Abstract
Here we present an entire temporal transcriptional profile of Lactococcus lactis subsp. cremoris UC509.9 undergoing lytic infection with two distinct bacteriophages, Tuc2009 and c2. Furthermore, corresponding high-resolution whole-phage genome tiling arrays of both bacteriophages were performed throughout lytic infection. Whole-genome microarrays performed at various time points postinfection demonstrated a rather modest impact on host transcription. The majority of changes in the host transcriptome occur during late infection stages; few changes in host gene transcription occur during the immediate and early infection stages. Alterations in the L. lactis UC509.9 transcriptome during lytic infection appear to be phage specific, with relatively few differentially transcribed genes shared between cells infected with Tuc2009 and those infected with c2. Despite the apparent lack of a coordinated general phage response, three themes common to both infections were noted: alternative transcription of genes involved in catabolic flux and energy production, differential transcription of genes involved in cell wall modification, and differential transcription of genes involved in the conversion of ribonucleotides to deoxyribonucleotides. The transcriptional profiles of both bacteriophages during lytic infection generally correlated with the findings of previous studies and allowed the confirmation of previously predicted promoter sequences. In addition, the host transcriptional response to lysogenization with Tuc2009 was monitored along with tiling array analysis of Tuc2009 in the lysogenic state. Analysis identified 44 host genes with altered transcription during lysogeny, 36 of which displayed levels of transcription significantly reduced from those for uninfected cells.
INTRODUCTION
Bacteriophages are biological entities that function as obligate parasites; they cannot multiply without the biosynthetic functions supplied by their bacterial hosts. In order to proliferate, they must infect their host strain(s) to produce progeny, which, in the majority of cases, results in cell death through lysis and the release of new virions (1). Alternatively, temperate bacteriophages are able to undergo lysogenic infection, whereby lytic functions are repressed and the viral genome integrates into the host chromosome (2). Some temperate phages impart a lysogenic conversion phenotype to their hosts, whereby the hosts benefit from lysogenic infection. This benefit can take the form of superinfection exclusion or virulence phenotypes (3).
Phage-host interactions have been thoroughly detailed in Escherichia coli and have been explored in a handful of other species (4). Generally, host gene transcription is inhibited, either by phage-encoded inhibitors of the host's RNA polymerase, as demonstrated for T7-like and ϕKMV-like phages (the latter infecting Pseudomonas aeruginosa) (5, 6), or by the modification of host RNA polymerase to redirect transcription exclusively toward phage genes, as in the case of phage T4 (7). In addition, T4 phage-encoded ADP-ribosyltransferases interact with host-encoded proteins, some of which are involved in translation processes (8). Recently, Lavigne and colleagues (9) have identified possible phage-directed posttranslational modifications of host proteins as another level of host modulation during lytic infection of P. aeruginosa.
Whole-genome transcriptomics studies have shown that host transcription shutdown or reprogramming is by no means a universal feature of bacteriophage infection (10–14). The majority of these studies, undertaken in E. coli, P. aeruginosa, Lactococcus lactis, and the archaeon Sulfolobus solfataricus, have revealed a remarkably small effect of phage infection on host transcriptomes, with more-pronounced changes occurring only during late infection and usually involving increased transcription of stress response-associated genes.
Analysis of the lactococcal host transcriptome during the early stages of phage infection has recently been carried out by Fallico and colleagues (11). The host response of Lactococcus lactis subsp. lactis IL1403 infected with phage c2 was monitored 10 min postinfection. Sixty-four genes displaying levels of transcription different from those for uninfected cells were identified. From these data, the authors hypothesized a complex four-strand host response to c2 infection, which targets membrane stress proteins, decoration of cell wall polysaccharides, energy conservation, and maintenance of the proton motive force.
L. lactis is used globally as a starter culture for cheese production. Bacteriophages infecting L. lactis can lead to slow or failed fermentations, negatively affecting the production process, as well as the quality of the product, and are therefore a major economic concern (15). Due to the scale of this problem, bacteriophages infecting lactic acid bacteria (LAB), such as L. lactis, have been thoroughly characterized, especially with regard to bacteriophage genomics and host defense mechanisms (16, 17).
Lactococcal phages are grouped into 10 recognized species (18) on the basis of morphology, genetics, and infection characteristics. Of these, 3 species are routinely isolated from dairy fermentations (19). These include the 936 species (small isometric headed), the c2 species (prolate headed), and the highly polythetic P335 species (small isometric headed). All three species are members of the double-stranded DNA (dsDNA) Caudovirales order and belong to the family Siphoviridae, possessing long, noncontractile tails. Apart from members of the P335 species, which can be lysogenic, these phages are strictly lytic.
L. lactis UC509.9, whose complete genome has recently been made available (20), is an Irish dairy starter strain isolated from a mixed starter culture in the 1980s (21). L. lactis UC509.9 is a lysogenic and lytic host to the well-characterized P335-species phage Tuc2009 (21–24) and is also able to undergo infection by the strictly lytic c2 species phage, which has been thoroughly characterized (25–27). Monitoring the host transcriptional response to infection by different bacteriophages will help us understand how bacteria cope with bacteriophage infection. In the current study, we demonstrate that infection of L. lactis UC509.9 by either c2 or Tuc2009 has a rather modest impact on host transcription, yet our results reveal a phage-specific host response to infection.
MATERIALS AND METHODS
Bacterial strains, bacteriophages, and growth conditions.
L. lactis UC509.9 and the Tuc2009-lysogenized derivative of L. lactis UC509.9, L. lactis UC509, were grown under static conditions in GM17 broth (M17 broth [Oxoid Ltd., Hampshire, United Kingdom] supplemented with 0.5% d-glucose). Bacteriophages c2 (26) and Tuc2009 (21) were propagated on L. lactis UC509.9 as described previously (28) and were maintained at 4°C. Bacteriophages were concentrated using 10% polyethylene glycol, as described previously (29).
DNA microarray and tiling array analysis.
DNA microarrays containing oligonucleotide primers representing each of the 2,066 annotated genes on the genome of L. lactis UC509.9 (GenBank accession number CP003157), as well as two complete genome tiling arrays of bacteriophages Tuc2009 (NC_002703) and c2 (NC_001706) at 4-bp resolution, were designed using eArray (Agilent)and chipD (30) and were obtained from Agilent Technologies (Palo Alto, CA, USA).
For sample collection, 600 ml of prewarmed GM17 broth (30°C) was inoculated with 2% of an overnight culture of L. lactis UC509.9. This culture was grown at 30°C under static conditions to an optical density at 600 nm (OD600) of 0.13, at which point CaCl2 was added to a final concentration of 10 mM. The culture was further incubated for 10 min to allow equilibration. At this point, the culture was split into two equal volumes. To one portion, 10 ml of a concentrated 1.5 × 1010-PFU/ml phage suspension (either c2 or Tuc2009) in TBT buffer (100 mM Tris [pH 7.5], 100 mM NaCl, 10 mM MgCl2) was added, producing a final multiplicity of infection (MOI) of 5. To the other portion, used as a control, a corresponding amount of TBT buffer without phage was added. Samples (30 ml) were collected at 2, 5, 10, 15, 25, 35, and, for Tuc2009 only, 45 min postinfection (p.i.) by centrifugation (performed in a Thermo Scientific SL16R centrifuge using a 75003658 rotor at 5,500 rpm for 1 min). Pellets were flash frozen in a −80°C ethanol (EtOH) bath. Samples were then maintained at −80°C until further processing and analysis.
Cells were disrupted, and total RNA was extracted, as described previously (31) using a High Pure RNA isolation kit (Roche, Germany). RNA quality and yield were assessed by observation of band integrity on a 1% agarose gel and determination of the OD260. For all RNA preparations, the OD260/OD280 ratio was >1.9 and the OD260/OD230 ratio was >2.1. For cDNA synthesis, 10 to 20 μg of total RNA was used in an annealing reaction with 1.6 ng μl−1 of random nonamers (MWG Biotech, Germany). For the tiling arrays, to prevent spurious second-strand synthesis, 0.1 μg μl−1 of actinomycin D (Sigma) was added in addition to 1 μl RNaseOUT (Invitrogen, USA). The annealing mixture was heated to 70°C for 5 min and was then cooled at room temperature for 10 min. Reverse transcription was performed using 10 μl of the annealing mixture and SuperScript III reverse transcriptase (Invitrogen, USA) for 16 h at 42°C. cDNA was purified using the Kreatech DSK-001 kit (Kreatech, Amsterdam, Netherlands). Twenty-five micrograms of cDNA was labeled with Cy3 or Cy5 by using the Kreatech DSK-001 labeling kit (Kreatech, Amsterdam, Netherlands). For the tiling array, samples containing bacteriophage cDNA were labeled with Cy3, while control samples were labeled with Cy5.
