Abstract
A novel flagellatropic phage of Salmonella enterica serovar Typhimurium, called iEPS5, was isolated and characterized. iEPS5 has an icosahedral head and a long noncontractile tail with a tail fiber. Genome sequencing revealed a double-stranded DNA of 59,254 bp having 73 open reading frames (ORFs). To identify the receptor for iEPS5, Tn5 transposon insertion mutants of S. Typhimurium SL1344 that were resistant to the phage were isolated. All of the phage-resistant mutants were found to have mutations in genes involved in flagellar formation, suggesting that the flagellum is the adsorption target of this phage. Analysis of phage infection using the ΔmotA mutant, which is flagellated but nonmotile, demonstrated the requirement of flagellar rotation for iEPS5 infection. Further analysis of phage infection using the ΔcheY mutant revealed that iEPS5 could infect host bacteria only when the flagellum is rotating counterclockwise (CCW). These results suggested that the CCW-rotating flagellar filament is essential for phage adsorption and required for successful infection by iEPS5. In contrast to the well-studied flagellatropic phage Chi, iEPS5 cannot infect the ΔfliK mutant that makes a polyhook without a flagellar filament, suggesting that these two flagellatropic phages utilize different infection mechanisms. Here, we present evidence that iEPS5 injects its DNA into the flagellar filament for infection by assessing DNA transfer from SYBR gold-labeled iEPS5 to the host bacteria.
INTRODUCTION
Bacteriophages (phages) are obligate intracellular parasites of bacteria. The basic life cycle of phages involves the use of bacterial cellular metabolism for production of new phage particles, release of those particles from their cellular confines, and then infection of new host cells (1). Phage infection is initiated by the phage binding to a specific receptor on the surface of the host bacterium, followed by the delivery of the phage genome to the host cell. Adsorption involves multiple steps, including reversible binding for the initial recognition and proper positioning, followed by irreversible binding to the final receptor. However, the molecular mechanisms by which phages deliver their genomes into a host are far from being elucidated.
Generally, a phage is able to infect a limited number of bacterial strains or related strains of the same species (2, 3). Interactions between a phage and the host bacteria is primarily dependent on the nature and structural characteristics of receptors on the bacterial cell surface (4). The representative surface components of the receptors include OmpA, FhuA, BtuB, and LamB (3, 5–8). In addition to membrane proteins, many phages use lipopolysaccharide (LPS) as their receptors (4, 9, 10), a fact that reflects the dominance and diversity of these molecules on the surface of Gram-negative bacteria. Another macromolecular structure known to bind phages is the flagellum. Phage χ1 (VIII.113), which was identified in 1936, is a flagellatropic phage with a very broad range of enteric bacterial hosts (11). Subsequently, phages of both Gram-negative and Gram-positive bacteria, including Chi (12) and PBS1 (13), have been shown to require the flagellum for infection of the host bacteria. A nut-on-a-bolt theory has been proposed to explain the requirement of CCW flagellar rotation for successful infection by flagellatropic phages such as Chi (12, 14). Recently, other flagellatropic phages, φCb13 and φCbK, infecting Caulobacter crescentus, have been shown to actively interact with the flagellum and subsequently attach to receptors on the cell pole (15). They bind to the CCW rotating flagella via the phage head filament, which increases the likelihood of final adsorption to the pilus portals by facilitating concentration of phage particles around the receptor.
The bacterial flagellum is a 20-nm-thick and 10- to 15-μm-long helical structure that protrudes from the cell body. Salmonella enterica serovar Typhimurium has 6 to 8 flagella per cell that are distributed around the cell surface (peritrichous). The flagellum consists of three components, a basal body (engine) by which it is anchored in the cell envelope, a hook (universal joint), which connects the basal body, and the filament (propeller) (4, 16). Bacterial flagellar function by rotation, which is driven by the proton-motive force (16). The integral membrane proteins MotA and MotB form the stator complex, which is necessary for motor rotation (17, 18). The rotor switches between the clockwise (CW) and CCW directions, controlled by a chemotactic signal transduction system that monitors chemical environmental cues (19, 20).
In this study, we isolated and characterized a novel bacteriophage, iEPS5, which infects Salmonella specifically using the flagellum as a receptor. The infection process of iEPS5 was compared to that of Chi. Here, we provide some evidence that these two flagellatropic phages utilize different infection mechanisms.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
The bacterial strains and plasmids used in this study are listed in Table S1 in the supplemental material. S. Typhimurium strains were derived from strain SL1344. Phage P22-mediated transductions were performed as previously described (21). The bacteria were grown at 37°C in LB medium (Difco) under aerobic conditions. Ampicillin (Duchefa Biochemie), chloramphenicol (Duchefa Biochemie), kanamycin (Duchefa Biochemie), and isopropyl-β-d-thio-galactopyranoside (IPTG; Gold Bio) were used at 50 μg/ml, 25 μg/ml, 50 μg/ml, and up to 2 mM, respectively.
Construction of strains.
The S. Typhimurium strain CH504, which is deleted in the motA gene, was constructed using the one-step gene inactivation method (22). The kanamycin resistance (Kmr) cassette from plasmid pKD13 (22) was amplified using primers motA-lamb-F and motA-lamb-R. Primer motA-lamb-F carries the sequence immediately upstream of the start codon of the motA gene following the priming site 1 sequence of pKD13 (22). Primer motA-lamb-R harbors the sequence immediately downstream of the stop codon of the motA gene linked to the priming site 4 sequence of pKD13 (22). The resulting PCR product was integrated into the motA region in strain SL1344, from which the Kmr cassette was removed using plasmid pCP20 (23). Deletion of the corresponding genes was verified by colony PCR. Strains CH501, 502, 505, and CH506, deleted in the fliR, flgK, cheY, and fliK genes, respectively, were constructed by using the same strategy. The primers used to construct the strains are listed in Table S2 in the supplemental material.
Construction of plasmids.
Several vectors were made for complementation of the mutant phenotypes. To construct plasmid pUHE-motA, in which the MotA protein is expressed from the lac promoter, the motA gene was amplified by PCR using primers motA-pUHE-F and motA-pUHE-R with chromosomal DNA from strain SL1344 as the template. The product was introduced between the EcoRI and BamHI restriction sites of pUHE21-2lacIq (24). The same vector and strategy were used to construct pUHE-cheY and pUHE-fliK. Plasmid pACYC-flgK, which contains the FlgK protein coding sequence and its promoter, was constructed for complementation of the flgK deletion mutant. The flgK gene was amplified by PCR using primers flgK-pACYC-F and flgK-pACYC-R with chromosomal DNA from strain SL1344 as the template. The product was introduced between the HindIII and SphI restriction sites of pACYC184 (25). The fliR gene was also cloned into the pACYC184 vector for complementation. The sequences of the motA, cheY, fliK, fliR, and flgK coding regions in the recombinant plasmids were confirmed by nucleotide sequencing. The primers used to construct the plasmids are listed in Table S2 in the supplemental material.
Propagation of bacteriophages.
