± 0.002 |
| Pectoral (right)a,b | 0.106 ± 0.001 | 0.107 ± 0.001 | 0.115 ± 0.001 | 0.118 ± 0.002 |
| Dorsala,b | 0.088 ± 0.003 | 0.088 ± 0.005 | 0.091 ± 0.003 | 0.095 ± 0.004 |
| Pelvic (left)a,b | 0.088 ± 0.001 | 0.088 ± 0.001 | 0.090 ± 0.003 | 0.096 ± 0.003 |
| Pelvic (right)a,b | 0.097 ± 0.001 | 0.097 ± 0.001 | 0.102 ± 0.001 | 0.104 ± 0.001 |
| Ventrala | 0.097 ± 0.001 | 0.099 ± 0.002 | 0.102 ± 0.001 | 0.104 0± 0. 02 |
| Caudal (upper)a | 0.105 ± 0.001 | 0.109 ± 0.002 | 0.109 ± 0.001 | 0.113 ± 0.001 |
| Caudal (lower)a | 0.103 ± 0.002 | 0.105 ± 0.001 | 0.109 ± 0.001 | 0.112 ± 0.002 |
Fin showed statistically significant differences between diet treatment groups (P < 0.05); no statistical differences were determined between the density treatment groups.
Fin showed statistically significant interaction (P < 0.05) between diet and density treatments.
Rainbow trout diet and rearing density alter processing attributes and product quality.
Fish from the grain-based-diet treatment groups had significantly greater dress yield than fish from the fishmeal diet groups (Table 2). There was a small but statistically significantly higher percentage of protein in fillets from fish fed a grain-based diet, and these fish also contained significantly higher fillet levels of eicosadienoic acid and total omega-6 fatty acids. However, fish fed fishmeal-based feed had fillets with significantly higher levels of EPA, DHA, and total omega-3 fatty acids. No statistical differences were noted between treatment groups for fillet contaminants (see Table S21 in the supplemental material). Among all pesticides examined, only DDE and PCBs were detected. Levels of DDE and total PCBs in both treatment groups were very low, and as measured, would be of little or no concern to human health (the maximum DDE levels detected were >750 times lower than FDA limits [5 ppm] for the edible portions of fish; PCB levels were >250 times lower than FDA limits [2 ppm] for food fish). No density effects (P < 0.05) were noted for any of the processing and product quality parameters investigated (Table 2). These results indicate that the tested variations in diet, not rearing density, had a marked impact on yield and nutrient content.
DISCUSSION
Diet composition and rearing density have been identified as environmental factors that can impact the health and physiology of rainbow trout. Furthermore, the diet type is known to impact the composition of the intestinal microbiota in a variety of animal species. The study reported here is the first to test whether diet and rearing density interact in rainbow trout to impact the gut microbiota composition, health, and fish performance metrics. Our results reveal consistent effects of diet composition on fish growth and product quality and novel interactions between diet and rearing density on fish welfare. Despite these marked changes in fish health and yield, the tested alterations in diet and rearing density were not sufficient to significantly alter an unexpectedly large core microbiota in the intestines of aquacultured rainbow trout. As discussed below, these results have important implications for aquaculture of rainbow trout and other finfish, as well as for our understanding of vertebrate gut microbial ecology.
Characterization of the microbial lineages ubiquitous in any habitat is an important step toward understanding the determinants of microbiota membership and the respective roles of its members and for developing effective approaches for managing and manipulating that microbial ecosystem. Deep sequencing of 16S rRNA genes from the intestines of humans, mice, and zebrafish sampled from different populations and geographic locations have suggested that very few bacterial OTUs are common among all individuals from a given host species and that they represent a minor portion of the overall community membership (20, 28, 46). In contrast, we found that all of the individual aquacultured rainbow trout analyzed in this study possessed very similar intestinal bacterial communities dominated by a large shared core microbiota comprised of 52 OTUs. Moreover, the relative abundances of most of these shared OTUs were largely unaffected by tested alterations in diet or rearing density. Since the sequencing depth of this study was not sufficient to saturate diversity in any sample (see Fig. S2 in the supplemental material), the size of this shared core microbiota may be even larger than our data indicate.