Labeled cDNA was hybridized using the Agilent Gene Transcription hybridization kit (part number 5188-5242) as described in the manual for Agilent two-color microarray-based gene expression analysis, version 4.0 (G4140-90050). Following hybridization, microarrays were washed in accordance with Agilent's standard procedures and were scanned using an Agilent DNA microarray scanner (model G2565A). The scanning results were converted to data files and were normalized with Agilent's Feature Extraction software (version 9.5) using default settings with the following modification: Loess normalization was performed using the probes representing L. lactis UC509.9 as a reference set, as in a previously described method (32). Differential transcription tests were performed with the Cyber-T implementation of a variant of the t test, as described previously (33). A gene was considered differentially transcribed if the P value was ≤0.001 and the ratio of the level of transcription to the level of transcription for the control was ≥2.5 (or ≥5 for lysogen experiments). For the tiling array analysis, interslide normalization was performed by determining a scaling factor per slide from the total signal of all UC509.9 probes and then dividing the signals of the tiling probes by the scaling factor. To determine transcription start sites (TSS), the melting temperature (Tm) of the portion of the tiling probe overlapping an area where the suspected transcript commenced was calculated. The TSS was determined to be at the position where the overlapping probe Tm was similar to the probe hybridization temperature.
qRT-PCR microarray validation.
cDNA was generated as described above. Ambion Turbo DNase (Invitrogen, USA) was used to ensure that RNA samples were DNA free prior to reverse transcription. Primers for quantitative reverse transcription-PCR (qRT-PCR) were designed using Primer3Plus (34). qRT-PCR was performed using SYBR green I Master Mix (Roche, USA), and the reactions were performed according to the manufacturer's instructions, in triplicate, using a LightCycler 480 II detection system (Roche, USA). Cycling conditions for all amplifications consisted of an initial activation step of 95°C for 10 min, followed by 45 cycles of 95°C for 15 s, 55°C for 5 s, and 72°C for 15 s. Transcription levels were determined using the 2−ΔΔCT method (35); the housekeeping gene alaS was used to normalize results (11).
Determination of chromosome integrity by quantitative PCR (qPCR).
Chromosome integrity after Tuc2009 infection was determined as described previously (9). L. lactis UC509.9 was infected with Tuc2009 and cells harvested as described above. Complete genomic DNA was extracted as described previously (29). Standard curves (data not shown) of DNA copy numbers were prepared using the pGEM-T Easy vector system (Promega, USA) with alaS and orf56 as amplification targets for the L. lactis UC509.9 and Tuc2009 genomes, respectively. Reactions and cycling conditions for targets were performed as described above.
Microarray data accession number.
All microarray and tiling array data determined in this study were deposited in NCBI's Gene Expression Omnibus (GEO) database (36) under the GEO series accession number GSE45655.
RESULTS
Transcriptional response of L. lactis UC509 to lysogeny compared with transcription by prophage-cured UC509.9.
To determine the effects of Tuc2009 lysogeny on its host, a Tuc2009-(re)lysogenized derivative of L. lactis UC509.9 was obtained and was designated L. lactis UC509. Transcriptomes of L. lactis UC509 and UC509.9, grown to early-exponential phase (OD600, ∼0.15), were obtained and compared. Analysis of microarray data demonstrated 44 chromosomal genes differentially expressed in L. lactis UC509 and UC509.9 (Table 1). Of these, the majority, i.e., 36 genes, exhibited lower transcription levels in UC509, while the remaining 9 genes exhibited higher transcription levels in UC509 than in its Tuc2009-free ancestor UC509.9. The downregulated genes encode proteins involved in nucleotide biosynthesis (ntd, dukA, nudH), amino acid metabolism (hisB, trpAD), and respiration (menE, hemH). In addition, four genes (represented by uc509_0204, uc509_0210, uc509_0211, and uc509_0212) presumed to be involved in the biosynthesis of the so-called lactococcal “pellicle,” a cell wall polysaccharide the structure of which has been determined in L. lactis MG1363 (37), exhibited lower levels of transcription in the lysogenic strain than in prophage-cured UC509.9.
Table 1.
L. lactis UC509 genes showing levels of transcription significantly differenta from those for Tuc2009 prophage-cured L. lactis UC509.9
| COG categoryb and gene | Functionc | Fold change | P |
|---|---|---|---|
| Amino acid transport and metabolism | |||
| trpA | Tryptophan synthase α-chain | −5.7 | 7.71E−08 |
| trpD | Anthranilate phosphoribosyltransferase | −8.9 | 8.79E−07 |
| Carbohydrate transport and metabolism, glpF2 | Glycerol uptake facilitator protein | 5.8 | 3.56E−08 |
| Cell wall/membrane/envelope biogenesis | |||
| uc509_0204 | Glycosyltransferase | −5.7 | 6.24E−11 |
| uc509_0210 | Glycosyltransferase | −5.1 | 2.49E−05 |
| uc509_0211 | β-1,3-Glucosyltransferase | −6.2 | 3.68E−05 |
| Coenzyme transport and metabolism | |||
| uc509_0212 | Lipopolysaccharide cholinephosphotransferase | −7.5 | 8.78E−06 |
| nadD | Nicotinate-nucleotide adenylyltransferase | −5.5 | 4.70E−05 |
| hemH | Ferrochelatase | −5.5 | 8.35E−14 |
| Energy production and conversion | |||
| nifJ | Pyruvate-flavodoxin oxidoreductase | 26.1 | 3.10E−06 |
| uc509_0612 | Inorganic pyrophosphatase | −5.4 | 3.04E−07 |
| uc509_1669 | Oxygen-insensitive NAD(P)H nitroreductase | −12.9 | 4.20E−05 |
| Function unknown | |||
| uc509_1435 | Putative VIT family iron transporter | 8 | 2.25E−09 |
| uc509_1436 | Putative VIT family iron transporter | 5.8 | 5.78E−12 |
| General function prediction only | |||
| uc509_1021 | Phosphohydrolase | −5.6 | 1.76E−07 |
| uc509_1700 | Hydrolase | −9.7 | 5.86E−09 |
| Inorganic ion transport and metabolism | |||
| cadA | Cadmium efflux ATPase | 20.9 | 3.84E−08 |
| amtB | Ammonium transporter | 6.7 | 6.58E−05 |
| uc509_0801 | Arsenate reductase | −5.1 | 1.93E−06 |
| cadD | Permease, cadmium resistance protein | −5.6 | 3.16E−06 |
| Lipid transport and metabolism | |||
| uc509_2209 | Activator of (R)-2-hydroxyglutaryl-CoA dehydratase | 12.1 | 9.16E−05 |
| menE | O-Succinylbenzoic acid-CoA ligase | −5.1 | 3.20E−05 |
| Nucleotide transport and metabolism | |||
| ntd | Nucleoside deoxyribosyltransferase-I | −10.1 | 3.11E−08 |
| dukA | Deoxyadenosine kinase | −6.5 | 5.78E−06 |
| Posttranslational modification, protein turnover, chaperones | |||
| uc509_1955 | U32 peptidase | 6.7 | 5.94E−09 |
| tpx | Thiol peroxidase | −6.1 | 7.44E−06 |
| Replication, recombination, and repair | |||
| uc509_0068 | Modification methylase ScrFII | −6 | 4.00E−06 |
| nudH | Adenosine (5′)-pentaphospho-(5″)-adenosine pyrophosphohydrolase | −10.5 | 7.60E−07 |
| xerD | Tyrosine recombinase XerD | −9.3 | 1.42E−06 |
| Secondary-metabolite biosynthesis, transport, and catabolism, uc509_1342 | rRNA (guanine-N1-)-methyltransferase | −13.1 | 2.81E−05 |
| Transcription, rmaD | Transcriptional regulator, MarR family | −5.7 | 1.55E−09 |
| Translation, ribosomal structure, and biogenesis | |||
| uc509_0317 | Transcriptional regulator, MarR family | −9.4 | 1.01E−11 |
| uc509_0396 | Transcriptional regulator, lysR family | −6.3 | 1.75E−06 |
| uc509_0448 | Transcriptional regulator | −10.4 | 1.17E−09 |
| uc509_0583 | Acetyltransferase | −6.6 | 1.73E−10 |
| uc509_0706 | Transcriptional regulator, MarR family | −5.5 | 5.01E−07 |
| uc509_1191 | Transcriptional regulator, MerR family | −5.3 | 1.12E−05 |
| fbpA | Fibronectin/fibrinogen-binding protein | −9.9 | 8.46E−06 |
| xylR | Xylose activator | −10.2 | 1.52E−05 |
| uc509_1462 | Transcriptional regulator, TetR family | −11.9 | 2.41E−07 |
| uc509_2032 | Transcriptional regulator, HTH type | −5.8 | 3.00E−06 |
| uc509_2125 | Transcriptional regulator, LacI family | −5.3 | 1.81E−06 |
| rpmG | LSU ribosomal protein | −5.6 | 9.16E−07 |
| uc509_1219 | Aminoglycoside phosphotransferase | −6.5 | 3.33E−06 |
At least 5-fold different; P, <0.001.