Phages were propagated by following the standard protocols for phage λ (26). Individual plaques were picked and eluted with sodium chloride-magnesium sulfate (SM) buffer (50 mM Tris-HCl, pH 7.5, 100 mM NaCl, and 10 mM MgSO4). After repicking and replating twice, the phages were inoculated into exponentially growing SL1344 at a multiplicity of infection (MOI) of 1 and incubated for 3 h at 37°C. Cellular debris was removed by centrifugation (8,000 × g for 20 min at 4°C), the phages were precipitated overnight by adding 10% polyethylene glycol (PEG) 6000, and then they were isolated by CsCl density-gradient ultracentrifugation (60,000 × g for 4 h at 4°C). The CsCl-purified phage suspension was dialyzed in SM buffer and stored at 4°C.
Bacteriophage host range.
Forty-two strains were used to test the host range of iEPS5. A 100-μl sample of an overnight culture of each tested bacterium was added to 5 ml of molten LB top agar (0.4% agar), and the mixture was overlaid on LB base agar (1.5% agar) plates. After solidification of the top agar, 10 μl of each serially diluted iEPS5 phage suspension was spotted on the overlaid plates. After incubation at 37°C for 18 h, spots forming single plaques were selected to determine the efficiency of plating (EOP). The EOP value was calculated by comparison of titers between the test bacterium and the propagation host (S. Typhimurium SL1344). The sensitivity of the test bacterium to iEPS5 was determined by the degree of clarity of the spots and EOP values. The bacterial strains used for host range are listed in Table 1.
Table 1.
The host range of iEPS5
| Bacterial isolate | Plaquea | Reference or sourceb |
|---|---|---|
| S. Typhimurium | ||
| SL1344 | T | NCTC |
| UK1 | T | 58 |
| 14028s | I | ATCC |
| LT2 | T | 33 |
| DT104 | T | 61 |
| ATCC 19586 | C | ATCC |
| ATCC 43147 | T | ATCC |
| 3068 | T | LC |
| ST1 | — | LC |
| ST2 | — | LC |
| ST3 | — | LC |
| ST4 | T | LC |
| ST5 | — | LC |
| ST6 | T | LC |
| ST8 | I | LC |
| ST10 | T | LC |
| S. Enteritidis | ||
| ATCC 13078 | — | ATCC |
| SE4 | TT | LC |
| Other Salmonella | ||
| S. Typhi Ty 2-b | — | IVI |
| S. Paratyphi A IB 211 | TT | IVI |
| S. Paratyphi B IB 231 | — | IVI |
| S. Paratyphi C IB 216 | — | IVI |
| S. Dublin IB 2973 | — | IVI |
| E. coli | ||
| MG1655 | — | 57 |
| DH5α | — | 59 |
| DH10B | — | 60 |
| E. coli O157:H7 | ||
| ATCC 35150 | — | ATCC |
| ATCC 43888 | — | ATCC |
| ATCC 43890 | — | ATCC |
| ATCC 43894 | — | ATCC |
| ATCC 43895 | — | ATCC |
| O157:NM 3204-92 | — | CDC |
| O157:NM H-0482 | — | CDC |
| Other Gram-negative bacteria | ||
| Shigella flexneri 2a strain 2457T | — | IVI |
| Shigella boydii IB 2474 | — | IVI |
| Vibrio fischeri ES-114 ATCC 700601 | — | ATCC |
| Pseudomonas aeruginosa ATCC 27853 | — | ATCC |
| Cronobacter sakazakii ATCC 29544 | — | ATCC |
| Other Gram-positive bacteria | ||
| Enterococcus faecalis ATCC 29212 | — | ATCC |
| Staphylococcus aureus ATCC 29213 | — | ATCC |
| Staphylococcus epidermis ATCC 35983 | — | ATCC |
| Bacillus subtilis ATCC 23857 | — | ATCC |
| Bacillus cereus ATCC 14579 | — | ATCC |
| Listeria monocytogenes ATCC 19114 | — | ATCC |
| Listeria innocua I | IVI |
C, clear plaque with EOP of 1 to 0.1; T, turbid plaque with EOP of 1 to 0.1; TT, very turbid plaque with EOP of 1 to 0.1; I, inhibition zone; —, nonspecific.
ATCC, American Type Culture Collection; CDC, Centers for Disease Control and Prevention; NCTC, National Collection of Type Cultures; IVI, International Vaccine Institute; LC, laboratory collection.
Transmission electron microscopy.
Bacteriophages were placed on carbon-coated copper grids and negatively stained with 2% aqueous uranyl acetate (pH 4.0) (Fisher Scientific). Transmission electron microscopy (TEM) was performed using an energy-filtering transmission electron microscope (LIBRA 120; Carl Zeiss) at an accelerating voltage of 120 kV at the National Instrumentation Center for Environmental Management (Seoul, South Korea).
One-step growth curves.
To determine the latent period, eclipse period, and burst size, we used the procedure previously described (27). Briefly, 50 ml of an exponential-phase (when the optical density at 600 nm was 1.0) culture of S. Typhimurium was harvested by centrifugation (10,000 × g for 5 min at 4°C). Phages were added at an MOI of 0.01 and allowed to adsorb for 5 min at room temperature. To remove the excess phage, the mixture was centrifuged as described above and the cell pellet was suspended with the same volume (50 ml) of fresh LB broth medium, followed by further aerobic incubation at 37°C. Two sets of samples were collected every 5 min for 1 h. The first sample was immediately diluted and plated without any treatment, and the second set of samples was plated after treatment with 1% chloroform (final concentration) to release intracellular phages to determine the eclipse period before phage titration. Based on the PFU counts per ml, the latent period, eclipse period, and burst size were determined.
Complete nucleotide sequencing and bioinformatics.
Phage genomic DNA was prepared as described elsewhere (28). A Genome Sequencer 20 system (454 Life Science Corp.) was initially used for whole-genome sequencing at Macrogen Korea. To reduce the contigs, a shotgun library was prepared using DNA that had been randomly sheared with a Hydroshear (part no. JHSH000000-1; Gene Machines). DNA fragments of the desired sizes (1 to 3 kb) were cloned into the pCR4Blunt-TOPO vector (Invitrogen, CA), and DNA sequencing was performed using Applied Biosystems BigDye v3.0 and an ABI Prism 3730 XL DNA analyzer. Finally, primer walking was used to fill the sequence gaps. The complete genome was assembled using SeqMan II sequence analysis software (DNASTAR, Madison, WI). The open reading frames (ORFs) were identified with the ORF Finder program at the National Center of Bioinformatics site (http://www.ncbi.nlm.nih.gov/gorf) and GenMark version 2.5 (http://opal.biology.gatech.edu/GeneMark/genemark_prok_gms_plus.cgi). The identified ORFs that are larger than 50 amino acids were selected to predict functions. Annotation of ORFs was conducted using BLASTP (29) and InterProScan programs (30). Sequence manipulations were performed using the CLC Genomics workbench version 3.6.1 software. The phylogenetic analysis of phage iEPS5 was performed using MEGA5 with the neighbor-joining method (31).
Random mutagenesis and identification of the phage receptor.