The factors underlying the large size of this shared core gut microbiota remain unknown and represent an important subject for future research. The aquacultured trout studied here were raised under identical husbandry conditions prior to the start of the experimental manipulations, and it is possible that early colonization events are strong determinants of bacterial community composition, greatly dampening the impact of the experimental manipulations. It is also possible that the large core microbiota might be due to rearing these animals in flowthrough tanks without water recirculation, likely limiting environmental variation among tanks and individuals during the experimental manipulations. Importantly, the aquacultured trout analyzed here were obtained from a single commercial supplier and raised in a single aquaculture facility, thereby limiting the environmental and host genetic variation and increasing the likelihood of similar microbiota membership. Previous studies have suggested that gut microbiota compositions can vary markedly among domesticated zebrafish and mice from different vivarium facilities (28, 47–49). We therefore expect that comparisons of gut microbiota from rainbow trout obtained from different aquaculture facilities or caught in the wild would reveal a smaller shared core microbiota than that reported here. Previous evaluations of gut microbiota composition in wild rainbow trout identified many of the bacterial genera that we observed in the shared core in this study (e.g., Aeromonas, Acinetobacter, Escherichia, Pseudomonas, Streptococcus, and Enterococcus) (50). These and many other genera observed within the shared core reported here have also been identified in culture-independent and culture-based evaluations of gut microbiota composition in aquacultured trout and other salmonids (9, 22, 30, 52–57). However, these previous reports did not identify these genera in all animals within the respective studies. This could be due at least in part to the limited sampling depths and the inherent limitations of the respective culture-based and culture-independent methods utilized in these studies.
To provide a more robust frame of reference for interpreting our observations, we compared our results with the only other published study that used deep sequencing to evaluate gut microbiota composition in aquacultured rainbow trout (30). This previous study by Desai and colleagues differed from ours in several ways, including the specific intestinal region analyzed (luminal contents of the distal intestine versus the whole midintestine in our study), the bacterial gene targeted for deep sequencing (cpn60 versus 16S rRNA gene in our study), the source of tank water (recirculating versus flowthrough in our study), and other aspects of animal husbandry. We detected no bacterial species or genera that were present in all animals across both studies; however, many of the genera within the shared core that we report here were frequently detected in the animals analyzed by Desai and colleagues. Of the 52 OTUs that comprise the shared core in our study, 44 were confidently assigned to one of 26 genera by RDP Classifier. Of those 26 genera, 8 (Weissella, Acidovorax, Citrobacter, Aeromonas, Enterococcus, Lactococcus, Pseudomonas, and Klebsiella) were observed by Desai and colleagues in at least half of the trout sampled in their study, and they observed an additional 10 genera (Erwinia, Leuconostoc, Escherichia/Shigella, Streptococcus, Veillonella, Acinetobacter, Bacillus, Sphingomonas, Chryseobacterium, and Pantoea) in at least one animal (30). This suggests that the shared core microbiota observed in our study is not a “true” core microbiota possessed by all rainbow trout. Although we have operationally defined the core microbiota as those OTUs possessed by 100% of the samples in a group, others have suggested that the criteria for core microbiota can be relaxed to include OTUs present in less than 100% of samples or deeper taxonomic levels (21, 58). Using this relaxed definition, the frequent detection of several bacterial genera in rainbow trout from diverse populations and locations suggests that these genera may be members of a true core microbiota shared by many or all aquacultured rainbow trout. Additional studies are needed to directly compare individual animals from different aquaculture facilities and wild fish to determine whether a true core microbiota exists in rainbow trout and to determine how variations in husbandry techniques and animal provenance impact the composition of the gut microbiota in rainbow trout.
Despite the dominance of the shared core we observed, analysis of the accessory core microbiotas—the set of OTUs present in all individuals in at least one experimental group but not in the shared core—revealed several significant differences between experimental conditions. For example, fish fed a grain-based diet were enriched for the genera Lactobacillus and Streptococcus compared to those fed a fishmeal-based diet. The relative abundance of the genus Streptococcus was the only one in the study to display a significant statistical interaction between diet and density, where the effect of diet was greater in fish raised at high density. Moreover, the relative abundance of one Streptococcus OTU—the only OTU in the shared core microbiota with a statistically significant variation among treatment groups—was increased in both groups fed the grain-based diet (see Fig. S3 in the supplemental material). The genera Lactobacillus and Streptococcus contain species that are used as probiotics in mammals and fish (58, 59). These diet-dependent differences in the gut microbial community structure raise the possibility that minority members may contribute to the physiological differences, such as growth rate, that we observed between fish raised on the fishmeal-based versus the grain-based diet.