COG, cluster of orthologous groups.
VIT, vacuolar iron transporter; CoA, coenzyme A.
Of the eight chromosomal genes showing increased transcription during lysogeny, four encode transport proteins (cadA, glpF2, uc509_1435, uc509_1436). Of these, uc509_1435 and uc509_1436 encode members of the VIT1 protein family, which is involved in iron and manganese transport (38).
Transcriptional analysis of Tuc2009 during lysogeny.
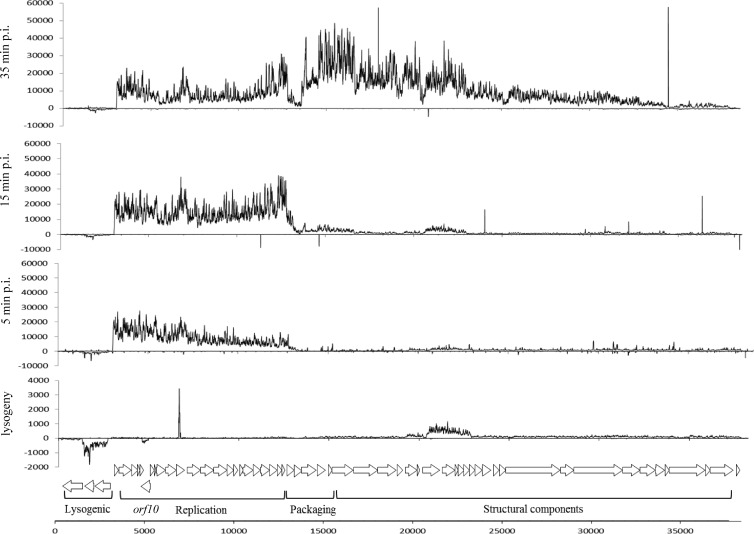
Transcriptional profiling of Tuc2009 during lysogeny was carried out by tiling array analysis in parallel with monitoring of the L. lactis UC509 response to lysogeny. In general, tiling array analysis displayed the expected profile for phages in the lysogenic state (Fig. 1). Transcription of cI2009, which encodes the Tuc2009 repressor, responsible for maintaining the lysogenic state (2), and sie2009, which specifies a superinfection exclusion protein, preventing infection by particular 936-species bacteriophage (28, 39), was clearly visible (Fig. 1). Previously, two transcripts, Y1, which covered sie2009 only, and Y2, which encompassed cI2009, sie2009, and int2009, were detected by Northern blot analysis and were mapped to promoters upstream of sie2009 and int2009 (23). In our data, the Y1 transcript is not discernible, likely due to read-through of the Y2 transcript. The previously determined transcriptional start site (TSS) for Y2 corresponds well with our tiling data. No obvious transcriptional activity was observed for the integrase-encoding gene int2009 during lysogeny. The intergenic region between sie2009 and int2009 contains both a possible transcriptional terminator (positions 1249 to 1291), as predicted using Transterm (40), and possible −10 (TAAACT) and −35 (TTGACG) sequences that could constitute a promoter.
Fig 1.
Tiling array signals of Tuc2009 infecting L. lactis UC509.9 at different stages postinfection (p.i.) and during lysogenic infection. To aid in the visualization of signals, scaling differs between plots. A schematic representation of the Tuc2009 genome, depicting putative protein-encoding regions, is shown at the bottom.
An unexpected observation was the transcription of orf10, which encodes a protein with no known function, during lysogeny. Low-level transcription was detected from a region beginning at a predicted TSS 27 bp upstream of orf10. Analysis using BPROM (SoftBerry) suggests a previously undetected promoter upstream of orf10 (with proposed −35 and [extended] −10 sequences that correspond to ATGTAT and TGCTATAAT, respectively) and a predicted TSS at nucleotide position 4987 (GenBank accession number NC_002703). The genomic position of orf10 is the exception to the general arrangement of Tuc2009 genes in two oppositely orientated clusters, for either a lytic or a lysogenic lifestyle; orf10 is located within the early replication region in the opposite orientation to the surrounding genes. The GC content of orf10 is 34.1%, similar to that of the rest of the replication module and typical of lactococcal phages and their hosts (23). The predicted product of orf10 does not contain any domains discernible by BLASTP (41), Pfam (42), and HHpred (43) analysis, and therefore, we could not assign any function to this gene.
Bacteriophage tiling arrays during lytic infection.
Temporal transcriptional profiling of Tuc2009 and c2 lytically infecting L. lactis UC509.9 was carried out using high-resolution tiling arrays. Previously, transcriptional analyses by Northern blotting and primer extension were performed in detail for both Tuc2009 (23) and c2 (27).
Of the six previously identified Tuc2009 transcripts, with determined start points, termed Y1, Y2, E1, E4, M1/L, and M2, only Y2, E1, and M1/L were clearly identifiable by tiling array analysis using mRNA isolated from Tuc2009-infected UC509.9 (Fig. 1). The Y2 transcript, previously predicted to encompass int2009, sie2009, and rep2009, was detected at all postinfection time points (Fig. 1). However, as with Tuc2009 during lysogeny, this transcript appears to cease upstream of int2009, 30 bp downstream of sie2009. Only at 15 min postinfection can transcription of int2009 be observed. From our tiling data it is not possible to discern whether the transcript encompassing int2009 at 15 min postinfection is (a processed) part of the Y2 transcript or an individual transcript. The expression of these transcripts that encompass the lysogeny module was low compared to the expression of the lytic genes carried in transcripts E1 and M1/L.
The E1 transcript was detected at 2 min postinfection, with signal intensity increasing at 5, 10, and 15 min postinfection, before its strength decreased at subsequent time points, although the transcript was still detectable during late infection. The M1/L transcript was clearly visible from 25 min postinfection and persisted until the end of infection, in agreement with previous results (23). The start position(s) of the E4 and M2 transcripts could not be detected, although these may have been masked due to possible high read-through of the preceding transcripts.
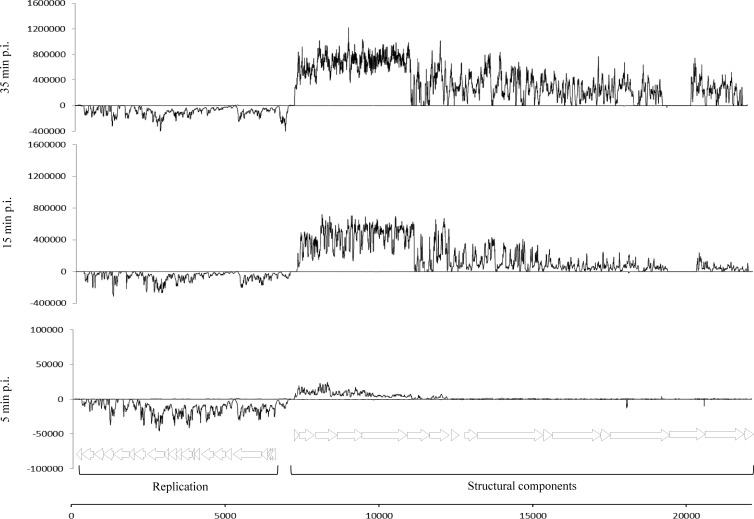
The transcription patterns of phage c2 during L. lactis UC509.9 infection were also monitored using tiling arrays (Fig. 2). As in a previous study using L. lactis MG1363 as a host (27), c2 transcripts were detected from all early promoters at all postinfection time points. Transcription directed by the late promoter was first detected 10 min postinfection and appeared to correspond to a transcript that encompassed l1 to l10, the latter encoding the predicted tail adsorption protein (26). The transcription pattern for l6, encoding a putative hypothetical protein, does not correlate with the transcription pattern for a functional gene. The transcript signal within l6 is interrupted several times, decreasing at around position 11402 (GenBank accession number NC_001706.1) before increasing and decreasing over the remainder of l6. No transcription signal was detected for l15, which encodes a host specificity determinant (44). No obvious explanation can be given for this finding, although the c2 phage in our collection may have an alternative host specificity determinant, different from that for the c2 phage sequenced and deposited in GenBank. In all cases, for both bacteriophages, the transcriptional start sites that had been identified previously by primer extension analysis were shown to be consistent with the transcription initiation points determined by our tiling data (23, 27) (data not shown).