To screen for the phage receptor of the host bacteria, random mutagenesis was performed using the EZ-Tn5 pMOD-3<R6Kγori/MCS> transposon system (Epicentre, Madison, WI) according to the manufacturer's instructions. The transposon construct was released from pMOD3 by restriction digestion with PvuII, separated in 1% agarose gels, and purified using a gel extraction kit (Promega, Madison, WI). The complexes were electroporated (at 1.8 kV) into the host cells, and transformants were selected on LB agar plates containing kanamycin (50 μg/ml). The resulting colonies were individually cultured and stored at −80°C in LB broth containing 15% (vol/vol) glycerol. The phage receptor was identified by plaque assays using these random mutant libraries. Each random mutant was overlaid on a 24-well plate containing 500 μl per well of base agar. Seven μl of phage at 106 PFU/ml, which was a sufficient titer for identifying infection, then was dotted into each well and incubated for 8 h at 37°C.
Determination of the transposon insertion site.
To localize the Tn5 insertion site, chromosomal DNA was extracted from candidate clones that were resistant to iEPS5. After self-ligation of restriction enzyme-digested DNA, a portion of the ligation mixture was electroporated into EC100D pir+, and the transformants were rescued on LB agar containing kanamycin (50 μg/ml). The self-ligated vector was recovered using a plasmid DNA purification kit (DNA-spin; iNtRON) and sequenced with Tn5-specific primers provided by the manufacturer (pMOD<MCS> forward sequencing primer and pMOD<MCS> reverse sequencing primer). The PCR product was sequenced and used for a BLAST search to determine the transposon insertion site.
Motility assay.
A 1-μl aliquot of an overnight culture was spotted in the middle of a swim plate (LB, 0.3% agar) and allowed to dry for 1 h at room temperature. The plates were incubated at 37°C for 8 h.
In vitro adsorption assay.
To investigate the adsorption of phage to various Salmonella strains, a phage-binding assay was performed. The host bacteria were cultivated in LB medium overnight. The cells were inoculated into 8 ml of LB medium and incubated at 37°C to a final concentration of 108 CFU/ml. After 2 h of incubation, the bacteria were infected with phages at an MOI of 0.01. Every minute, samples were collected and the bacterial cells were removed by centrifugation at 16,000 × g for 1 min at 4°C and filtered using 0.22-μm filters (Millipore). Finally, the number of PFU in the collected supernatant samples (nonadsorbed phage) was determined by serial dilution and standard plate counting using a reference strain. The phage titer at time zero was defined as 100%.
Phage staining and epifluorescence microscopy.
Phages were labeled with SYBR gold (Invitrogen) as previously described (32). The SYBR gold stock solution (10,000 ×) was diluted with TE (10 mM Tris-HCl, 1 mM EDTA, pH 7.5) buffer. Phage lysates (approximately 1011 PFU/ml) were stained with 5× (final concentration) SYBR gold solution for 20 min in the dark at room temperature. The phage solution was diluted with SM buffer and was filtered through an Amicon Ultra-15 10,000 molecular-weight-cutoff centrifugal filter (Millipore) at 1,500 × g for 90 min at 25°C to remove the free SYBR gold particles. SYBR gold-labeled phages were added to cells at an MOI of approximately 100 and incubated with shaking in the dark for up to 1 h. One-milliliter samples, taken at the various times, were centrifuged at 4,000 × g for 5 min. The pellets were washed with motility solution (0.01 M KPO4, 0.067 M NaCl, 10−4 M EDTA [pH 7.0]) and resuspended in a small volume of motility solution for observation under a fluorescence microscope.
Nucleotide sequence accession number.
The GenBank accession number for the complete genome sequence and annotation information for Salmonella phage iEPS5 is KC677662.
RESULTS AND DISCUSSION
Isolation and characterization of iEPS5.
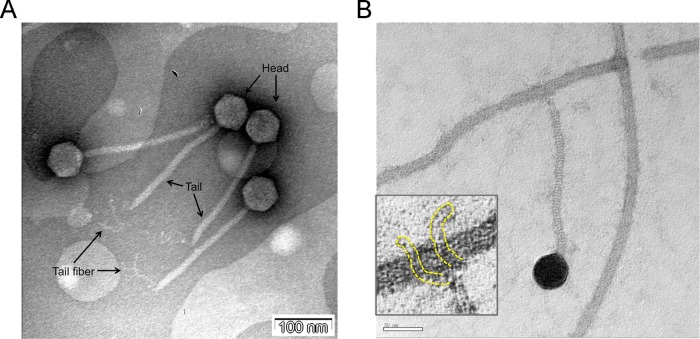
The overall aim of this study was to assess the interaction between the bacteriophage and its host bacterium during infection. We initially isolated a phage, iEPS5, infecting S. Typhimurium SL1344 from a sewage sample. TEM analysis revealed that it belongs to the Siphoviridae family and featured an isometric capsid (59 ± 2 nm) and noncontractile tail (216 ± 3 nm) (Fig. 1A). At the end of the tail, there is a tail fiber that is involved in the phage binding to the bacterial cell (Fig. 1A). The plaque morphology of iEPS5 was turbid and small, and the lysis activity in a broth culture was relatively low (data not shown). iEPS5 has a narrow host range that is restricted to several of the strains of Salmonella that were tested (Table 1). All of the S. Typhimurium type strains and four of the nine S. Typhimurium isolates tested were sensitive to iEPS5. However, iEPS5 could not infect other Gram-negative bacteria, including Escherichia coli, or Gram-positive bacteria. The high host specificity of iEPS5 is unusual among the flagellatropic phages, as other flagellatropic phages, such as χ1 (11) and Chi (12), have relatively broad host ranges.
Fig 1.
(A) Transmission electron micrograph of iEPS5. Head, tail, and tail fiber are marked with arrows. Scale bar, 100 nm. (B) Transmission electron micrograph of the iEPS5 adsorption to the host bacterial flagella. The inset shows long flexible tail fibrils, outlined with yellow dotted lines, the phage utilizes for attachment to a flagellum. Scale bar, 50 nm.
General characteristics of the genome of iEPS5.