Early studies report an association between alternative protein diets and decreased fish growth (60–62), likely in response to intestinal inflammation brought about by dietary antinutritional factors (38, 60). Subsequent research has demonstrated improved performance with newer plant-based diet formulations (7, 10, 63) with reduced antinutritional factors (64). We did not detect differences in intestinal inflammation between treatment groups, and furthermore, the grain-based-feed treatment groups, despite slower growth, had better feed conversion than groups fed a fishmeal-based diet. Fin condition is an indicator of fish welfare (65), but its etiology is a complex, multifactorial process (66). We found that fin indices were significantly better in the grain-based-diet treatment groups. Barrows and Lellis (67) suggest an association between fin health and elements within the protein and/or mineral fraction(s) of diets; however, in our study, it is difficult to identify specific dietary components' impacts on fin condition. Lower fin indices were found in the high-density treatment groups, as has been previously noted by others (41, 68–70), underscoring the importance of maintaining an appropriate density range.
In summary, we find that variations in rearing density and diet composition within the context of a single aquaculture facility are sufficient to interactively alter rainbow trout growth, performance, fillet quality, and welfare. However, these tested variations in rainbow trout husbandry had only minor effects on the gut microbiota composition and did not markedly alter a surprisingly large core microbiota shared among all animals in the study. Although the shared core microbiota we observed in this cohort of aquacultured rainbow trout may not be a “true” core microbiota shared among all aquacultured or wild rainbow trout, our results do reveal that rainbow trout gut microbiota composition can achieve remarkable consistency within the context of a single aquaculture facility. This should encourage additional research and implementation of alternative diets and husbandry practices for trout production by reducing concerns over the potential impact on the structure and function of the gut microbiota.
Supplementary Material
ACKNOWLEDGMENTS
We are grateful to the Core for Applied Genomics and Ecology at the University of Nebraska—Lincoln for 454 pyrosequencing services. We are grateful to Ian Carroll and the Microbiome Core Facility at the University of North Carolina—Chapel Hill for bioinformatics assistance.
This research was supported by the USDA Agricultural Research Service under Agreement No. 59-1930-5-510 and National Institutes of Health grants R01 DK081426, R01 GM095385, and P30 DK34987.
The use of trade names does not imply endorsement by the U.S. government.
Footnotes
Published ahead of print 14 June 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.00924-13.
REFERENCES
- 1. FAO 2010. The state of world fisheries and aquaculture 2010. FAO Fisheries and Aquaculture Department, Rome, Italy [Google Scholar]
- 2. Barrows FT, Hardy RW. 2001. Nutrition and feeding. p 483–558 In Wedemeyer GA. (ed), Fish hatchery management, 2nd ed John Wiley & Sons, New York, NY [Google Scholar]
- 3. Gatlin DM, III, Barrows FT, Brown P, Dabrowski K, Gaylord TG, Hardy RW, Herman E, Hu G, Krogdahl Å, Nelson R, Overturf K, Rust M, Sealey W, Skonberg D, Souza EJ, Stone D, Wilson R, Wurtele E. 2007. Expanding the utilization of sustainable plant products in aquafeeds: a review. Aquac. Res. 38:551–579 [Google Scholar]