Fig 2.
Tiling array signals of c2 infecting L. lactis UC509.9 at different stages postinfection. To aid in the visualization of signals, scaling differs between plots. A schematic representation of the c2 genome, depicting putative protein-encoding regions, is shown at the bottom.
General features of the L. lactis UC509.9 response to Tuc2009 lytic infection.
To examine the temporal transcriptional response of L. lactis UC509.9 to lytic infection, Tuc2009 was added to early-log-phase cells at an MOI of 5. Samples for RNA extraction were then harvested at regular intervals until 45 min postinfection. To determine the integrity of the genomic DNA of L. lactis UC509.9 throughout infection, qPCR was performed at regular intervals postinfection. The host genome does not appear to be targeted for degradation, since copies of the host's alaS gene remain at a constant level throughout Tuc2009 infection (see Fig. S1 in the supplemental material). This is in line with expectations, since Tuc2009 has no gene whose product could provide such nuclease activity (23). Over the 45-min time course of Tuc2009 infection, 108 genes of the L. lactis UC509.9 genome exhibited transcription levels significantly altered from those of uninfected cells (at least 2.5-fold differential transcription; P, ≤0.001) (Fig. 3A). These represent approximately 5.5% of the total number of L. lactis UC509.9 genes identified (20). Of these 108 genes, 64 exhibited increased transcription, while 44 exhibited reduced transcription from that of uninfected cells. Of the 108 differentially transcribed genes, 55% showed differential transcription at more than one time point during the infection time course. No gene was observed to display a transcriptional level different from that in uninfected cells at all postinfection time points analyzed. During early Tuc2009 infection (i.e., 2 to 10 min postinfection), 19 genes were differentially regulated, compared to 109 genes at later time points (15 to 45 min postinfection). Microarray results were validated for selected genes at 35 min postinfection (Table 2), verifying that the microarray data obtained were a true reflection of phage infection-mediated differential transcription patterns.
Fig 3.
Heat maps of L. lactis UC509.9 genes significantly differentially transcribed (at least 2.5-fold differential transcription; P, <0.001) during lytic Tuc2009 (A) or c2 (B) infection at various times postinfection and in uninfected cells. The color scale represents the fold change in transcription (red, increased; blue, decreased).
Table 2.
qRT-PCR validation of selected L. lactis UC509.9 genes displaying significantly altered transcription at 35 min after infection with Tuc2009
| Gene | Function | Fold change by: |
|
|---|---|---|---|
| qRT-PCR | Microarray | ||
| argS | Arginyl-tRNA synthetase | 6.6 | 3.12 |
| dacA | d-Alanyl-d-alanine carboxypeptidase | 5.6 | 7.46 |
| holB | DNA polymerase III delta prime subunit | 5.5 | 3.76 |
| hsdS | Type I restriction-modification system, specificity subunit | 2.2 | 2.40 |
| metC | Cystathionine gamma-lyase | 3.2 | 3.14 |
| nrdD | Ribonucleotide reductase class III, large subunit | 6.5 | 7.16 |
| nrdE | Ribonucleotide reductase class Ib, α subunit | 4.7 | 4.38 |
| nrdF | Ribonucleotide reductase class Ib, β subunit | 6.7 | 4.00 |
| nrdG | Ribonucleotide reductase class III, activating protein | 4.3 | 5.82 |
| nrdI | Ribonucleotide reduction protein NrdI | 5.0 | 4.08 |
| purK | Phosphoribosylaminoimidazole carboxylase ATPase | 5.3 | NSDa |
| uc509_0426 | rRNA small subunit methyltransferase I | 2.5 | 3.66 |
| uc509_0481 | Hypothetical protein | 17.5 | 2.58 |
| uc509_1772 | UDP-glucose 4-epimerase | 2.5 | 3.25 |
| xylH | 4-Oxalocrotonate tautomerase | 5.1 | 5.90 |
| yabA | DNA replication initiation control protein | 6.1 | 3.51 |
NSD, not significantly different.
Early Tuc2009 infection (2 to 10 min).
During the early stages of L. lactis UC509.9 infection by Tuc2009, host genes involved in purine uptake, synthesis and salvage (purM, add, pbuX, and xpt) showed reduced transcription (Fig. 3A). purM is part of the well-characterized PurR-controlled regulon (45). While other PurR regulon genes do not show significant differential transcription by microarray analysis, qRT-PCR of purK, a member of the PurR regulon that encodes a phosphoribosylaminoimidazole carboxylase ATPase subunit, does suggest that other members of the PurR regulon also exhibit reduced expression during early infection (Table 2).
Increased transcription of noxE, encoding a NADH oxidase that enables the regeneration of NAD+ from NADH in the presence of oxygen (46), at 5 min p.i. suggests increased energy production via glycolysis due to increased availability of NAD+. This would result in an increase in pyruvate levels, which is reflected in the increased transcription of ldh (l-lactate-dehydrogenase) during late infection. Transcription of the nifJ and uc509_2209 genes, whose products are functional under strictly anaerobic conditions, is also reduced during early infection (Fig. 3A).
Late Tuc2009 infection.
More-pronounced differences between the transcriptomes of infected and uninfected cells occur from 15 min p.i. onward, with a total of 96 genes differentially transcribed during late infection (Fig. 3A). Only a small number of genes that exhibited transcription levels different from those for uninfected cells during early infection remained differentially transcribed during late infection. The genes that continued to exhibit altered transcription (nifJ, uc509_1358, uc509_1955, uc509_2146) did so only until 25 min p.i.
The increased transcription of ldh (encoding l-lactate-dehydrogenase) during late infection suggests increased production of lactate from pyruvate. This process requires the reduction of NADH to NAD+. As with the action of noxE during early infection, increased ldh activity would lead to increased availability of NAD+ for energy production via glycolysis.
Genes involved in translation, including argS, encoding arginyl-tRNA synthetase, and tgt, encoding a tRNA guanine transglycosylase, show increased transcription from 25 to 45 min p.i. This may be a phage-mediated change, since high-level translation of the phage's structural proteins is occurring at this time.
Furthermore, L. lactis UC509.9 appears to modify its peptidoglycan structure during infection with Tuc2009. From 10 min p.i. through the remainder of the infection time course, dacA, encoding d-Ala-d-Ala carboxypeptidase, predicted to convert a pentapeptide side chain to a tetrapeptide side chain (47), exhibits increased transcription.
A significant increase in the transcription of nrdGD, specifying the anaerobic ribonucleotide reductase, also occurs during late infection. In addition, increased transcription of nrdHIEF, encoding the class Ib ribonucleotide reductase, which is functional under aerobic and anaerobic conditions (48), is also detectable. Ribonucleotide reductases catalyze the conversion of ribonucleotides to deoxyribonucleotides, thus making them available for DNA repair and replication. The increased expression of these genes is likely a response to diminished levels of nucleotides in infected cells relative to those in uninfected cells due to ongoing bacteriophage genome replication.
General features of the L. lactis UC509.9 response to c2 infection.
During the 35-min time course of c2 infection, 157 genes of the L. lactis UC509.9 genome were shown to be transcribed at levels different from those for uninfected cells (at least 2.5-fold differential transcription; P, ≤0.001) (Fig. 3B). These represent approximately 7.5% of the L. lactis UC509.9 gene complement. Of these 157 genes, 108 exhibited increased transcription, while 49 exhibited reduced transcription from that in uninfected cells. Seventy genes exhibited differential transcription at more than one time point postinfection. No genes were differentially transcribed throughout infection, although nrdGD, encoding anaerobic ribonucleotide reductases, was downregulated from 5 min p.i. onward. The majority of differentially transcribed genes were detected toward the end (15 to 35 min) of infection. Of the 157 genes showing levels of transcription different from those for uninfected cells, 40 encoded (hypothetical) proteins of no known function.
Early c2 infection.