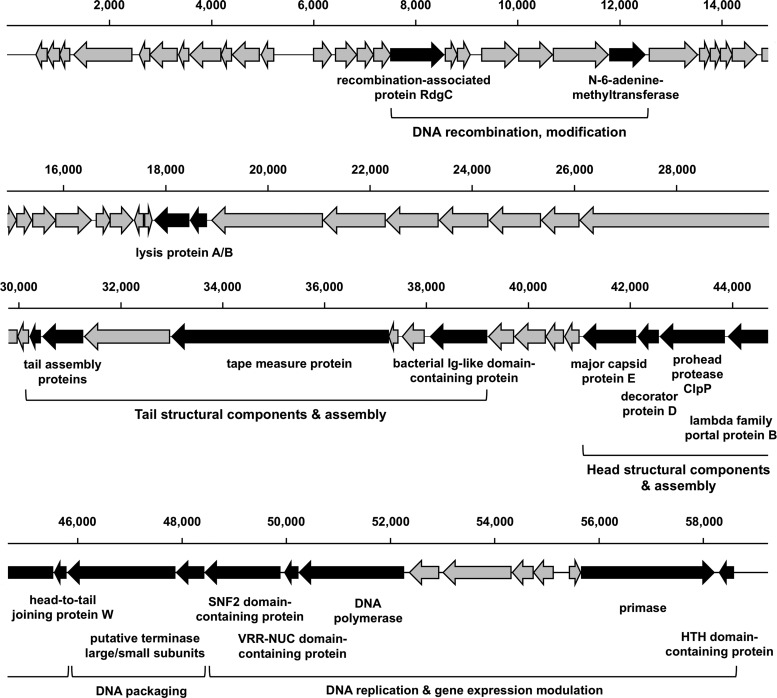
The iEPS5 genome comprises 59,254 bp, with an overall G+C content of 56.31%, which is higher than that of the Salmonella genome (52%) (33). The genome of iEPS5 has 73 ORFs clustered into five functional modules (Fig. 2; also see Table S3 in the supplemental material). The structural module was the largest of them and can be divided into three submodules, the packaging, head structure, and tail morphogenesis submodules. The terminase of the bacteriophage, consisting of a large (ORF62) and a small (ORF63) subunit, is essential for the initiation of DNA packaging. The head morphogenesis submodule, clustered in the region of ORF57 to ORF61, contains the genes for the capsid protein, the prohead protease ClpP, and the portal protein that is required for DNA entering the capsid. The tail module appeared to extend from ORF46 to ORF52. A large number of genes encode tail assembly proteins, and their levels of homology with those of other phages indicate that iEPS5 has an unusual tail. The other two modules are linked to DNA replication, modification, and expression. These modules had lower levels of identities to the known phage genes than the structural modules. BLASTP analysis using predicted ORFs of iEPS5 revealed that its genome is relatively close to that of phage Enc34 (GenBank accession no. JQ340774) (34), targeting Enterobacter cancerogenus, and phage Redjac (GenBank accession no. JX296113) (35), targeting Providencia stuartii, suggesting that these phages have a common ancestor (see Table S3). Prediction of genomic termini of phage iEPS5 conducted as described by Casjens and Gilcrease (36) revealed that phage iEPS5 is classified in the λ-like 5′-extended COS ends group (see Fig. S6).
Fig 2.
Schematic representation of the iEPS5 genome. The predicted ORFs, with their directions of transcription and tRNA-coding sequences, are indicated by arrows. Scale bars mark genome positions at 2,000-bp intervals. Gray arrows indicate ORFs coding for hypothetical proteins, and black arrows indicate predicted functional ORFs.
Adsorption, latent period, and burst size.
To investigate iEPS5's in vitro adsorptive ability to the host bacterium, S. Typhimurium SL1344 was infected with iEPS5 at an MOI of 0.01. More than 50% of the phages were adsorbed onto the host bacteria in 5 min, and at least 90% were absorbed in 15 min (see Fig. S1A in the supplemental material). One-step growth curve analysis revealed that the eclipse and latent periods of iEPS5 were 15 and 30 min, respectively (see Fig. S1B). The eclipse period was consistent with the in vitro adsorption assay results. The burst size was more than 100 PFU per infected host cell.
iEPS5 uses the flagellum as a receptor.
To identify the receptor for iEPS5, a transposon-mediated random mutant library was constructed, and resistant clones were screened. From 1,700 clones, we found five mutants that were resistant to iEPS5 infection (no plaque formation) and one mutant that yielded highly turbid plaques (Table 2). The transposon insertion sites were determined by sequencing the transposon-chromosome boundaries. All six of the resistant mutants had insertions in genes involved in flagellar biosynthesis and included the fliA gene, encoding flagella-specific sigma factor, the fliR and fliP genes, encoding the flagellar export apparatus, the flgK gene, encoding a hook-filament junction, and the hnr gene, encoding a response regulator of RpoS (Table 2). Most of the structural components of the flagellum of Salmonella are exported via a flagellum-specific pathway, which is a member of the family of type III secretary pathways (16, 37). Salmonella strains with mutations in the flagellar export apparatus (FliR and FliP) still express flagellin monomers, but they cannot be secreted to assemble functional flagella, so the mutant is nonmotile (38, 39). Without FlgK, monomeric flagellin can be secreted but not assembled (16, 40). FliA is responsible for the transcription of the class III flagellar genes, which include the filament structure genes and the genes of the chemosensory pathway (41). As expected, these resistant mutants were nonmotile on swim plates due to the absence of flagella on their cell surfaces (see Fig. S2A in the supplemental material). The hnr mutant also showed very low motility on the swim plate (see Fig. S2A), because flagellar expression was reduced due to the higher than normal intracellular concentration of RpoS that occurs in the absence of Hnr (42). Hnr modulates the intracellular RpoS concentration by promoting RpoS degradation by the ClpXP protease (43, 44); thus, RpoS is stabilized in the absence of Hnr. The reduced motility of the hnr mutant resulted in iEPS5 infection, producing very turbid plaques.
Table 2.
iEPS5-resistant isolates
| Candidate | Sequencing result | Motility |
|---|---|---|
| 3-B12 | fliR; flagellar export apparatus | No |
| 3-D2 | fliR; flagellar export apparatus | No |
| 6-H10 | hnr; response regulator of RpoS | Reduced |
| 12-B9 | flgK; hook-filament junction protein | No |
| 14-A7 | fliA; flagellar biosynthesis sigma factor | No |
| 15-D9 | flip; flagellar export apparatus | No |
The validity of the results described above was further assessed using two representative mutants with the fliR or flgK gene deleted by the one-step gene inactivation method (22). These two mutants were nonmotile (Fig. 3A), and iEPS5 did not adsorb to these two strains (Fig. 4C). The motility and sensitivity to iEPS5 of the ΔfliR and the ΔflgK strains were fully restored upon complementation with pACYC-fliR and pACYC-flgK, respectively (Fig. 3A and B). All of these results suggest that the flagellum is a receptor of iEPS5. Salmonella has two flagellar types and switches between two alternative antigenic forms of its flagellin protein, either type B (FljB) or C (FliC) (45, 46). Mutants deleted in either fliC or fljB were motile (see Fig. S2A in the supplemental material) and showed similar sensitivities to iEPS5 (see Fig. S2B), indicating that there is no preference of flagellar types used by iEPS5 as a receptor.
Fig 3.
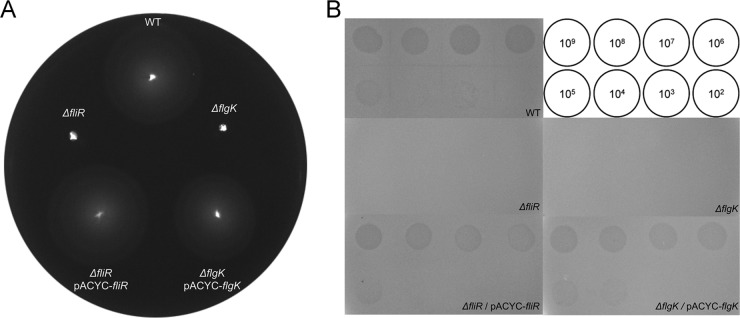
Motility and plaque assays of flagellar mutants and their complementation strains. (A) One μl of an overnight culture was spotted in the middle of a swim plate and incubated at 37°C for 8 h. (B) Serially diluted phages were spotted onto plates of the wild type (SL1344), its isogenic flagellar mutants (ΔfliR [CH501] and ΔflgK [CH502]), and their complementation strains (ΔfliR/pACYC-fliR [CH511] and ΔflgK/pACYC-flgK [CH512]), respectively. The serially diluted phages from 102 to 109 PFU/ml were used as indicated inside the circle.
Fig 4.