- 4. Gaylord TG, Barrows FT, Teague AM, Johansen KA, Overturf KE, Shepherd B. 2007. Supplementation of taurine and methionine to all-plant protein diets. Aquaculture 269:514–524 [Google Scholar]
- 5. Stickney RR, Hardy RW, Koch K, Harrold R, Seawright D, Massee KC. 1996. The effects of substituting selected oilseed protein concentrates for fish meal in rainbow trout, Oncorhynchus mykiss diets. J. World Aquac. Soc. 27:57–63 [Google Scholar]
- 6. Gaylord TG, Barrows FT. 2008. Apparent digestibility of gross nutrients from feedstuffs in extruded feeds for rainbow trout Oncorhynchus mykiss. J. World Aquac. Soc. 39:827–834 [Google Scholar]
- 7. Davidson J, Good C, Barrows FT, Welsh C, Kenney PB, Summerfelt ST. 2013. Comparing the effects of feeding a grain- or a fish meal-based diet on water quality, waste production, and rainbow trout Oncorhynchus mykiss performance within low exchange water recirculating aquaculture systems. Aquac. Eng. 52:45–57 [Google Scholar]
- 8. Krogdahl Å, Bakke-McKellep AM, Baeverfjord G. 2003. Effects of graded levels of standard soybean meal on intestinal structure, mucosal enzyme activities, and pancreatic response in Atlantic salmon (Salmo salar L.). Aquac. Nutr. 9:361–371 [Google Scholar]
- 9. Mansfield GS, Desai AR, Nilson SA, Van Kessel AG, Drew MD, Hill JE. 2010. Characterization of rainbow trout (Oncorhynchus mykiss) intestinal microbiota and inflammatory marker gene expression in a recirculating aquaculture system. Aquaculture 307:95–104 [Google Scholar]
- 10. Barrows F, Gaylord TG, Stone DJ, Smith CE. 2007. Effect of protein source and nutrient density on growth efficiency of rainbow trout (Oncorhynchus mykiss). Aquac. Res. 38:1747–1758 [Google Scholar]
- 11. Ellis T, North B, Scott AP, Bromage NR, Porter M, Gadd D. 2002. Review paper: the relationships between stocking density and welfare in farmed rainbow trout. J. Fish. Biol. 61:493–531 [Google Scholar]
- 12. Rawls JF, Mahowald MA, Ley RE, Gordon JI. 2006. Reciprocal gut microbiota transplants from zebrafish and mice to germ-free recipients reveal host habitat selection. Cell 127:423–433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Turnbaugh PJ, Ley RE, Mahowald MA, Magrini V, Mardis ER, Gordon JI. 2006. An obesity-associated gut microbiome with increased capacity for energy harvest. Nature 444:1027–1031 [DOI] [PubMed] [Google Scholar]
- 14. Turnbaugh PJ, Backhed F, Fulton L, Gordon JI. 2008. Diet-induced obesity is linked to marked but reversible alterations in the mouse distal gut microbiome. Cell Host Microbe 3:213–223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Vijay-Kumar M, Aitken JD, Carvalho FA, Cullender TC, Mwangi S, Srinivasan S, Sitaraman SV, Knight R, Ley RE, Gewirtz AT. 2010. Metabolic syndrome and altered gut microbiota in mice lacking Toll-like receptor 5. Science 328:228–231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Nayak SK. 2010. Role of gastrointestinal microbiota in fish. Aquac. Res. 41:1553–1573 [Google Scholar]
- 17. Heijtz RD, Wang S, Anuar F, Qian Y, Bjorkholm B, Samuelsson A, Hibberd ML, Forssberg H, Pettersson S. 2011. Normal gut microbiota modulates brain development and behavior. Proc. Natl. Acad. Sci. U. S. A. 108:3047–3052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Archie EA, Theis KR. 2011. Animal behavior meets microbial ecology. Anim. Behav. 82:425–436 [Google Scholar]
- 19. Semova I, Carten JD, Stombaugh J, Mackey LC, Knight R, Farber SA, Rawls JF. 2012. Microbiota regulate intestinal absorption and metabolism of fatty acids in the zebrafish. Cell Host Microb. 12:277–288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Shade A, Handelsman J. 2012. Beyond the Venn diagram: the hunt for a core microbiome. Environ. Microbiol. 14:4–12 [DOI] [PubMed] [Google Scholar]