From the tiling array data (Fig. 2), transcripts directed by early c2 promoters were detected 2 min after infection of L. lactis UC509.9. During early infection (2 to 10 min p.i.), very few host genes showed differential transcription; only 10 of the 157 significantly differentially transcribed genes identified were detected during this period, and most of these encode hypothetical proteins with no known function. No L. lactis UC509.9 genes displayed differential transcription at 2 min p.i., while just 3 host genes (nrdGD and uc509_2146, predicted to encode subunits of a ribonucleotide reductase and an Fe-S cluster protein, respectively) were differentially transcribed at 5 min p.i. At 10 min p.i., 10 L. lactis UC509.9 genes, including the genes differentially transcribed at 5 min p.i., exhibited differential transcription. icaB, encoding a polysaccharide deacetylase, was shown to be downregulated from 10 min p.i. through the remainder of the time course.
Late c2 infection.
Transcriptional activity originating from the late c2 promoter, PL1, can be detected in L. lactis UC509.9 after 10 min p.i. (Fig. 2); therefore, 15, 25, and 35 min p.i. are considered to be the late infection stages. During this time, 52, 34, and 122 host genes (at 15, 25, and 35 min p.i., respectively) displayed levels of transcription different from those for uninfected cells. The nrdDG and icaB genes, as mentioned above, continued to exhibit transcription levels lower than those for the uninfected control throughout late infection.
During late infection, differential transcription of genes involved in the metabolism of pyruvate is detected. Components of the pyruvate dehydrogenase complex (PDH), encoded by pdhD and pdhC, display levels of transcription lower than those for uninfected cells from 15 min p.i. onward. Furthermore, at 15 and 25 min p.i., adhE shows levels of transcription lower than those for uninfected cells. The reduction in the level of adhE transcription is likely due to the decreased abundance of acetyl coenzyme A (acetyl-CoA), produced by PDH. These alterations in the cellular catabolic flux suggest an increased tendency toward the homofermentative production of lactate, resulting in increased NAD+ production.
Transcription appears to be limited during late infection, since rpoC, encoding the beta prime chain of RNA polymerase, exhibits reduced transcription levels at 15 and 35 min p.i., while rpoD, specifying the primary sigma factor for L. lactis (49), also displays transcriptional downregulation at 15 min p.i.
It is during late infection that genes involved in the stress response exhibit levels of transcription different from those in uninfected cells. The uc509_1816 and uc509_2207 genes, encoding proteins belonging to the universal stress response family (UspA) (50), show increased transcription at 15 min p.i. and at both 15 and 25 min p.i., respectively. Interestingly, genes involved in regulating the cell envelope stress response (CesSR) (51), encoded by uc509_0928 and uc509_0929 in L. lactis UC509.9, showed no differential transcription until 25 min p.i., when their transcription was diminished from that for uninfected cells.
Of the total number of differentially transcribed genes throughout the c2 infection time course, 77% (122 genes) are differentially transcribed at 35 min postinfection. At this time, various genes involved in amino acid biosynthesis and nitrogen metabolism (Fig. 3B) are upregulated. Since the cells are grown in a nutrient-rich medium (52), the elevated level of transcription over that for uninfected cells is unlikely to be due to nutrient limitation. A possible explanation is that the high level of bacteriophage protein production causes depletion of the intracellular supply of free amino acids.
Plasmid transcriptional response to Tuc2009 and c2 infection.
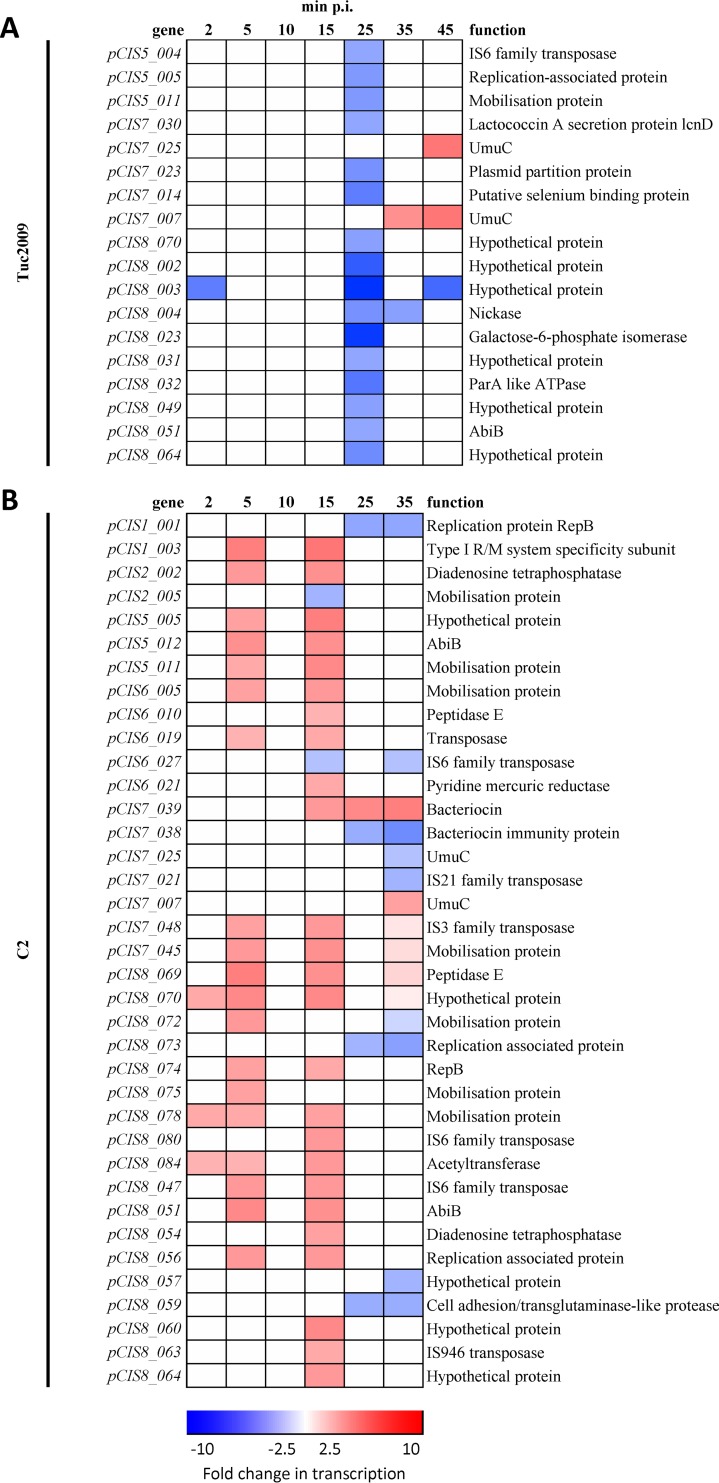
L. lactis UC509.9 contains 8 plasmids, termed pCIS1 to pCIS8, encoding a total of 167 extrachromosomal genes, with functions required for the strain's adaptation to the dairy environment, such as lactose and casein metabolism (20). In keeping with the minimal effects on the host genome, only a small number of plasmid-borne genes displayed differential transcription during the infection time course with either phage: 19 and 33 genes showed alternative transcription with Tuc2009 and c2, respectively (Fig. 4). The majority of changes occurred during late infection, in genes that specify hypothetical proteins located on the largest plasmids, pCIS7 (52 kb) and pCIS8 (80 kb).
Fig 4.
Heat maps of L. lactis UC509.9 plasmid genes significantly differentially regulated (at least 2.5-fold differential transcription; P, <0.001) during lytic Tuc2009 (A) or c2 (B) infection at various times postinfection and in uninfected cells. The color scale represents the fold change in transcription (red, increased; blue, decreased).
Of the phage defense mechanisms, only one gene, pCIS8_051, exhibited increased transcription 5 and 15 min following the initiation of c2 phage infection. This gene encodes a protein with high similarity to AbiB, an abortive infection protein previously shown to be active against c2 and 936-species phages (53). The fact that we can propagate c2 on L. lactis UC509.9 without difficulty suggests that our c2 phage is insensitive to AbiB. Differential transcription of the stress response-induced, low-fidelity polymerase encoded by umuC (54), 2 copies of which are present on pCIS7, occurs during Tuc2009 infection. The level of transcription of these two umuC genes is increased at 35 min p.i. During c2 infection, in contrast, they show alternative transcription relative to that of the uninfected control at 25 min p.i.
Comparative features of L. lactis UC509.9 infection by Tuc2009 and c2.
Only 20 genes exhibited differential transcription during the infection of L. lactis UC509.9 by Tuc2009 as well as c2 (Table 3). Of these, only uc509_1462 and uc509_1503, encoding predicted hypothetical proteins, displayed similar differential transcription during the two infections. The uc509_1503 gene is predicted to encode a protein with five transmembrane helices that is upregulated at 35 min p.i. during c2 and Tuc2009 infection, while uc509_2146, encoding a hypothetical protein with flagellin N-methylase (FliB) family domains, displays increased transcription during early infection by both c2 and Tuc2009.