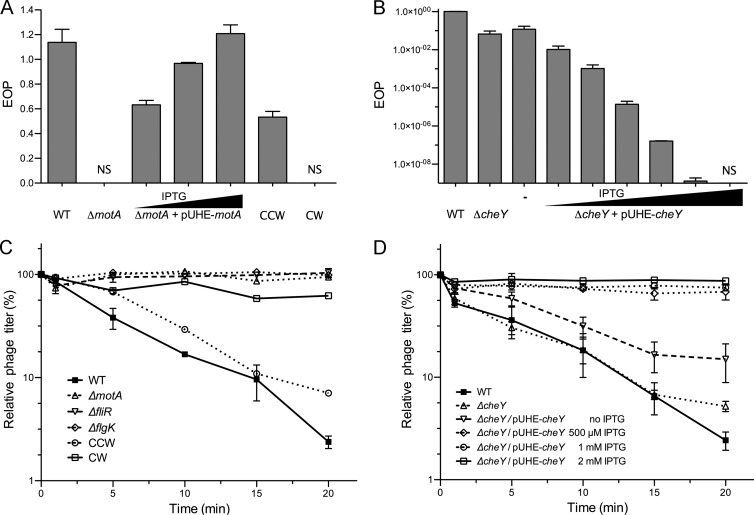
iEPS5 requires CCW-rotating flagella for adsorption to and final infection of the host bacteria. The plaque-forming ability (A and B) and adsorption (C and D) of iEPS5 against each strain was measured. For panels A and B, the wild type (WT; SL1344), its isogenic mutants, and two reported chemotaxis mutant strains (SJW3076 [CCW-biased mutant] and SJW2811 [CW-biased mutant]) were grown exponentially and plated on the top agar and infected with serially diluted iEPS5. The concentrations of IPTG are 100, 200, and 500 μM for motA complementation and 100, 200, 500, 1,000, 1,500, and 2,000 μM for cheY complementation, respectively. For panels C and D, exponentially growing cells were infected with iEPS5 (MOI, 0.01) and incubated at 37°C. After centrifugation and filtration, the phage titer in the filtrate was determined by standard overlay assay. The results are expressed as means and standard deviations from triplicate assays. The concentrations of IPTG are 100, 200, 500, 1,000, 1,500, and 2,000 μM, respectively. NS, nonsensitive.
TEM analysis revealed a direct interaction between iEPS5 and flagella. After incubation of iEPS5 with its host bacteria, followed by several washing steps to remove free phages, we could observe the phage bound to the flagellar filament via its tail fiber (Fig. 1B).
Requirement of flagellar rotation for iEPS5 infection.
It is known that the flagellatropic phage Chi requires flagella rotating CCW for successful infection (12). The requirement for flagellar rotation for iEPS5 infection was tested using the motA mutant, which assembles a normal flagellar filament and basal body but cannot rotate its flagella due to a lack of torque (18). iEPS5 could not adsorb and infect this paralyzed flagellar mutant (Fig. 4A and C). Complementation of the mutant with a plasmid harboring motA fused to the lac promoter restored flagellar motility and its sensitivity to iEPS5 in the presence of IPTG, indicating that motile flagella are essential for iEPS5 infection (Fig. 4A). Assessment using diverse flagellar mutants with different motilities revealed that the infectivity of iEPS5 was reliant on the degree of motility; iEPS5 presented low EOP against the mutants with low motility (see Fig. S3 in the supplemental material). Taken together, the data suggest that a rotating flagellum is required for iEPS5 infection.
CCW flagellar rotation is required for iEPS5 infection.
Although iEPS5 can infect only motile strains of Salmonella, one exception was the SJW3076 strain (see Fig. S3 in the supplemental material). SJW3076 was highly sensitive to iEPS5, even though it is nonmotile due to its CCW-biased rotation of flagella, which results from the deletion of chemotaxis genes cheA through cheZ (47). These results imply that the direction of flagellar rotation is critical for iEPS5 infection. Alternating directions of flagellar rotation is crucial for bacterial movement (48). Switching from CCW to CW rotation of the flagella occurs as a result of the binding of phosphorylated CheY protein to the base of the flagellum (20); therefore, deletion of cheY of Salmonella makes the flagellar rotation CCW biased. Similar to SJW3076, the ΔcheY mutant was also nonmotile but highly sensitive to iEPS5 in the spotting assay (data not shown), confirming the requirement of CCW rotation of flagella for iEPS5 infection. The direction of flagellar rotation can be changed from CCW biased in the ΔcheY mutant strain to CW biased by overexpressing CheY protein (14, 49). A plasmid harboring a cheY gene under the control of the lac promoter was introduced into the ΔcheY mutant, and the EOPs of iEPS5 were measured with increasing amounts of IPTG. As expected, the bacterial resistance to iEPS5 increased in an IPTG-dependent manner. When we added 2 mM IPTG, iEPS5 could no longer absorb to and infect its host (Fig. 4B and D).
Interestingly, iEPS5 had defects on EOPs in both SJW3076 and ΔcheY mutants (Fig. 4B). Successful phage multiplication needs various biological processes after injection of phage genome to the host bacterial cells. The bacterial metabolic activity may not be optimal for phage multiplication in SJW3076 or in ΔcheY mutants, even though phage adsorption was not affected by the mutations in these strains (Fig. 4B and D).
Further experiments using the CW-biased strain SJW2811, in which the deletion of the region of the fliG gene encoding amino acid residues 169 to 171 results in an extremely strong CW motor bias (50, 51), confirmed that iEPS5 could not adsorb to and infect the CW-biased strains (Fig. 4B and D). These results indicate that iEPS5 infects its host bacteria using a CCW-rotating flagellum as a receptor; thus, the direction of flagellar rotation is critical for iEPS5 infection.
Importance of the flagellar filament for iEPS5 infection.