- 21. Li K, Bihan M, Methe BA. 2013. Analyses of the stability and core taxonomic memberships of the human microbiome. PLoS One 8:e63139. 10.1371/journal.pone.0063139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Ringo E, Strom E, Tabachek J-A. 1995. Intestinal microflora of salmonids: a review. Aquac. Res. 26:773–789 [Google Scholar]
- 23. Kim DH, Brunt J, Austin B. 2007. Microbial diversity of intestinal contents and mucus in rainbow trout (Oncorhyncus mykiss). J. Appl. Microbiol. 102:1654–1664 [DOI] [PubMed] [Google Scholar]
- 24. Dimitroglou A, Merrifield DL, Moate R, Davies SJ, Spring P, Sweetman J, Bradley G. 2009. Dietary mannan oligosaccharide supplementation modulates intestinal microbial ecology and improves gut morphology of rainbow trout, Oncorhyncus mykiss (Walbaum). J. Anim. Sci. 87:3226–3234 [DOI] [PubMed] [Google Scholar]
- 25. Huber I, Spanggaard B, Appel KF, Rossen L, Nielsen T, Gram L. 2004. Phylogenetic analysis and in situ identification of the intestinal microbial community of rainbow trout (Oncorhyncus mykiss, Walbaum). J. Appl. Microbiol. 96:117–132 [DOI] [PubMed] [Google Scholar]
- 26. Navarrete P, Magne F, Mardones P, Riveros M, Opazo R, Suau A, Pochart P, Romero J. 2010. Molecular analysis of intestinal microbiota of rainbow trout (Oncorhyncus mykiss). FEMS Microbiol. Ecol. 71:148–156 [DOI] [PubMed] [Google Scholar]
- 27. Navarrete P, Magne F, Araneda C, Fuentes P, Barros L, Opazo R, Espejo R, Romero J. 2012. PCR-TTGE analysis of 16S rRNA from rainbow trout (Oncorhyncus mykiss) gut microbiota reveals host-specific communities of active bacteria. PLoS One 7:e31335. 10.1371/journal.pone.0031335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Roeselers G, Mittge EK, Stephens WZ, Parichy DM, Cavanaugh CM, Guillemin K, Rawls JF. 2011. Evidence for a core microbiota in the zebrafish. ISME J. 5:1595–1608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Sullam KE, Essinger SD, Lozupone CA, O'Connor MP, Rosen GL, Knight R, Kilham SS, Russell JA. 2012. Environmental and ecological factors that shape the gut bacterial communities of fish: a meta-analysis. Mol. Ecol. 21:3363–3378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Desai AR, Links MG, Collins SA, Mansfield GS, Drew MD, Van Kessel AG, Hill JE. 2012. Effects of plant-based diets on the distal gut microbiome of rainbow trout (Oncorhyncus mykiss). Aquaculture 353:134–142 [Google Scholar]
- 31. Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, Fierer N, Peña AG, Goodrich JK, Gordon JI, Huttley GA, Kelley ST, Knights D, Koenig JE, Ley RE, Lozupone CA, McDonald D, Muegge BD, Pirrung M, Reeder J, Sevinsky JR, Turnbaugh PJ, Walters WA, Widmann J, Yatsunenko T, Zaneveld J, Knight R. 2010. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 7:335–336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Reeder J, Knight R. 2010. Rapidly denoising pyrosequencing amplicon reads by exploiting rank-abundance distributions. Nat. Methods 7:668–669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Caporaso JG, Bittinger K, Bushman FD, DeSantis TZ, Andersen GL, Knight R. 2010. PyNAST: a flexible tool for aligning sequences to a template alignment. Bioinformatics 26:266–267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Wang Q, Garrity GM, Tiedje JM, Cole JR. 2007. Naïve Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl. Environ. Microbiol. 73:5261–5267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Segata N, Izard J, Waldron L, Gevers D, Miropolsky L, Garrett WS, Huttenhower C. 2011. Metagenomic biomarker discovery and explanation. Genome Biol. 12:R60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Kindschi GA. 1987. Method for quantifying degree of fin erosion. Prog. Fish Cult. 49:314–315 [Google Scholar]