Table 3.
L. lactis UC509.9 genes showing levels of transcription during either Tuc2009 or c2 infection significantly differenta from those for uninfected cells
| Gene | Product | Direction of transcriptional changeb (min p.i.) for: |
|
|---|---|---|---|
| c2 | Tuc2009 | ||
| add | Adenosine deaminase | + (35) | − (2) |
| cadA | Cadmium efflux ATPase | + (10) | − (2, 5, 10) |
| dppA | Oligopeptide ABC transporter | − (15) | + (2, 15) |
| gltD | Glutamate synthase [NADPH] small chain | − (15) | + (25, 35, 45) |
| hsdS | Type I restriction-modification system, specificity subunit S | + (35) | − (35) |
| nrdD | Ribonucleotide reductase of class III (anaerobic), large subunit | − (5, 10, 15, 25, 35) | + (15, 25, 35, 45) |
| nrdG | Ribonucleotide reductase of class III (anaerobic), activating protein | − (5, 10, 15, 25, 35) | + (15, 25, 35, 45) |
| telA1 | Tellurite resistance protein | − (25, 35) | + (45) |
| uc509_0277 | Hypothetical protein | − (10, 15) | + (15, 25, 35, 45) |
| uc509_1016 | Transcriptional regulator, TetR family | + (35) | − (25) |
| uc509_1462 | Transcriptional regulator, TetR family | + (35) | + (15, 25, 35, 45) |
| uc509_1503 | Hypothetical protein | + (35) | + (35) |
| uc509_1771 | Rrf2 family transcriptional regulator | + (25, 35) | − (25, 35) |
| uc509_1772 | UDP-glucose 4-epimerase | + (15, 25) | − (25, 35, 45) |
| uc509_1879 | Hypothetical protein | − (25, 35) | + (45) |
| uc509_1880 | Transcriptional regulator | − (15, 35) | + (45) |
| uc509_1927 | Hypothetical protein | + (15) | − (25) |
| uc509_2146 | Hypothetical protein | + (5, 10, 25, 35) | − (2, 5, 10, 15) |
| uc509_2210 | Transcriptional regulator, TetR family | + (35) | − (2, 5, 10) |
| uc509_2223 | Cation-transporting ATPase | − (10, 15, 25, 35) | + (35) |
Differences of >2.5 fold (P < 0.001).
−, reduced transcription; +, increased transcription.
DISCUSSION
This study analyzed the temporal transcriptional response of L. lactis UC509.9 undergoing infection with either Tuc2009 or c2, representing phages of two different species (P335 and c2, respectively) of the family Siphoviridae. DNA microarrays of the host and high-resolution tiling arrays of each phage were used to provide corresponding data sets of the entire transcriptome at various points postinfection. An MOI of 5 was used in these experiments, which is not sufficient to infect every cell in the culture simultaneously yet is sufficient to ensure lysis of the experimental culture after 1 h, when viable phage particles are detected by plaque assay (data not shown). This level was applied because we did not want to place nonphysiological stress on the cells; high MOIs have been known to cause “lysis from without” (55) due to the muralytic enzymes associated with bacteriophage tails (56). It was presumed that there would be a subpopulation of uninfected cells in the phage-infected cultures at this MOI. We surmised that this would have minimal effect on our results, since the cells would behave as uninfected cells, apart from the possible dampening of the transcription profiles of genes in the infected samples.
Our data show that the response of L. lactis UC509.9 to infection is phage specific; relatively few differentially regulated genes are shared by transcriptomes obtained following infection by Tuc2009 and those obtained after infection with c2 (Table 3). Also, it was observed that infection with either phage has relatively minimal effects on the L. lactis UC509.9 transcriptome. Apparently, there is no need for major reprogramming of the cell at the transcriptional level in order to establish infection and produce phage progeny. The increased transcription of genes, many of which are involved in amino acid metabolism and nucleotide conversion, during late infection by both bacteriophages may be a cellular response to a perceived lack of metabolites due to depletion resulting from the rapid production of phage progeny.
The apparent lack of widespread cellular reprogramming or lack of a cellular transcriptional response has been noted previously for E. coli, where global changes in whole-genome transcription were moderate during infection with phage PRD1 (10). In the PRD1 model, most changes were shown to occur after bacteriophage synthesis, and the most highly induced host genes were stress related. It has been hypothesized previously that it may be in the interest of the phage not to elicit a defensive host response (10) and that this is therefore a specific strategy followed by the phage. It is worth noting that phages of the family Tectiviridae, of which PRD1 is a member, are not tailed phages and are genetically distinct from Caudovirales yet elicit apparently similar host responses. The minimal transcriptional response in Lactococcus species, and indeed in other species, also suggests that cellular adaptation for the production of phage progeny is likely to be more active at the translational or possibly the posttranslational level (9).
Despite the minimal impact on the host transcriptome and the apparent lack of a coordinated general phage response, Tuc2009 and c2 infections have three effects in common. These are (i) alternative transcription of genes involved in catabolic flux and energy production, (ii) differential transcription of genes involved in cell wall modification, and (iii) differential transcription of genes involved in the conversion of ribonucleotides to deoxyribonucleotides. These effects are similar to responses observed by Fallico et al. (11), where genes involved in the decoration of cell wall polysaccharides and energy conservation displayed differential regulation during c2 infection of L. lactis IL1403. Studies of phage genome replication in the c2-related prolate-headed phage c6A suggest that phage genome replication continues until lysis (57). Therefore, the increase in the level of transcription of genes encoding ribonucleotide reductases is likely to enable continued rapid bacteriophage replication.
The alternative transcription of genes encoding proteins involved in cell wall modification during bacteriophage infection has been noted previously. Notably, L. lactis IL1403, when infected with c2, showed increased transcription of the dltACBD operon, responsible for d-alanyl modification of cell wall components (11). Moreover, modification of peptidoglycan has been linked to reduced susceptibility to 936-species phages in L. lactis MG1363 (58). There have also been suggestions that phage proteins target cell wall biosynthesis and modification in other species. Several phage-encoded proteins, such as Kil, encoded by the E. coli prophage Rac (59), have been shown to affect cell division. In the present study, during c2 infection, icaB, containing a polysaccharide deacetylase family domain (Polysacc_deac_1), exhibited reduced transcription from 10 min p.i., whereas during Tuc2009 infection, dacA transcription was increased. Knockout mutants of a polysaccharide deacetylase-encoding gene, pgdA, in L. lactis IL1403 exhibited fully acetylated peptidoglycan and increased autolysis (60), whereas knockout of dacA in L. lactis MG1363 caused increased lysozyme resistance (47). The increase in dacA transcription during bacteriophage infection may possibly be a Tuc2009-induced effect that aids in phage progeny escape.
Apart from perceived functional similarities, our results are remarkably different from those of the previous study by Fallico et al. (11), in which L. lactis IL1403 was infected with c2. Only 10 genes identified by Fallico and colleagues are differentiated during c2 infection in both studies. This is likely accounted for by the differences in experimental setup, such as the mid-log growth phase (OD600, 0.4) and the very high MOI of 800 used in the previous study. Cellular physiology during mid-log phase would be markedly different from that of L. lactis UC509.9 during early-exponential phase (61). It is possible, therefore, that host infection profiles at different cell growth phases show alternative responses to phage infection.
In addition to the host response, we monitored the transcriptional profiles of both Tuc2009 and c2 throughout infection and that of Tuc2009 during lysogeny. Transcriptional profiles generally correlated with the findings of previous transcriptional studies of these two phages (23, 27). The most notable feature of the tiling array data is the detection of orf10 transcription during lysogeny. The majority of gene sequences found in the bacteriophage genetic repertoire are novel sequences with no known function (62). Because most of these genetic sequences are completely unexplored, speculation on functions is difficult. The transcription of orf10 during lysogeny suggests that it plays a role in the establishment or maintenance of the lysogenic state of Tuc2009. However, supplying orf10 on the constitutive lactococcal-promoter-containing multicopy vector pNZ44 (63) in L. lactis UC509.9 did not affect the efficiency of plaquing (EOP) or the frequency of lysogeny during infection with Tuc2009 (data not shown). Nevertheless, the suggestion that orf10 is a “moron” gene, whereby expression of the gene confers an advantage on the host, cannot be ruled out. orf10 seems to be conserved in some P335 phages, and homologs of this gene (98 to 99% sequence identity) are found in phage ul36 (64) and within prophages found in lactococcal genomes, such as L. lactis ÌL1403 prophages bIL285 and bIL286 (65) and the L. lactis MG1363 prophage MG-3 (66).