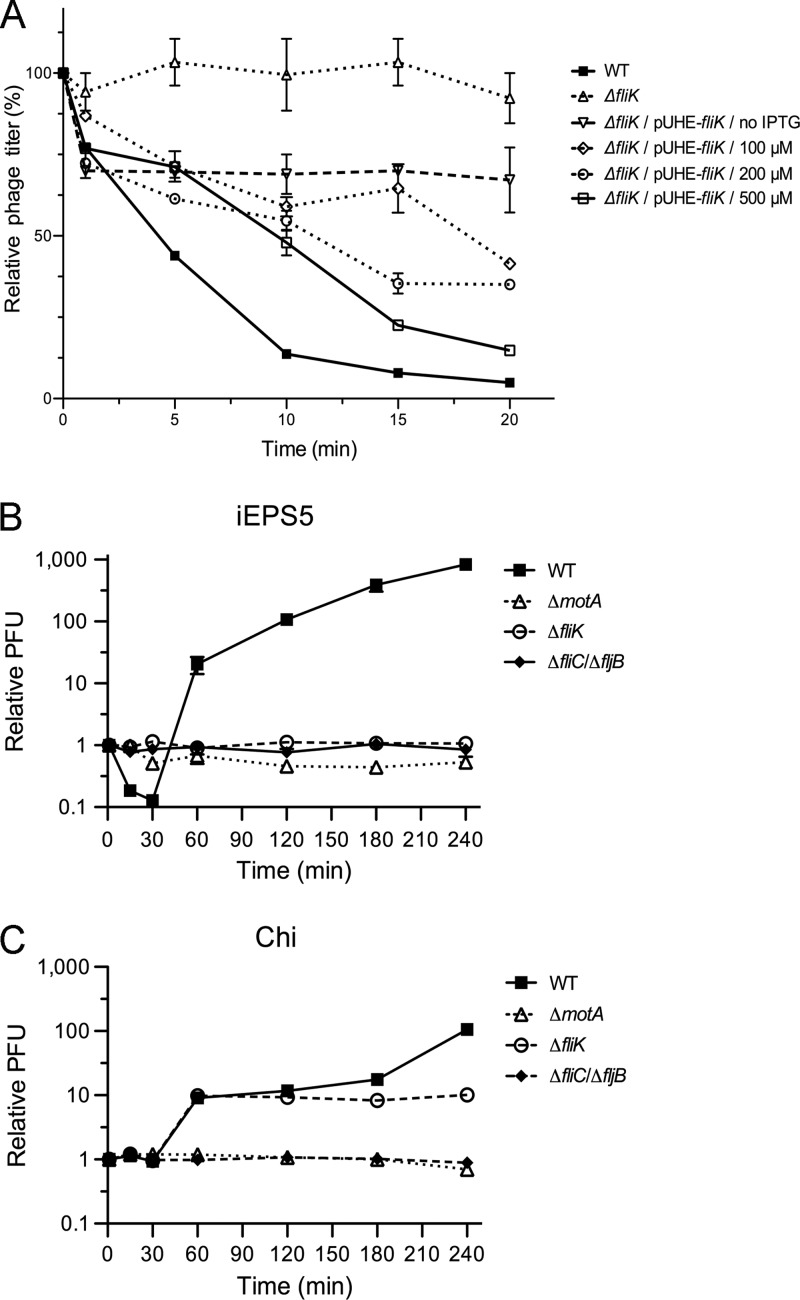
According to the bolt-and-nut model of Chi, the hook structure is also important for flagellatropic phage infection; the phage must traverse the hook to reach the filament base (14). A molecular ruler, FliK, controls the length of the flagellar hook (normally ∼55 nm). FliK catalyzes the secretion substrate specificity switch from rod-hook substrate specificity to late substrate secretion, which involves the filament subunits. The ΔfliK mutant has a polyhook structure with a wide length distribution of up to approximate 1 μm but does not produce flagellar filaments (52–54).
We found interesting differences in the infection of the ΔfliK strain by iEPS5 and Chi; Chi could infect the ΔfliK strain, albeit with lower efficiency than it infected the wild-type bacterium, but iEPS5 could not. Neither phage could infect the Salmonella mutants lacking flagella (ΔfliC and ΔfljB strains) or the ΔmotA strain (Fig. 5). TEM analysis of the ΔfliK strain indicated that iEPS5 could not bind to the polyhook structure by their tail fibers (see Fig. S4 in the supplemental material). These results suggested that iEPS5 uses the flagellar filament as a receptor, but the role of flagellar filament in the iEPS5 infection process may be different from that of Chi, which uses the flagellar filament as a guide to reach the bacterial surface (12, 14).
Fig 5.
Requirement of flagellar filament for iEPS5 infection. iEPS5 adsorption to the wild-type strain (WT; SL1344), polyhook strain (ΔfliK; CH506), and its complementation strain (ΔfliK/pUHE-fliK; CH516). (A) Exponentially growing cells were infected with iEPS5 (MOI, 0.01) and incubated at 37°C. After centrifugation and filtration, the phage titer in the filtrate was determined by standard overlay assay. The results are expressed as means and standard deviations from triplicate assays. The concentrations of IPTG are 100, 200, and 500 μM for the complementation strain. (B and C) Bacterial challenge assay with iEPS5 (B) and Chi (C) to diverse flagellar types, including normal flagella (WT; SL1344), nonmotile flagella (ΔmotA; CH504), normal hook without filament (ΔfliC/ΔfljB; CH509), and polyhook without filament (ΔfliK; CH506). Each phage was added at an MOI of 0.1 to the bacterial culture after 1.5 h of incubation (time point 0).
The flagellar filament might be the DNA injection site of iEPS5.
Phages inject their DNA into the host bacteria only when they remain bound to their specific receptor on the bacterial surface (3, 4). To elucidate the differences in the infection processes of Chi and iEPS5, their phage DNA injection processes were compared using SYBR gold-labeled phages. SYBR gold was selected from several DNA staining materials for its ability to penetrate the phage capsid (32, 55, 56). SYBR gold is an unsymmetrical cyanine dye that exhibits ∼1,000-fold fluorescence enhancement upon binding to RNA, single-stranded DNA, or double-stranded DNA (56). Binding a fluorophore such as SYBR gold to the phage DNA may not alter the phage infection process (47). The fluorescent phages appeared as small dots when viewed under an epifluorescence microscope (Fig. 6; also see Fig. S5 in the supplemental material). Moreover, after injection inside the host, the fluorophore molecules bound to phage DNA retain their fluorescence (32, 56).
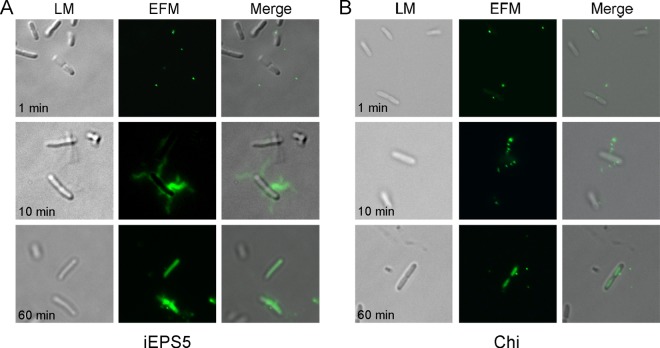
Fig 6.
Visualization of bacterial cells during infection by SYBR gold-labeled phages. Pictures were taken at 1, 10, and 60 min after infection. LM, light microscopy; EFM, epifluorescence microscopy; merge, LM and EFM. Magnification, 1,000×.
The SYBR gold-labeled phages were mixed with the host bacteria and incubated for 1 h under the same conditions as those used for the in vitro adsorption assay. Interestingly, from between 1 and 10 min after infection, some of the flagella became fluorescent, but most of the fluorescence seemed to move to the cytosol of the bacteria 1 h after infection (Fig. 6A). The most probable explanation for the appearance of fluorescent flagellar filaments is that the filaments were filled with SYBR gold-labeled iEPS5 DNA, because SYBR gold moves exclusively with DNA, which is the genome of iEPS5 in this case. Because iEPS5 can infect bacteria only when the flagella rotate CCW, these results imply that the phage can inject its DNA through the flagellar filaments into the interior of the bacteria only when the flagella rotate CCW. Fluorescent flagellar filaments were not seen when Chi was used but the cytosol appeared fluorescent (Fig. 6B), demonstrating that Chi and iEPS5 employ different infection processes. Flagellar filament is a 20-nm-thick and 10- to 15-μm-long hollow tube with a narrow channel of 2.5 to ∼3.0 nm in diameter (16, 37). The diameter of DNA is approximately 2 nm; therefore, the flagellin channel could be a passageway for phage DNA injection. The detailed mechanism of DNA injection through the flagellar filament should be elucidated for better understanding of the process.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by a Midcareer Researcher Program (No. 20090078983) and the WCU Program (R32-2008-000-10183-0) through an NRF grant funded by the MEST.