- 37. Aussanasuwannakul A, Kenney PB, Brannan RG, Slider SD, Salem M, Yao J. 2010. Relating instrumental texture, determined by variable-blade and Allo-Kramer shear attachments, to sensory analysis of rainbow trout, Oncorhynchus mykiss, fillets. J. Food Sci. 75:S365–S374 [DOI] [PubMed] [Google Scholar]
- 38. Association of Official Analytical Chemists 1990. Official methods of analysis, 15th ed Association of Official Analytical Chemists, Washington, DC [Google Scholar]
- 39. Bligh EG, Dyer WJ. 1959. A rapid method of lipid extraction and purification. Biochem. Physiol. 37:911–917 [DOI] [PubMed] [Google Scholar]
- 40. Fritshe KL, Johnston PU. 1990. Effect of dietary-linolenic acid on growth, metastasis, fatty acid profile and prostaglandin production of two murine mammary adenocarcinomas. J. Nutr. 120:1601–1609 [DOI] [PubMed] [Google Scholar]
- 41. Ramette A. 2007. Multivariate analyses in microbial ecology. FEMS Microbiol. Ecol. 62:142–160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Baeverfjord G, Krogdahl A. 1996. Development and regression of soybean meal induced enteritis in Atlantic salmon, Salmo salar L., distal intestine: a comparison with the intestines of fasted fish. J. Fish Dis. 19:375–387 [Google Scholar]
- 43. Bakke-McKellep AM, Press CM, Baeverfjord LG, Krogdahl A, Landsverk T. 2000. Changes in immune and enzyme histochemical phenotypes of cells in the intestinal mucosa of Atlantic salmon, Salmo salar L., with soybean meal-induced enteritis. J. Fish Dis. 23:115–127 [Google Scholar]
- 44. St-Hilaire S, Ellis T, Cooke A, North BP, Turnbull JF, Knowles T, Kestin S. 2006. Fin erosion on commercial rainbow trout farms in the UK. Vet. Rec. 159:446–450 [DOI] [PubMed] [Google Scholar]
- 45. Adams CE, Turnbull JF, Bell A, Bron JE, Huntingford FA. 2007. Multiple determinants of welfare in farmed fish: stocking density, disturbance, and aggression in Atlantic salmon (Salmo salar). Can. J. Fish Aquat. Sci. 64:336–344 [Google Scholar]
- 46. Durban A, Abellan JJ, Jimenez-Hernandez N, Latorre A, Moya A. 2012. Daily follow-up of bacterial communities in the human gut reveals stable composition and host-specific patterns of interaction. FEMS Microbiol. Ecol. 81:427–437 [DOI] [PubMed] [Google Scholar]
- 47. Alexander DA, Orcutt RP, Henry JC, Baker J, Jr, Bissahoya AC, Threadgill DW. 2006. Quantitative PCR assays for mouse enteric flora reveal strain-dependent differences in composition that are influenced by the microenvironment. Mamm. Genome 17:1093–1104 [DOI] [PubMed] [Google Scholar]
- 48. Ivanov II, Atarashi K, Manel N, Brodie EL, Shima T, Karaoz U, Wei D, Goldfarb KC, Santee CA, Lynch SV, Tanoue T, Imaoka A, Itoh K, Takeda K, Umesaki Y, Honda K, Littman DR. 2009. Induction of intestinal Th17 cells by segmented filamentous bacteria. Cell 139:495–498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Friswell MK, Gika H, Stratford IJ, Theodoridis G, Telfer B, Wilson ID, McBain AJ. 2010. Site and strain-specific variation in gut microbiota profiles and metabolism in experimental mice. PLoS One 5:e8584. 10.1371/journal.pone.0008584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Trust TJ, Sparrow RAH. 1974. The bacterial flora in the alimentary tract of freshwater salmonid fishes. Can. J. Microbiol. 20:1219–1228 [DOI] [PubMed] [Google Scholar]
- 51. Reference deleted.