In conclusion, we have demonstrated a temporal transcriptional host response of Lactococcus to infection with different species of bacteriophage. This has revealed similarly low impacts on overall host transcription, with phage-specific transcriptional responses, suggesting that there is no common host strategy in response to infection with bacteriophages from different species.
Supplementary Material
ACKNOWLEDGMENT
This research was funded by a Science Foundation Ireland (SFI) Principal Investigatorship award (08/IN.1/B1909) to D.V.S.
Footnotes
Published ahead of print 31 May 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.01197-13.
REFERENCES
- 1. Ackermann HW. 2007. 5500 Phages examined in the electron microscope. Arch. Virol. 152:227–243 [DOI] [PubMed] [Google Scholar]
- 2. Kenny JG, Leach S, de la Hoz AB, Venema G, Kok J, Fitzgerald GF, Nauta A, Alonso JC, van Sinderen D. 2006. Characterization of the lytic-lysogenic switch of the lactococcal bacteriophage Tuc2009. Virology 347:434–446 [DOI] [PubMed] [Google Scholar]
- 3. Basu S, Ghosh RK. 2005. Isolation and characterization of unserotypable lysogens of Vibrio cholerae phage PS166. Med. Sci. Monit. 11:BR335–BR342 [PubMed] [Google Scholar]
- 4. Wagemans J, Lavigne R. 2012. Phages and their hosts, a web of interactions—applications to drug design, p 119–133 In Hyman P, Abedon ST. (ed), Bacteriophages in health and disease. CABI Press, Wallingford, United Kingdom [Google Scholar]
- 5. Klimuk E, Akulenko N, Makarova KS, Ceyssens PJ, Volchenkov I, Lavigne R, Severinov K. 2013. Host RNA polymerase inhibitors encoded by ϕKMV-like phages of Pseudomonas. Virology 436:67–74 [DOI] [PubMed] [Google Scholar]
- 6. Hesselbach BA, Nakada D. 1977. “Host shutoff” function of bacteriophage T7: involvement of T7 gene 2 and gene 0.7 in the inactivation of Escherichia coli RNA polymerase. J. Virol. 24:736–745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Svenson SB, Karlstrom OH. 1976. Bacteriophage T4-induced shut-off of host-specific translation. J. Virol. 17:326–334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Depping R, Lohaus C, Meyer HE, Ruger W. 2005. The mono-ADP-ribosyltransferases Alt and ModB of bacteriophage T4: target proteins identified. Biochem. Biophys. Res. Commun. 335:1217–1223 [DOI] [PubMed] [Google Scholar]
- 9. Lavigne R, Lecoutere E, Wagemans J, Cenens W, Aertsen A, Schoofs L, Landuyt B, Paeshuyse J, Scheer M, Schobert M, Ceyssens PJ. 2013. A multifaceted study of Pseudomonas aeruginosa shutdown by virulent podovirus LUZ19. mBio 4:e00061–13. 10.1128/mBio.00061-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Poranen MM, Ravantti JJ, Grahn AM, Gupta R, Auvinen P, Bamford DH. 2006. Global changes in cellular gene expression during bacteriophage PRD1 infection. J. Virol. 80:8081–8088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Fallico V, Ross RP, Fitzgerald GF, McAuliffe O. 2011. Genetic response to bacteriophage infection in Lactococcus lactis reveals a four-strand approach involving induction of membrane stress proteins, d-alanylation of the cell wall, maintenance of proton motive force, and energy conservation. J. Virol. 85:12032–12042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Osterhout RE, Figueroa IA, Keasling JD, Arkin AP. 2007. Global analysis of host response to induction of a latent bacteriophage. BMC Microbiol. 7:82. 10.1186/1471-2180-7-82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Ortmann AC, Brumfield SK, Walther J, McInnerney K, Brouns SJ, van de Werken HJ, Bothner B, Douglas T, van de Oost J, Young MJ. 2008. Transcriptome analysis of infection of the archaeon Sulfolobus solfataricus with Sulfolobus turreted icosahedral virus. J. Virol. 82:4874–4883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Ravantti JJ, Ruokoranta TM, Alapuranen AM, Bamford DH. 2008. Global transcriptional responses of Pseudomonas aeruginosa to phage PRR1 infection. J. Virol. 82:2324–2329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Garneau JE, Moineau S. 2011. Bacteriophages of lactic acid bacteria and their impact on milk fermentations. Microb. Cell Fact. 10(Suppl. 1):S20. 10.1186/1475-2859-10-S1-S20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Labrie SJ, Samson JE, Moineau S. 2010. Bacteriophage resistance mechanisms. Nat. Rev. Microbiol. 8:317–327 [DOI] [PubMed] [Google Scholar]
- 17. Mahony J, Murphy J, van Sinderen D. 2012. Lactococcal 936-type phages and dairy fermentation problems: from detection to evolution and prevention. Front. Microbiol. 3:335. 10.3389/fmicb.2012.00335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Deveau H, Labrie SJ, Chopin MC, Moineau S. 2006. Biodiversity and classification of lactococcal phages. Appl. Environ. Microbiol. 72:4338–4346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Labrie S, Moineau S. 2000. Multiplex PCR for detection and identification of lactococcal bacteriophages. Appl. Environ. Microbiol. 66:987–994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Ainsworth S, Zomer A, de Jager V, Bottacini F, van Hijum SAFT, Mahony J, van Sinderen D. 2013. Complete genome of Lactococcus lactis subsp. cremoris UC509.9, host for a model lactococcal P335 bacteriophage. Genome Announc. 1:e00119–12. 10.1128/genomeA.00119-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Arendt EK, Daly C, Fitzgerald GF, van de Guchte M. 1994. Molecular characterization of lactococcal bacteriophage Tuc2009 and identification and analysis of genes encoding lysin, a putative holin, and two structural proteins. Appl. Environ. Microbiol. 60:1875–1883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Kenny JG, McGrath S, Fitzgerald GF, van Sinderen D. 2004. Bacteriophage Tuc2009 encodes a tail-associated cell wall-degrading activity. J. Bacteriol. 186:3480–3491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Seegers JFML, Mc Grath S, O'Connell-Motherway M, Arendt EK, van de Guchte M, Creaven M, Fitzgerald GF, van Sinderen D. 2004. Molecular and transcriptional analysis of the temperate lactococcal bacteriophage Tuc2009. Virology 329:40–52 [DOI] [PubMed] [Google Scholar]
- 24. Vegge CS, Vogensen FK, Mc Grath S, Neve H, van Sinderen D, Brondsted L. 2006. Identification of the lower baseplate protein as the antireceptor of the temperate lactococcal bacteriophages TP901-1 and Tuc2009. J. Bacteriol. 188:55–63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Geller BL, Ivey RG, Trempy JE, Hettinger-Smith B. 1993. Cloning of a chromosomal gene required for phage infection of Lactococcus lactis subsp. lactis C2. J. Bacteriol. 175:5510–5519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Lubbers MW, Waterfield NR, Beresford TP, Le Page RW, Jarvis AW. 1995. Sequencing and analysis of the prolate-headed lactococcal bacteriophage c2 genome and identification of the structural genes. Appl. Environ. Microbiol. 61:4348–4356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Lubbers MW, Schofield K, Waterfield NR, Polzin KM. 1998. Transcription analysis of the prolate-headed lactococcal bacteriophage c2. J. Bacteriol. 180:4487–4496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Mahony J, McGrath S, Fitzgerald GF, van Sinderen D. 2008. Identification and characterization of lactococcal-prophage-carried superinfection exclusion genes. Appl. Environ. Microbiol. 74:6206–6215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Maniatis T, Sambrook J, Fritsch EF. 1982. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY [Google Scholar]
- 30. Dufour YS, Wesenberg GE, Tritt AJ, Glasner JD, Perna NT, Mitchell JC, Donohue TJ. 2010. chipD: a web tool to design oligonucleotide probes for high-density tiling arrays. Nucleic Acids Res. 38:W321–W325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. van Hijum SA, de Jong A, Baerends RJ, Karsens HA, Kramer NE, Larsen R, den Hengst CD, Albers CJ, Kok J, Kuipers OP. 2005. A generally applicable validation scheme for the assessment of factors involved in reproducibility and quality of DNA-microarray data. BMC Genomics 6:77. 10.1186/1471-2164-6-77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. van Hijum SA, Baerends RJ, Zomer AL, Karsens HA, Martin-Requena V, Trelles O, Kok J, Kuipers OP. 2008. Supervised Lowess normalization of comparative genome hybridization data—application to lactococcal strain comparisons. BMC Bioinformatics 9:93. 10.1186/1471-2105-9-93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Baldi P, Long AD. 2001. A Bayesian framework for the analysis of microarray expression data: regularized t-test and statistical inferences of gene changes. Bioinformatics 17:509–519 [DOI] [PubMed] [Google Scholar]