Footnotes
Published ahead of print 7 June 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.00706-13.
REFERENCES
- 1. Hyman P, Abedon ST. 2010. Bacteriophage host range and bacterial resistance. Adv. Appl. Microbiol. 70:217–248 [DOI] [PubMed] [Google Scholar]
- 2. Evans TJ, Crow MA, Williamson NR, Orme W, Thomson NR, Komitopoulou E, Salmond GP. 2010. Characterization of a broad-host-range flagellum-dependent phage that mediates high-efficiency generalized transduction in, and between, Serratia and Pantoea. Microbiology 156:240–247 [DOI] [PubMed] [Google Scholar]
- 3. Rakhuba DV, Kolomiets EI, Dey ES, Novik GI. 2010. Bacteriophage receptors, mechanisms of phage adsorption and penetration into host cell. Pol. J. Microbiol. 59:145–155 [PubMed] [Google Scholar]
- 4. Lindberg AA. 1973. Bacteriophage receptors. Annu. Rev. Microbiol. 27:205–241 [DOI] [PubMed] [Google Scholar]
- 5. Bonhivers M, Plancon L, Ghazi A, Boulanger P, le Maire M, Lambert O, Rigaud JL, Letellier L. 1998. FhuA, an Escherichia coli outer membrane protein with a dual function of transporter and channel which mediates the transport of phage DNA. Biochimie 80:363–369 [DOI] [PubMed] [Google Scholar]
- 6. Hong J, Kim KP, Heu S, Lee SJ, Adhya S, Ryu S. 2008. Identification of host receptor and receptor-binding module of a newly sequenced T5-like phage EPS7. FEMS Microbiol. Lett. 289:202–209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Mondigler M, Vogele RT, Heller KJ. 1995. Overproduced and purified receptor-binding protein Pb5 of bacteriophage-T5 binds to the T5 receptor protein FhuA. FEMS Microbiol. Lett. 130:293–300 [DOI] [PubMed] [Google Scholar]
- 8. Kim M, Ryu S. 2011. Characterization of a T5-like coliphage, SPC35, and differential development of resistance to SPC35 in Salmonella enterica serovar Typhimurium and Escherichia coli. Appl. Environ. Microbiol. 77:2042–2050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Incardona NL, Selvidge L. 1973. Mechanism of adsorption and eclipse of bacteriophage phi X174. II. Attachment and eclipse with isolated Escherichia coli cell wall lipopolysaccharide. J. Virol. 11:775–782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Quirk AV, Sletten A, Hignett RC. 1976. Properties of phage-receptor lipopolysaccharide from Pseudomonas morsprunorum. J. Gen. Microbiol. 96:375–381 [DOI] [PubMed] [Google Scholar]
- 11. Edwards S, Meynell GG. 1968. The widespread occurrence of enteric flagellar phages. J. Gen. Virol. 2:443–444 [DOI] [PubMed] [Google Scholar]
- 12. Schade SZ, Adler J, Ris H. 1967. How bacteriophage chi attracts motile bacteria. J. Virol. 1:599–609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Joys TM. 1965. Correlation between susceptibility to bacteriophage PBS1 and motility in Bacillus subtilis. J. Bacteriol. 90:1575–1577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Samuel AD, Pitta TP, Ryu WS, Danese PN, Leung EC, Berg HC. 1999. Flagellar determinants of bacterial sensitivity to chi-phage. Proc. Natl. Acad. Sci. U. S. A. 96:9863–9866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Guerrero-Ferreira RC, Viollier PH, Ely B, Poindexter JS, Georgieva M, Jensen GJ, Wright ER. 2011. Alternative mechanism for bacteriophage adsorption to the motile bacterium Caulobacter crescentus. Proc. Natl. Acad. Sci. U. S. A. 108:9963–9968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Chevance FF, Hughes KT. 2008. Coordinating assembly of a bacterial macromolecular machine. Nat. Rev. Microbiol. 6:455–465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Macnab RM. 1986. Proton-driven bacterial flagellar motor. Methods Enzymol. 125:563–581 [DOI] [PubMed] [Google Scholar]
- 18. Muramoto K, Macnab RM. 1998. Deletion analysis of MotA and MotB, components of the force-generating unit in the flagellar motor of Salmonella. Mol. Microbiol. 29:1191–1202 [DOI] [PubMed] [Google Scholar]
- 19. Mariconda S, Wang Q, Harshey RM. 2006. A mechanical role for the chemotaxis system in swarming motility. Mol. Microbiol. 60:1590–1602 [DOI] [PubMed] [Google Scholar]
- 20. Silversmith RE, Bourret RB. 1999. Throwing the switch in bacterial chemotaxis. Trends Microbiol. 7:16–22 [DOI] [PubMed] [Google Scholar]
- 21. Hughes KT, Maloy SR. 2007. Advanced bacterial genetics: use of transposons and phage for genomic engineering. Academic Press, San Diego, CA [Google Scholar]
- 22. Datsenko KA, Wanner BL. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. U. S. A. 97:6640–6645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Doublet B, Douard G, Targant H, Meunier D, Madec JY, Cloeckaert A. 2008. Antibiotic marker modifications of lambda Red and FLP helper plasmids, pKD46 and pCP20, for inactivation of chromosomal genes using PCR products in multidrug-resistant strains. J. Microbiol. Methods 75:359–361 [DOI] [PubMed] [Google Scholar]
- 24. Soncini FC, Vescovi EG, Groisman EA. 1995. Transcriptional autoregulation of the Salmonella typhimurium phoPQ operon. J. Bacteriol. 177:4364–4371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Chang AC, Cohen SN. 1978. Construction and characterization of amplifiable multicopy DNA cloning vehicles derived from the P15A cryptic miniplasmid. J. Bacteriol. 134:1141–1156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Miller JH. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 27. Park M, Lee JH, Shin H, Kim M, Choi J, Kang DH, Heu S, Ryu S. 2012. Characterization and comparative genomic analysis of a novel bacteriophage, SFP10, simultaneously inhibiting both Salmonella enterica and Escherichia coli O157:H7. Appl. Environ. Microbiol. 78:58–69 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Sambrook J, Russell WD. 2001. Molecular cloning: a laboratory manual, 3rd ed Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 29. Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403–410 [DOI] [PubMed] [Google Scholar]
- 30. Zdobnov EM, Apweiler R. 2001. InterProScan–an integration platform for the signature-recognition methods in InterPro. Bioinformatics 17:847–848 [DOI] [PubMed] [Google Scholar]
- 31. Kumar S, Nei M, Dudley J, Tamura K. 2008. MEGA: a biologist-centric software for evolutionary analysis of DNA and protein sequences. Brief. Bioinform. 9:299–306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Kunisaki H, Tanji Y. 2010. Intercrossing of phage genomes in a phage cocktail and stable coexistence with Escherichia coli O157:H7 in anaerobic continuous culture. Appl. Microbiol. Biotechnol. 85:1533–1540 [DOI] [PubMed] [Google Scholar]