- 52. Cahill MM. 1990. Bacterial flora of fishes: a review. Microb. Ecol. 19:21–41 [DOI] [PubMed] [Google Scholar]
- 53. Merrifield DL, Burnard D, Bradley G, Davies SJ, Baker RTM. 2009. Microbial community diversity associated with the intestinal mucosa of farmed rainbow trout (Oncoryhnchus mykiss Walbaum). Aquac. Res. 40:1064–1072 [Google Scholar]
- 54. Austin B, Al-Zahrani AMJ. 1988. The effect of antimicrobial compounds on the gastrointestinal microflora of rainbow trout, Salmo gairdneri Richardson. J. Fish Biol. 33:1–14 [Google Scholar]
- 55. Nieto TP, Toranzo AE, Barja JL. 1984. Comparison between the bacterial flora associated with fingerling rainbow trout cultured in two different hatcheries in the north-west of Spain. Aquaculture 42:193–206 [Google Scholar]
- 56. Sugita H, Shibuya K, Shimooka H, Deguchi Y. 1996. Antibacterial abilities of intestinal bacteria in freshwater cultured fish. Aquaculture 156:195–203 [Google Scholar]
- 57. Holben WE, Williams P, Saarinen M, Särkilahti LK, Apajalahti JHA. 2002. Phylogenetic analysis of intestinal microflora indicates a novel mycoplasma phylotype in farmed and wild salmon. Microb. Ecol. 44:175–185 [DOI] [PubMed] [Google Scholar]
- 58. Ashraf R, Shah NP. 2011. Selective and differential enumerations of Lactobacillus delbrueckii subsp. bulgaris, Streptococcus thermophilus, Lactobacillus acidophilus, Lactobacillus casei and Bifidobacterium spp. in yoghurt—a review. Int. J. Food Microbiol. 149:194–208 [DOI] [PubMed] [Google Scholar]
- 59. Kim SK, Bhatnagar I, Kang KH. 2012. Development of marine protiobiotics: prospects and approach. Adv. Food Nutr. Res. 65:353–362 [DOI] [PubMed] [Google Scholar]
- 60. Rumsey GL, Siwicki AK, Anderson DP, Bowser PR. 1994. Effect of soybean protein on serological response, non-specific defense mechanisms, growth and protein utilization in rainbow trout. Vet. Immunol. Immunopathol. 41:323–339 [DOI] [PubMed] [Google Scholar]
- 61. Olli J, Krogdahl A. 1995. Alcohol soluble components of soybeans seem to reduce fat digestibility in fish-meal-based diets for Atlantic salmon, Salmo salar L. Aquac. Res. 26:831–835 [Google Scholar]
- 62. Olli JJ, Krogdahl A, Vabeno A. 1995. Dehulled, solvent-extracted soybean meal as a protein source in diets for Atlantic salmon, Salmo salar L. Aquac. Res. 26:167–177 [Google Scholar]
- 63. Gaylord TG, Teague AM, Barrows FT. 2006. Taurine supplementation of all-plant protein diets for rainbow trout (Onchorhyncus mykiss). J. World Aquac. Soc. 37:509–517 [Google Scholar]
- 64. Buttle LG, Burrells AC, Good JE, Williams PD, Southgate PJ, Burrells C. 2001. The binding of soybean agglutinin (SBA) to the intestinal epithelium of Atlantic salmon, Salmo salar and rainbow trout, Oncorhynchus mykiss, fed high levels of soybean meal. Vet. Immunol. Immunopathol. 80:237–244 [DOI] [PubMed] [Google Scholar]
- 65. Ellis T, Oidtmann B, St-Hillaire S, Turnbull JF, North BP, MacIntyre CM, Nikolaidis J, Hoyle I, Kestin SC, Knowles TG. 2008. Fin erosion in farmed fish, p 121–149 In Branson EJ. (ed), Fish welfare. Blackwell Publishing Ltd., Oxford, United Kingdom [Google Scholar]
- 66. Latremouille DN. 2003. Fin erosion in aquaculture and natural environments. Rev. Fish. Sci. 11:315–335 [Google Scholar]
- 67. Barrows FT, Lellis WA. 1999. The effect of dietary protein and lipid source on dorsal fin erosion in rainbow trout, Oncorhynchus mykiss. Aquaculture 180:167–175 [Google Scholar]
- 68. Wedemeyer G. 1996. Physiology of fish in intensive culture systems, p 114 Chapman Hall, New York, NY [Google Scholar]
- 69. Turnbull J, Bell A, Adams C, Bron J, Huntingford FA. 2005. Stocking density and welfare of cage farmed Atlantic salmon: application of multivariate analysis. Aquaculture 243:121–132 [Google Scholar]
- 70. Brockmark S, Neregard L, Bohlin T, Bjornsson BT, Johnsson JI. 2007. Effects of rearing density and structural complexity on the pre- and postrelease performance of Atlantic salmon. Trans. Am. Fish. Soc. 136:1453–1462 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.