- 34. Untergasser A, Nijveen H, Rao X, Bisseling T, Geurts R, Leunissen JA. 2007. Primer3Plus, an enhanced web interface to Primer3. Nucleic Acids Res. 35:W71–W74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Livak KJ, Schmittgen TD. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 25:402–408 [DOI] [PubMed] [Google Scholar]
- 36. Edgar R, Domrachev M, Lash AE. 2002. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 30:207–210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Chapot-Chartier MP, Vinogradov E, Sadovskaya I, Andre G, Mistou MY, Trieu-Cuot P, Furlan S, Bidnenko E, Courtin P, Pechoux C, Hols P, Dufrene YF, Kulakauskas S. 2010. Cell surface of Lactococcus lactis is covered by a protective polysaccharide pellicle. J. Biol. Chem. 285:10464–10471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Kim SA, Punshon T, Lanzirotti A, Li L, Alonso JM, Ecker JR, Kaplan J, Guerinot ML. 2006. Localization of iron in Arabidopsis seed requires the vacuolar membrane transporter VIT1. Science 314:1295–1298 [DOI] [PubMed] [Google Scholar]
- 39. McGrath S, Fitzgerald GF, van Sinderen D. 2002. Identification and characterization of phage-resistance genes in temperate lactococcal bacteriophages. Mol. Microbiol. 43:509–520 [DOI] [PubMed] [Google Scholar]
- 40. Jacobs GH, Chen A, Stevens SG, Stockwell PA, Black MA, Tate WP, Brown CM. 2009. Transterm: a database to aid the analysis of regulatory sequences in mRNAs. Nucleic Acids Res. 37:D72–D76 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389–3402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Bateman A, Birney E, Durbin R, Eddy SR, Howe KL, Sonnhammer EL. 2000. The Pfam protein families database. Nucleic Acids Res. 28:263–266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Soding J, Biegert A, Lupas AN. 2005. The HHpred interactive server for protein homology detection and structure prediction. Nucleic Acids Res. 33:W244–W248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Stuer-Lauridsen B, Janzen T, Schnabl J, Johansen E. 2003. Identification of the host determinant of two prolate-headed phages infecting Lactococcus lactis. Virology 309:10–17 [DOI] [PubMed] [Google Scholar]
- 45. Jendresen CB, Martinussen J, Kilstrup M. 2012. The PurR regulon in Lactococcus lactis—transcriptional regulation of the purine nucleotide metabolism and translational machinery. Microbiology 158:2026–2038 [DOI] [PubMed] [Google Scholar]
- 46. Neves AR, Ramos A, Costa H, van Swam II, Hugenholtz J, Kleerebezem M, de Vos W, Santos H. 2002. Effect of different NADH oxidase levels on glucose metabolism by Lactococcus lactis: kinetics of intracellular metabolite pools determined by in vivo nuclear magnetic resonance. Appl. Environ. Microbiol. 68:6332–6342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Courtin P, Miranda G, Guillot A, Wessner F, Mezange C, Domakova E, Kulakauskas S, Chapot-Chartier MP. 2006. Peptidoglycan structure analysis of Lactococcus lactis reveals the presence of an l,d-carboxypeptidase involved in peptidoglycan maturation. J. Bacteriol. 188:5293–5298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Jordan A, Pontis E, Aslund F, Hellman U, Gibert I, Reichard P. 1996. The ribonucleotide reductase system of Lactococcus lactis. Characterization of an NrdEF enzyme and a new electron transport protein. J. Biol. Chem. 271:8779–8785 [DOI] [PubMed] [Google Scholar]
- 49. Gansel X, Hartke A, Boutibonnes P, Auffray Y. 1993. Nucleotide sequence of the Lactococcus lactis NCDO 763 (ML3) rpoD gene. Biochim. Biophys. Acta 1216:115–118 [DOI] [PubMed] [Google Scholar]
- 50. Kvint K, Nachin L, Diez A, Nystrom T. 2003. The bacterial universal stress protein: function and regulation. Curr. Opin. Microbiol. 6:140–145 [DOI] [PubMed] [Google Scholar]
- 51. Martinez B, Zomer AL, Rodriguez A, Kok J, Kuipers OP. 2007. Cell envelope stress induced by the bacteriocin Lcn972 is sensed by the lactococcal two-component system CesSR. Mol. Microbiol. 64:473–486 [DOI] [PubMed] [Google Scholar]
- 52. Terzaghi BE, Sandine WE. 1975. Improved medium for lactic streptococci and their bacteriophages. Appl. Microbiol. 29:807–813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Cluzel PJ, Chopin A, Ehrlich SD, Chopin MC. 1991. Phage abortive infection mechanism from Lactococcus lactis subsp. lactis, expression of which is mediated by an Iso-ISS1 element. Appl. Environ. Microbiol. 57:3547–3551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Yamada M, Nunoshiba T, Shimizu M, Gruz P, Kamiya H, Harashima H, Nohmi T. 2006. Involvement of Y-family DNA polymerases in mutagenesis caused by oxidized nucleotides in Escherichia coli. J. Bacteriol. 188:4992–4995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Delbrück M. 1940. The growth of bacteriophage and lysis of the host. J. Gen. Physiol. 23:643–660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Stockdale SR, Mahony J, Courtin P, Chapot-Chartier MP, van Pijkeren JP, Britton RA, Neve H, Heller KJ, Aideh B, Vogensen FK, van Sinderen D. 2013. The lactococcal phages Tuc2009 and TP901-1 incorporate two alternate forms of their tail fiber into their virions for infection specialization. J. Biol. Chem. 288:5581–5590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Powell IB, Tulloch DL, Hillier AJ, Davidson BE. 1992. Phage DNA synthesis and host DNA degradation in the life cycle of Lactococcus lactis bacteriophage c6A. J. Gen. Microbiol. 138:945–950 [DOI] [PubMed] [Google Scholar]
- 58. Roces C, Courtin P, Kulakauskas S, Rodriguez A, Chapot-Chartier MP, Martinez B. 2012. Isolation of Lactococcus lactis mutants simultaneously resistant to the cell wall-active bacteriocin Lcn972, lysozyme, nisin, and bacteriophage c2. Appl. Environ. Microbiol. 78:4157–4163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Conter A, Bouche JP, Dassain M. 1996. Identification of a new inhibitor of essential division gene ftsZ as the kil gene of defective prophage Rac. J. Bacteriol. 178:5100–5104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Meyrand M, Boughammoura A, Courtin P, Mezange C, Guillot A, Chapot-Chartier MP. 2007. Peptidoglycan N-acetylglucosamine deacetylation decreases autolysis in Lactococcus lactis. Microbiology 153:3275–3285 [DOI] [PubMed] [Google Scholar]
- 61. Larsen N, Boye M, Siegumfeldt H, Jakobsen M. 2006. Differential expression of proteins and genes in the lag phase of Lactococcus lactis subsp. lactis grown in synthetic medium and reconstituted skim milk. Appl. Environ. Microbiol. 72:1173–1179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Hatfull GF. 2008. Bacteriophage genomics. Curr. Opin. Microbiol. 11:447–453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. McGrath S, Fitzgerald GF, van Sinderen D. 2001. Improvement and optimization of two engineered phage resistance mechanisms in Lactococcus lactis. Appl. Environ. Microbiol. 67:608–616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Labrie S, Moineau S. 2002. Complete genomic sequence of bacteriophage ul36: demonstration of phage heterogeneity within the P335 quasi-species of lactococcal phages. Virology 296:308–320 [DOI] [PubMed] [Google Scholar]
- 65. Chopin A, Bolotin A, Sorokin A, Ehrlich SD, Chopin M. 2001. Analysis of six prophages in Lactococcus lactis IL1403: different genetic structure of temperate and virulent phage populations. Nucleic Acids Res. 29:644–651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Ventura M, Zomer A, Canchaya C, O'Connell-Motherway M, Kuipers O, Turroni F, Ribbera A, Foroni E, Buist G, Wegmann U, Shearman C, Gasson MJ, Fitzgerald GF, Kok J, van Sinderen D. 2007. Comparative analyses of prophage-like elements present in two Lactococcus lactis strains. Appl. Environ. Microbiol. 73:7771–7780 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.