- 33. McClelland M, Sanderson KE, Spieth J, Clifton SW, Latreille P, Courtney L, Porwollik S, Ali J, Dante M, Du F, Hou S, Layman D, Leonard S, Nguyen C, Scott K, Holmes A, Grewal N, Mulvaney E, Ryan E, Sun H, Florea L, Miller W, Stoneking T, Nhan M, Waterston R, Wilson RK. 2001. Complete genome sequence of Salmonella enterica serovar Typhimurium LT2. Nature 413:852–856 [DOI] [PubMed] [Google Scholar]
- 34. Kazaks A, Dislers A, Lipowsky G, Nikolajeva V, Tars K. 2012. Complete genome sequence of the Enterobacter cancerogenus bacteriophage Enc34. J. Virol. 86:11403–11404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Onmus-Leone F, Hang J, Clifford RJ, Yang Y, Riley MC, Kuschner RA, Waterman PE, Lesho EP. 2013. Enhanced de novo assembly of high throughput pyrosequencing data using whole genome mapping. PLoS One 8:e61762. 10.1371/journal.pone.0061762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Casjens SR, Gilcrease EB. 2009. Determining DNA packaging strategy by analysis of the termini of the chromosomes in tailed-bacteriophage virions. Methods Mol. Biol. 502:91–111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Macnab RM. 2004. Type III flagellar protein export and flagellar assembly. Biochim. Biophys. Acta 1694:207–217 [DOI] [PubMed] [Google Scholar]
- 38. Fan F, Ohnishi K, Francis NR, Macnab RM. 1997. The FliP and FliR proteins of Salmonella typhimurium, putative components of the type III flagellar export apparatus, are located in the flagellar basal body. Mol. Microbiol. 26:1035–1046 [DOI] [PubMed] [Google Scholar]
- 39. Ohnishi K, Fan F, Schoenhals GJ, Kihara M, Macnab RM. 1997. The FliO, FliP, FliQ, and FliR proteins of Salmonella typhimurium: putative components for flagellar assembly. J. Bacteriol. 179:6092–6099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Aldridge P, Karlinsey J, Hughes KT. 2003. The type III secretion chaperone FlgN regulates flagellar assembly via a negative feedback loop containing its chaperone substrates FlgK and FlgL. Mol. Microbiol. 49:1333–1345 [DOI] [PubMed] [Google Scholar]
- 41. Ohnishi K, Kutsukake K, Suzuki H, Iino T. 1990. Gene fliA encodes an alternative sigma factor specific for flagellar operons in Salmonella typhimurium. Mol. Gen. Genet. 221:139–147 [DOI] [PubMed] [Google Scholar]
- 42. Dong T, Yu R, Schellhorn H. 2011. Antagonistic regulation of motility and transcriptome expression by RpoN and RpoS in Escherichia coli. Mol. Microbiol. 79:375–386 [DOI] [PubMed] [Google Scholar]
- 43. Hengge-Aronis R. 2002. Signal transduction and regulatory mechanisms involved in control of the sigma(S) (RpoS) subunit of RNA polymerase. Microbiol. Mol. Biol. Rev. 66:373–395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Galperin MY. 2006. Structural classification of bacterial response regulators: diversity of output domains and domain combinations. J. Bacteriol. 188:4169–4182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Andrewes FW. 1922. Studies in group-agglutination. I. The Salmonella group and its antigenic structure. J. Pathol. Bacteriol. 25:505–521 [Google Scholar]
- 46. Bonifield HR, Hughes KT. 2003. Flagellar phase variation in Salmonella enterica is mediated by a posttranscriptional control mechanism. J. Bacteriol. 185:3567–3574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Magariyama Y, Yamaguchi S, Aizawa S. 1990. Genetic and behavioral analysis of flagellar switch mutants of Salmonella typhimurium. J. Bacteriol. 172:4359–4369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Harshey RM. 2003. Bacterial motility on a surface: many ways to a common goal. Annu. Rev. Microbiol. 57:249–273 [DOI] [PubMed] [Google Scholar]
- 49. Ravid S, Matsumura P, Eisenbach M. 1986. Restoration of flagellar clockwise rotation in bacterial envelopes by insertion of the chemotaxis protein CheY. Proc. Natl. Acad. Sci. U. S. A. 83:7157–7161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Togashi F, Yamaguchi S, Kihara M, Aizawa SI, Macnab RM. 1997. An extreme clockwise switch bias mutation in fliG of Salmonella typhimurium and its suppression by slow-motile mutations in motA and motB. J. Bacteriol. 179:2994–3003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Yamaguchi S, Aizawa S, Kihara M, Isomura M, Jones CJ, Macnab RM. 1986. Genetic evidence for a switching and energy-transducing complex in the flagellar motor of Salmonella typhimurium. J. Bacteriol. 168:1172–1179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Hirano T, Yamaguchi S, Oosawa K, Aizawa S. 1994. Roles of FliK and FlhB in determination of flagellar hook length in Salmonella typhimurium. J. Bacteriol. 176:5439–5449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Minamino T, Gonzalez-Pedrajo B, Yamaguchi K, Aizawa SI, Macnab RM. 1999. FliK, the protein responsible for flagellar hook length control in Salmonella, is exported during hook assembly. Mol. Microbiol. 34:295–304 [DOI] [PubMed] [Google Scholar]
- 54. Waters RC, O'Toole PW, Ryan KA. 2007. The FliK protein and flagellar hook-length control. Protein Sci. 16:769–780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Wu D, Van Valen D, Hu QC, Phillips R. 2010. Ion-dependent dynamics of DNA ejections for bacteriophage lambda. Biophys. J. 99:1101–1109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Mosier-Boss PA, Lieberman SH, Andrews JM, Rohwer FL, Wegley LE, Breitbart M. 2003. Use of fluorescently labeled phage in the detection and identification of bacterial species. Appl. Spectrosc. 57:1138–1144 [DOI] [PubMed] [Google Scholar]
- 57. Hayashi K, Morooka N, Yamamoto Y, Fujita K, Isono K, Choi S, Ohtsubo E, Baba T, Wanner BL, Mori H, Horiuchi T. 2006. Highly accurate genome sequences of Escherichia coli K-12 strains MG1655 and W3110. Mol. Syst. Biol. 2:2006.0007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Zhang X, Kelly SM, Bollen WS, Curtiss R., III 1997. Characterization and immunogenicity of Salmonella typhimurium SL1344 and UK-1 delta crp and delta cdt deletion mutants. Infect. Immun. 65:5381–5387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Hanahan D. 1983. Studies on transformation of Escherichia coli with plasmids. J. Mol. Biol. 166:557–580 [DOI] [PubMed] [Google Scholar]
- 60. Durfee T, Nelson R, Baldwin S, Plunkett G, III, Burland V, Mau B, Petrosino JF, Qin X, Muzny DM, Ayele M, Gibbs RA, Csorgo B, Posfai G, Weinstock GM, Blattner FR. 2008. The complete genome sequence of Escherichia coli DH10B: insights into the biology of a laboratory workhorse. J. Bacteriol. 190:2597–2606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Poppe C, Smart N, Khakhria R, Johnson W, Spika J, Prescott J. 1998. Salmonella typhimurium DT104: a virulent and drug-resistant pathogen. Can. Vet. J. 39:559–565 [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.